-
PDF
- Split View
-
Views
-
Cite
Cite
Yasuhiro Takenaka, Atsushi Yamaguchi, Yasushi Shigeri, A light in the dark: ecology, evolution and molecular basis of copepod bioluminescence, Journal of Plankton Research, Volume 39, Issue 3, May-June 2017, Pages 369–378, https://doi.org/10.1093/plankt/fbx016
Close - Share Icon Share
Abstract
Within the calanoid copepods, the bioluminescent species comprise 5–59% of the abundance and 10–15% of the biomass in the world's oceans. Most of the luminous species belong to the superfamily Augaptiloidea. The composition of bioluminescent species within the calanoid copepods shows latitudinal patterns; 5–25% of total calanoid copepods are found in high-latitude oceans, while 34–59% are in low-latitude oceans, reflecting a prey-predator relationship. Bioluminescent species of calanoid copepods are able to produce the light-emitting substrate coelenterazine. It is then transferred to higher predators through the food chain, and might be used for bioluminescence in other luminous organisms. A notable feature of copepod bioluminescence is the secreted-type, and its major function may be as an antipredatory response or a defensive behavior. Identification of more than 20 luciferase genes from calanoid copepods has revealed the highly conserved sequences of those genes. This leads us to the speculation that the genes for luciferase within the group of calanoid copepods have evolved independently of comparable genes outside of this group. We discuss here the ecological and biological functions of copepod bioluminescence, the significant diversity in luminous intensity, which might be evolutionarily relevant to their motility and habitat depth, and the promising future directions of bioluminescence studies.
INTRODUCTION
Many luminous organisms exhibit bioluminescence from photophores, the light-emitting organs that contain their own photocytes or symbiotic luminous bacteria (Shimomura, 2006). Bioluminescence has been identified in a wide variety of organisms, such as mollusks, cnidarians, insects and fishes (Shimomura, 2006; Haddock et al., 2010; Moline et al., 2013). Since Raphaël Dubois (1833–1877) demonstrated the first example of a luciferin-luciferase reaction (L-L reaction, Dubois, 1885), the understanding of bioluminescence at the molecular level has increased exponentially. In the late 20th century, the crystallization and structural determination of luciferin, the luminescent substrate, from several organisms has been reported (Shimomura et al., 1957; White et al., 1961, 1963; Shimomura and Johnson, 1972; Inoue et al., 1975), and the efficient chemical synthesis of luciferin exploited (Moline et al., 2013). With the advent of molecular biology, the gene cloning of luciferases, enzymes that catalyze the photogenic oxidation of luciferin, has been performed using fireflies (Wood et al., 1984) and luminescent bacteria (Cohn et al., 1985). To date, a number of different primary structures of the luciferases have been isolated from various luminous organisms, from bacteria and protista to insects and fishes (Shimomura, 2006).
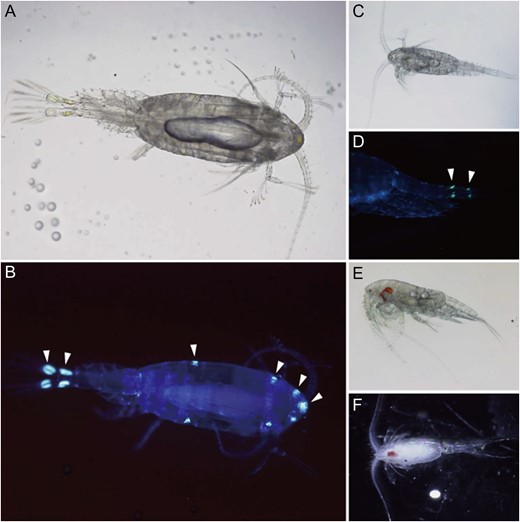
Bioluminescent calanoid copepods. (A) microscopic image of Metridia okhotensis; (B) fluorescent image of M. okhotensis illuminated by UV light. Arrowheads indicate the possible position of luminous glands; (C) bright-field image of Metridia pacifica; (D) fluorescent image of the urosome of M. pacifica. Arrowheads indicate the possible luminous glands on the caudal rami and anal segment; (E) Pleuromamma abdominalis; (F) Pleuromamma xiphias. (Photos: Y. Takenaka).
