-
PDF
- Split View
-
Views
-
Cite
Cite
Daniel D. Scott, Christian Trahan, Pierre J. Zindy, Lisbeth C. Aguilar, Marc Y. Delubac, Eric L. Van Nostrand, Srivathsan Adivarahan, Karen E. Wei, Gene W. Yeo, Daniel Zenklusen, Marlene Oeffinger, Nol12 is a multifunctional RNA binding protein at the nexus of RNA and DNA metabolism, Nucleic Acids Research, Volume 45, Issue 21, 1 December 2017, Pages 12509–12528, https://doi.org/10.1093/nar/gkx963
Close - Share Icon Share
Abstract
To counteract the breakdown of genome integrity, eukaryotic cells have developed a network of surveillance pathways to prevent and resolve DNA damage. Recent data has recognized the importance of RNA binding proteins (RBPs) in DNA damage repair (DDR) pathways. Here, we describe Nol12 as a multifunctional RBP with roles in RNA metabolism and genome maintenance. Nol12 is found in different subcellular compartments—nucleoli, where it associates with ribosomal RNA and is required for efficient separation of large and small subunit precursors at site 2; the nucleoplasm, where it co-localizes with the RNA/DNA helicase Dhx9 and paraspeckles; as well as GW/P-bodies in the cytoplasm. Loss of Nol12 results in the inability of cells to recover from DNA stress and a rapid p53-independent ATR-Chk1-mediated apoptotic response. Nol12 co-localizes with DNA repair proteins in vivo including Dhx9, as well as with TOPBP1 at sites of replication stalls, suggesting a role for Nol12 in the resolution of DNA stress and maintenance of genome integrity. Identification of a complex Nol12 interactome, which includes NONO, Dhx9, DNA-PK and Stau1, further supports the protein's diverse functions in RNA metabolism and DNA maintenance, establishing Nol12 as a multifunctional RBP essential for genome integrity.
INTRODUCTION
In eukaryotic cells, the DNA damage response (DDR) comprises a network of overlapping cellular signaling pathways that detect varied insults to DNA and direct their timely and accurate resolution (1). To achieve this, the DDR must coordinate DNA repair itself with various replicative processes including DNA replication, cell growth, cell cycle progression and apoptosis/senescence (1–4). Mutations fin DDR components cause genomic instability and a broad spectrum of heritable and spontaneous human diseases (5). Implementation of much of the DDR program is achieved through transcriptional regulation, both by key effector transcription factors such as TP53 and through direct regulation of RNA polymerases I, II and III (2,6,7). However, the DDR additionally modulates a large array of RNA binding proteins (RBPs) to control the synthesis, maturation and decay of cellular RNAs (8–11). The DDR regulates both constitutive and transcript-specific splicing through targeting of spliceosomal components and of individual RBPs such as hnRNP K, Sam68, EWSR1, DDX54 and SRSF10, respectively (7,12,13). RBPs such as HuR, AUF1 and TIAR modulate mRNA stability in response to DDR signaling, as do various miRNAs whose maturation is controlled by the DDR via Dicer (1). HuR also promotes translation of the TP53 mRNA (17). Consistent with these diverse roles, a number of large-scale genetic and proteomic studies of proteins involved in the DDR have shown enrichment for RBPs (2).
More evidence is emerging, however, that RBPs can go beyond the paradigm of being DDR effectors and can themselves participate directly in DNA repair and the DDR (9,10). Key RNA-regulatory structures within the cell, most notably the nucleolus and paraspeckles, act as platforms for the regulation and/or assembly of DDR complexes and pathways; fundamental reorganization of these organelles is a hallmark of the DDR (3). Several RBPs including RRP6/EXOSC10, Xrn2, DDX1 and DDX19 are required for preventing the formation of, or resolving aberrant RNA:DNA hybrids (R-loops) within the genome (4,5). The multifunctional transcription/translation factor YB-1 is able to bind directly to sites of nucleotide damage and to coordinate repair complex assembly and/or metabolize the sites directly (28), while the nucleolar, ribosome biogenesis proteins NPM1/B23 and NCL/C23 act as histone chaperones across several DNA repair pathways (6). Numerous other RBPs including FUS/TLS, SFPQ/PSF, NONO/p54nrb, RBM14, RBMX, PRP19, RPS3 and Dicer—many of which are components of the nucleolus and/or paraspeckles—are also recruited to sites of, and participate in DNA damage/repair, though the precise mechanisms of their action(s) have not been fully elucidated (7–9).
In addition to DNA damage, disruption of the accurate maturation and assembly of ribosomes in the nucleolus, a process that involves more than 300 proteins (10), is known to induce G1/S cell cycle arrest via a process termed the ‘nucleolar stress response’ in response to diverse cellular insults including transcriptional inhibitors, nutritional stress, confluency, as well as the depletion or mutation of various components of the assembling or mature ribosomes (11,12). In this process, perturbation of pre-ribosomal RNA (pre-rRNA) synthesis, processing and/or assembly with trans-acting factors or ribosomal proteins can result in major structural arrangements of the nucleolus and proteasomal degradation of most mature ribosomal proteins, with the notable exception of RpL5/uL18 and RpL11/uL5 (13,14). These two proteins form a tertiary subcomplex with 5S rRNA which complex protects them from degradation; this complex subsequently accumulates in the non-ribosomal nuclear fraction where it interacts with Mdm2 and prevents its constitutive ubiquitination and consequent degradation of p53 (12–16). While several early papers suggested the existence of independent pathways for other RPs or ribosome biogenesis factors to regulate p53 accumulation, subsequent work demonstrated that these pathways in fact act via RpL5/RpL11/5S and have underlined the central role of this complex in the induction of cell cycle arrest in response to perturbation of ribosomal biogenesis (14,16,17).
In this paper, we identify the human protein Nol12 as a member of this growing class of RBPs that simultaneously function in RNA metabolism and the DDR. Previously, the Drosophila melanogaster Nol12 homologue Viriato was shown to modulate signaling during eye development, and its interactors were overwhelmingly involved in development of the nervous system (18,19). Loss of Viriato resulted in cell proliferation, developmental delay and apoptosis. Moreover, Viriato, as well as Nop25, the Nol12 homologue in mice, were both shown to affect nucleolar integrity (19,20), while its yeast homologue, Rrp17, was identified as a putative 5′-3′ exonuclease with function in ribosome biogenesis (21); a potential role for Nol12 in ribosome maturation during processing of the Internal Transcribed Spacer 1 (ITS1), which excised during ribosomal RNA maturation, has also been suggested in HeLa cells, concurrent with that of Rrp17 (22).
Here, we show that the human protein Nol12 is an RNA binding protein required for both ribosome maturation and genome integrity in higher eukaryotes. Nol12 is part of early 90S and pre-60S ribosomal subunits in vivo, where it is required for efficient separation of the large and small subunit precursors via cleavage at site 2. Knockdown of NOL12 impairs cellular proliferation by inducing a G1/S cell cycle arrest, but does so outside of a nucleolar stress response and in a p53-independent manner. In addition, Nol12 has a putative role in the resolution of DNA stress as loss of the protein leads to a rapid activation of the DNA damage response kinase ATR and apoptotic response. Moreover, Nol12 interacts with chromatin-associated factors, in particular, proteins implicated in DNA damage repair such as the paraspeckle component SFPQ and the RNA/DNA DEAD-box helicase Dhx9 involved in genome instability prevention, and, furthermore, localizes to sites of replication stress and DNA insults in vivo, suggesting a role for Nol12 in the maintenance of genome integrity.
MATERIALS AND METHODS
DNA constructs
NOL12 cDNA was amplified by PCR from pRS414–3xHA-Nol12 (42) with oligonucleotides NdeI-Nol12-F and XhoI-Nol12-R. The PCR product was cloned into NdeI-XhoI sites of pET21a to generate plasmid pET21a-Nol12–6xHis, which was used as template to create a siRNA-resistant cDNA of Nol12 by introducing silent mutations by site-directed mutagenesis using the QuickChange Multi Site Mutagenesis Kit (Agilent Technologies) and oligos siRNA5-mut, siRNA3–8-mut, siRNA6-mut and Link+HA.
In order to construct a PrA-NOL12 encoding plasmid for Flip-in recombination Flp-In T-REx system, the Nol12 cDNA was amplified by PCR from the same template as above with oligonucleotides NcoI-Nol12-F and NotI-Nol12-R, cloned pENTR4 to generate pENTR4-Nol12. Protein A was amplified by PCR from pBXA (53) and cloned into pFRT-TO-DEST-GFP to generate pFRT-TO-DEST-PrA. pENTR4-Nol12 and pFRT-TO-DEST-PrA were recombined with LR clonase to generate pFRT-TO-PrA-Nol12 according to the Gateway system instructions (Life Technologies; see Supplementary Material and Methods). All DNA constructs in this study were sequenced. Oligos used for cloning and mutagenesis are shown in Supplementary Table S10. Plasmids are listed in Supplementary Table S11.