DISTRIBUTION AND ECOLOGICAL ASPECTS OF CALANOID COPEPOD BIOLUMINESCENCE
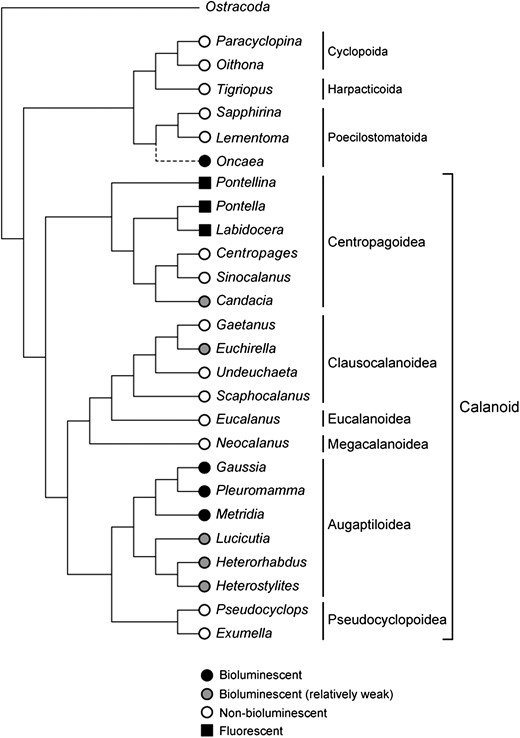
Taxa of bioluminescent and non-bioluminescent copepods depicted based on molecular phylogeny of 18 S rRNA nucleotide sequences. The Augaptiloidea superfamily contains a number of bioluminescent species. Most luciferases were isolated from species in the Metridinidae family, comprising Gaussia, Pleuromamma and Metridia genera. Some species in different phyla are also luminous, but their distribution is patchy. The fluorescent proteins were identified from several species in the Centropagoidea superfamily. Oncaea conifera is known as a bioluminescent species and belongs to the Poecilostomatoida superfamily (Herring et al., 1993). Although the complete 18 S rRNA sequence of Oncaea was not available in the database, it was depicted with a dashed line in the tree based on the partial 18 S and ITS1 sequences.
(A) Composition of bioluminescent families relative to total calanoid copepod abundance (circles) or biomass (in dry mass [DM] = squares or carbon [C] = triangles) from cold-water region (Arctic-subarctic region, left) and warm-water region (temperate-tropical region, right). Data sources are 1Deevey and Brooks (1977), 2Ward and Shreeve (1999), 3Auel and Hagen (2002), 4Yamaguchi et al. (2002), 5Koppelmann and Weikert (2007), 6Homma and Yamaguchi (2010) and 7Kosobokova and Hopcroft (2010). (B) Prey-predator linkage between bioluminescent copepod families and their predators (cephalopods, myctophids, gonostomatids and cnidarians). Cn: percentage composition in number of bioluminescent copepods in their food items. Data sources are 8Lancraft et al. (1988), 9Passarella and Hopkins (1991) and 10Williams et al. (2001).
The percentage of the four families within the bioluminescent copepods also shows a different latitudinal pattern. Thus, only Metridinidae dominated in high-latitude oceans (Fig. 3A). While in low-latitude oceans, in addition to Metridinidae, Augaptilidae and Lucicutiidae were also abundant. Especially in the eastern Mediterranean Sea, Augaptilidae outnumbered Metridinidae. Since the feeding mode of Augaptilidae is carnivorous (Arashkevich, 1969; Boxshall, 1985), its dominance, along with the other families in low-latitude oceans, may be related to high species diversity in low-latitude oceans (Woodd-Walker et al., 2002; Rombouts et al., 2009).
In summary, for pelagic calanoid copepods, the composition of bioluminescent species is much higher than for the other marine pelagic and benthic biomes. Bioluminescent copepods are also important food sources for cephalopods and myctophid and gonostomatid fishes, accounting for more than 31–89% of their food items (Fig. 3B). This suggests that bioluminescent copepods play the role of coelenterazine couriers within the marine pelagic realm. The composition of the bioluminescent species in calanoid copepods also showed latitudinal patterns; they are high in number in low-latitude oceans, which may be related to the high species diversity of these regions.
ROLE OF CALANOID COPEPOD BIOLUMINESCENCE IN THE TRANSFER OF LUMINESCENCE SUBSTRATE THROUGH THE TROPHIC CHAIN
The substrate for luminescence in calanoid copepods is an imidazopyrazinone luciferin, coelenterazine. The luciferins containing the imidazopyrazinone structure are found in various other luminous marine organisms besides calanoid copepods, such as fishes, decapod shrimps, protists, molluscs, cnidarians, chaetognaths, ostracods and radiolarians. Similar chemical structures of luciferins found in phylogenetically distant bioluminescent organisms may be derived from dietary transfer of luminescence substrates through the trophic chain (Cormier et al., 1967; Thompson et al., 1987, 1988a, b, 1995a; Thompson and Rees, 1995). It has been suggested that some luminous marine organisms are unable to produce their own luciferins and depend on a dietary supply of them for bioluminescence (Mallefet and Shimomura, 1995; Haddock et al., 2001). Metridia pacifica was found to retain the biosynthetic pathway for the light-emitting substrate coelenterazine (Oba et al., 2009), and can be a prey for higher consumers as the ultimate source of coelenterazine. Considering the high percentage of bioluminescent calanoid copepod species, they might have an essential role as suppliers of coelenterazine within the marine environments (Mallefet and Shimomura, 1995) (Fig. 3B). For example, the firefly squid, Watasenia scintillans, indigenous to northern Japan, feeds predominantly on calanoid copepods, including M. pacifica (Hayashi and Hirakawa, 1997). Liver tissues of W. scintillans contain coelenterazine and show strong chemiluminescence (Inoue et al., 1975). Although the light-emitting substrate of W. scintillans, coelenterazine disulfate, is not identical to coelenterazine, it is probable that W. scintillans synthesizes its own luciferin by utilizing coelenterazine from M. pacifica as a starting molecule (Inoue et al., 1976). Furthermore, a specialized prey-predator linkage has been reported between Augaptiloidea copepods and mesopelagic myctopid and gonostomatid fishes, both having coelenterazine (Merrett and Roe, 1974; Gordon et al., 1985; Lancraft et al., 1988; Hopkins and Sutton, 1998) (Fig. 3B). Except for copepods, the decapod Systellaspis debilis is also probably able to synthesize coelenterazine (Thomson et al., 1995b), although its biosynthesis has not yet been elucidated. Thus, the possibility exists that calanoid copepods may have a profound influence on marine bioluminescence with the functions of producers and couriers of coelenterazine to luminous organisms utilizing imidazopyrazinone luciferins.
FUNCTIONS OF CALANOID COPEPOD BIOLUMINESCENCE
The major function of copepod bioluminescence may be an antipredatory response or a defensive behavior, although currently this is just speculation (Mauchline, 1998). There is a notable relationship, for example, between bioluminescence and the jump response in the calanoid copepod Pleuromamma xiphias (Augaptiloidea) when tested with a quantified hydrodynamic disturbance. Weak stimuli elicited only a jump response, whereas supra-threshold stimuli resulted in both a jump and bioluminescent discharge (Hartline et al., 1999). This suggests that copepods save their bioluminescence for what is perceived to be a stronger threat by a predator.
There is a large diversity in luminosity between each Augaptiloidea species (Takenaka et al., 2012; Fig. 2). This may be another clue as to how calanoid copepods developed their bioluminescence. The Megacalanoidea and Clausocalanoidea, which are considered as lately diverged superfamilies after Augaptiloidea in the evolution of calanoid copepods (Bradford-Grieve et al., 2010), show little or no bioluminescence (Takenaka et al., 2012; Fig. 2). In these species, where they have developed myelinated nerve fibers (Davis et al., 1999) and chemosensory organs, they have greater motility and faster escape responses (~2 ms) from predators (Lenz and Hartline, 1999; Lenz et al., 2000; Waggett and Buskey, 2008). Whereas the Augaptilodea organisms, which have only unmyelinated axons, show significantly slower responses (~6 ms) to initiate an escape behavior (Davis et al., 1999). Thus, differences in an escape response among calanoid copepod superfamilies could be relevant to the function of their bioluminescence as a defensive mechanism to escape from surrounding predators. Bioluminescent species in the Augaptiloidea family have evolved several defensive behaviors, including a strong escape jump and bioluminescence, to avoid predation, whereas species in the Megacalanoidea and Clausocalanoidea families may have developed a reliance on improved speed and strength of the escape response itself, made possible by the evolution of myelinated axons. For the ancestral species in the Megacalanoidea and Clausocalanoidea, bioluminescence as a defensive function might have been less essential than for the non-myelinated Augaptiloidea superfamily, including the Metridinidae family, resulting in a gradual loss of luminescence activity as myelination evolved.
The habitat depth of calanoid copepods also seems to be an important factor that can affect the intensity of bioluminescence. The Heterorhabdidae family also belong to the Augaptiloidea superfamily, but inhabits mesopelagic and bathypelagic zones (200–3000 m) (Yamaguchi and Ikeda, 2000), and shows much weaker bioluminescence than Metridinidae species (Takenaka et al., 2012, 2013; Fig. 2). Compared with Metridinidae species, such as M. pacifica that inhabits the epipelagic layer (0–200 m), Heterorhabdidae would be exposed to lower predation pressure by mesopelagic and bathypelagic fishes. Furthermore, counter-illumination is not available at such great and dark depths. These ecological aspects might have led to the evolution of diversified bioluminescence in the Augaptiloidea superfamily.
In addition to the defensive function of bioluminescence in copepods, it could also be used for communication as a warning signal between individuals of Metridia longa, as was proposed by Buskey and Swift (1985); however there are no reports of a possible function for bioluminescence in sexual recognition by copepod species. A potential antioxidative function of bioluminescence proposed for other organisms (Rees et al., 1998) also remains unsupported in copepods.
FUNCTIONS OF COPEPOD BIOLUMINESCENCE INFERRED FROM THE STRUCTURE OF COPEPOD LUCIFERASES
The biological functions of copepod bioluminescence could be interpreted by comparing structures of luciferases isolated from different copepod species and how the luciferase gene has evolved from a common ancestral origin in each group of luminous species. More than 20 genes coding for luciferase have been isolated so far from luminous calanoid copepods. All natural copepod luciferases identified so far contain about 20 amino acids of an N-terminal signal sequence for extracellular secretion. Even recombinant copepod luciferases expressed in mammalian cultured cells were also efficiently secreted into the culture medium (Markova et al., 2004; Tannous et al., 2005; Takenaka et al., 2008, 2012). This property of the protein can provide calanoid copepods with the ability to produce the secreted-type luminescence. When they are secreted with luciferin from the luminous glands (Fig. 1B and D), the display of a cloud of sparkles may serve as counter-illumination to hide the silhouette of the body and/or as a smoke screen in a bioluminescent defense (Haddock et al., 2010). Another function of bioluminescence in copepods is startling the approaching predators by a strong flash. Many of the copepod luciferases exhibit spike-like, strong light when they are mixed with the substrate luciferin. This flash-type bioluminescence can act effectively as a repellent or distractor to predators. In contrast, stable and long-lasting luminescence is more suitable for counter-illumination and smoke-screen roles.
In Metridia and Pleuromamma species, multiple luciferase genes were cloned from the same cDNA library (Takenaka et al., 2008, 2012; Borisova et al., 2008). The MpLuc1 and MpLuc2 from M. pacifica would be transcribed from two different loci or alleles, since a survey of their genomic sequences revealed the presence of different lengths of the introns at different positions in two MpLuc genes (Takenaka et al., 2008), and also in MoLuc genes (Takenaka et al., 2012). There are distinct differences in the enzymatic properties of MpLuc1 and MpLuc2, as seen in their luminescence kinetics. MpLuc1 sharply emits light upon mixing with the substrate coelenterazine, whereas MpLuc2 shows a much slower build-up and gradual decay of luminescence (Takenaka et al., 2008). Although details about the expression and localization of MpLuc1 and MpLuc2 in the body of M. pacifica are still unclear, it is plausible that the copepod uses two isoforms with different enzymatic characteristics for different defensive roles, such as startling and counter-illumination.
EVOLUTION OF COPEPOD BIOLUMINESCENCE
The multiple sequence alignments of copepod luciferases isolated so far have revealed the presence of two tandem consensus sequences of C-x(3)-C-L-x(2)-L-x(4)-C-x(8)-P-x-R-C (x, amino acid residue) (Takenaka et al., 2013), which suggests that all known calanoid copepod luciferases originated from the same ancestral gene. Each consensus sequence was thought to act as a core of the functional domain. It has been reported that each of two domains in GLuc exhibited weak, but notable, luciferase activity when they were expressed in E. coli and characterized independently (Inouye and Sahara, 2008). As is obvious from the presence of four cysteine residues in the consensus sequence, there are a lot of cysteine residues in the full amino acid sequence of copepod luciferases. The presence of tandem consensus sequences in catalytic domains mentioned above, and the abundant cysteine residues, can be the characteristic benchmarks for the identification of copepod luciferases.
Among the many luciferases, beetle luciferases are probably the most studied in the field of evolution (Viviani, 2002). Notable similarities in catalytic mechanisms (McElroy et al., 1967) and amino acid sequences (Suzuki et al., 1990) between a luciferase from the North American firefly and acyl-CoA synthetase have been reported. It was proven that firefly luciferase possesses both catalytic activities of the bioluminescence reaction (ATP-dependent monooxygenase) and the fatty acyl-CoA synthetic reaction (Oba et al., 2003). In marine organisms, it has been suggested that aequorin and obelin, the coelenterate photoproteins, originated from ancestral calcium-binding proteins, such as calmodulin, based on the amino acid sequence similarity and their reactivity to calcium ion (Tsuji et al., 1995). In copepods, the BLAST search uncovered no protein showing higher similarity to the overall structure of any copepod luciferase or consensus sequences described above. Thus, this strategy, which was successful in studies of other organisms to reveal evolutionary origins, has not yet produced this outcome for copepods. The lack of significant homologies between copepod luciferase sequences and any other luciferase sequences suggests that copepod luciferases evolved from their ancestral origins independent of non-copepod luciferases. This strongly supports the idea that the capacity to generate light-emission has arisen through convergent evolution of bioluminescence-related genes among different lineages.
Takenaka et al. tried to estimate an amino acid sequence of an ancestral copepod luciferase by in silico phylogenetic analyses of all known luciferases derived from the Augaptiloidea superfamily (Takenaka et al., 2013). As a consequence of the ancestral sequence reconstruction, two candidates for the ancestral genes, aCopLuc43 and aCopLuc48, were predicted. The aCopLuc43 and aCopLuc48 were the putative common ancestors for the Metridinidae/Lucicutiidae families and for the Heterorhabdidae family, respectively. The aCopLuc43 and aCopLuc48 expressed in mammalian cells exhibited remarkable luciferase activities in the intracellular lysates and culture media. In addition, they only reacted with coelenterazine, not with Cypridina luciferin, just as seen in native copepod luciferases. The results suggest that ancestral species acquired the ability to produce bioluminescence before evolutionary diversification of related copepod species.
FUTURE DIRECTIONS
Although the application of copepod luciferases as a reporter in biomedical studies has developed rapidly over the last 10 years (Tannous et al., 2005; Remy and Michnick, 2006; Morlighem et al., 2007; Wu et al., 2007; Wurdinger et al., 2008; Kim et al., 2011), mechanisms of production, secretion and regulation of copepod bioluminescence remain almost entirely unknown. A future, basic biological study of copepod bioluminescence could be the morphological recognition of copepod bioluminescence with techniques such as immunostaining or whole mount in situ hybridization; these will provide further information about the expression and localization of bioluminescent-related proteins, such as luciferases and other unknown accessory proteins. Besides these in vitro studies, observation of bioluminescent behaviors in copepod culture will give us clues to understand how bioluminescence works in nature. Development of a long-term copepod culture system equipped with a high-speed camera may afford researchers in bioluminescence and zooplankton studies a tool to observe live copepod subjects directly, with or without predators. The system also would enable us to sample copepods in good condition for molecular and morphological analyses in the lab.
To further clarify the evolution and origin of copepod luciferases, we may need more information about the primary structure of luciferase and luciferase-like proteins from a variety of marine organisms, including copepods. For example, the tiny poecilostomatoid copepod Oncaea is known to have bioluminescence (Herring et al., 1993) (Fig. 2), but there is no information about its luciferase gene structure. Structural and enzymatic comparison of luciferase genes isolated from calanoid and poecilostomatoid copepods might provide phylogenetic and evolutionary insights for understanding marine bioluminescence. Furthermore, metagenomic approaches will also uncover the evolutionary trait of bioluminescence in marine organisms. On the other hand, we cannot rule out the possibility that non-bioluminescent calanoid copepods, such as the Megacalanoidea and Clausocalanoidea, still possess luciferase genes that previously coded the active luciferase, but lost function subsequently. Luciferase gene loss was suggested in the dinoflagellate Gonyaulax spinifera, which shows very weak luminescence (Baker et al., 2008). These kinds of pseudogenes, or the remnants of the genes, may be the clues to how luciferase genes evolved in copepods during myelination.
Another approach to reveal the evolution of copepod bioluminescence is identification of the pathways of coelenterazine synthesis in calanoid copepods. The de novo synthesis of coelenterazine from two L-tyrosines and an L-phenylalanine has been shown by feeding isotopically labeled L-amino acids to M. pacifica (Oba et al., 2009; Markova and Vysotski, 2015). The chemistry of coelenterazine biosynthesis and features of related enzymes will be a promising area for future bioluminescence studies. Phylogeny of the biosynthetic proteins might shed light on the relationship among marine bioluminescent and/or non-bioluminescent organisms. Also, as a biotechnological application, if genes encoding the metabolic enzymes involved in luciferin biosynthesis were identified, we could develop autoluminescent organisms equipped with luciferases and luciferin biosynthesis systems.
ACKNOWLEDGEMENTS
We thank Prof. Leslie Sargent Jones (Appalachian State University) for her careful reading of the manuscript and her important comments.
FUNDING
This work was supported by JSPS Bilateral Researcher Exchange Program in 2012, 2013 and 2015 (YS) and an AIST research grant.
REFERENCES
Author notes
Corresponding editor: Roger Harris


![(A) Composition of bioluminescent families relative to total calanoid copepod abundance (circles) or biomass (in dry mass [DM] = squares or carbon [C] = triangles) from cold-water region (Arctic-subarctic region, left) and warm-water region (temperate-tropical region, right). Data sources are 1Deevey and Brooks (1977), 2Ward and Shreeve (1999), 3Auel and Hagen (2002), 4Yamaguchi et al. (2002), 5Koppelmann and Weikert (2007), 6Homma and Yamaguchi (2010) and 7Kosobokova and Hopcroft (2010). (B) Prey-predator linkage between bioluminescent copepod families and their predators (cephalopods, myctophids, gonostomatids and cnidarians). Cn: percentage composition in number of bioluminescent copepods in their food items. Data sources are 8Lancraft et al. (1988), 9Passarella and Hopkins (1991) and 10Williams et al. (2001).](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/plankt/39/3/10.1093_plankt_fbx016/2/m_fbx016f03.jpeg?Expires=1716403590&Signature=O7eYZs6bXgHOEmrgIsV~bj8iq91VpU0JQk26bUKgw7Fuf8813XL11~lm4GiNv~ua2IBQhX4kHwNrSxnABrWi3Tw46lYQF9QQW90PxqpGs5OA66zkbc26Y~REomZsPziZ3IlGF2BY0OTlpg3OGtvETKjsMFu8c-GaW13VQbsyKrMYN4TSKczj31kjOwZQe5x2QOqLfqHoGkOGVXzNxBQJ2KCN5x7QCXOLTpfT6avEkubX3BZCt5UEZ2N2g5ercI~K-wQ1jU261fN8MhytNLZxriu1bcPPbKu5E~XSbGdWt1zZLx6dEp5f5-lxFipA3bJFKEpUOmgK0H9a-4KgieF~dQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