Cell culture and drug treatment
HCT116 WT, HCT116 p53−/− and HEK293T cells were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% Fetal Calf Serum (FCS) and Penicillin/Streptomycin (1000 IU, 1 mg/ml respectively; Wisent) at 37°C with 5% CO2. Synthetic siRNAs against NOL12 (#5 and #8), XRN2, p21, p27 and p53 and a scrambled (SCR) control were supplied by Sigma-Aldrich. siRNAs were transfected at a concentration of 10nM using Lipofectamine RNAiMAX (Life Technologies) in a reverse transfection protocol according to the manufacturer's instructions. Between 7.5 × 104 and 5.4 × 105 cells/ml were transfected, depending on the duration of knockdown. siRNA sequences are listed in Supplementary Table S8. Caffeine (2 or 5 mM), VE822 (1 μM), KU55933 (1μM), ChiR-124 (100 nM), NSC109555 (5 μM) or NU7441 (0.5 μM) were added to the above growth media for the duration of the time course; these supplemented media were changed every 24 h throughout the time course to maintain an effective dose. Actinomycin D (0.05 or 1 μg/ml) was applied to cells for 3 h, arsenite (0.5 mM), hydroxyl urea (1 mM; HU) for 1 h and etoposide (25 or 50 μM) for 3 or 1 h, immediately prior to downstream processing.
Western blotting
siRNA-transfected cells were harvested at the indicated time points by trypsin-treatment, washed with PBS and lysed by sonication (5 min 30’ on/off, 4°C) in RIPA buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40, pH 7.5) supplemented with cOmplete protease inhibitor, EDTA-free (Roche). Soluble fractions were resolved on a 4–12% gradient Bis-Tris NuPAGE® gel according to the manufacturer's instructions, transferred to a PVDF membrane which was blocked with 5% milk or BSA in TBS+T (50 mM Tris, 150 mM NaCl, 0.05% Tween-20, pH 7.6) and probed with the indicated primary and secondary antibodies according to Supplementary Table S7. Secondary antibody-conjugated fluorescence was detected using the Li-Cor Odyssey Infrared scanner. Representative images of biological replicates are shown.
Sucrose gradient
HCT 116 cells transfected with either scramble or Nol12#5 siRNAs were harvested 36 h post-transfection, lysed and cleared using the hot needle method and centrifuged at 600 × g for 30 s. 6 O.D. at 260 nm were loaded onto 10–45% (w/v) gradient and spun at 22.3k RPM for 16 h at 4°C in a SW 41 Ti rotor. Sucrose gradients were fractionated in 500 μl fractions using a piston gradient fractionator (BioComp), and whole fractions were precipitated by TCA. Pellets were directly resuspended in 2× Leammli buffer, the pH was adjusted, and the samples were entirely loaded on a 4–12% NuPAGE Bis–Tris gels. The gels were transferred to PVDF membranes, and probed with both RPL5/ul18 and RPL11/uL5 antibodies (Supplementary Table S7) as well as Nol12 antibody to assess knockdown efficiency.
ssAP-MS/MS and western blotting analysis of Nol12-containing complexes
For ssAP purification of Nol12-containing complexes, HEK293T Flp-In T-REx PrA-Nol12 cells were grown to ∼50% confluency, induced for 24 h with 1μg/ml doxycycline and lysed by cryogrinding (23). Purifications form PrA-Nol12cells and negative controls (PrA-containing or untagged) purifications were performed in duplicate with five volumes of extraction buffer supplemented with 100 mM NaCl (±100 μg/ml RNAse A) or 30 mM NaCl (+100 μg/ml RNAse A) as previously described (23). One tenth of each affinity purification sample was eluted with 0.5 M NH4OH and resolved on 4–12% gradient Bis-Tris NuPAGE® gels for silver staining. 50 μg of the ‘input’ samples, and 0.25 volumes of each ‘eluate’ sample, were resolved on a 4–12% gradient Bis–Tris NuPAGE® gels and Nol12-interacting proteins analyzed by western blotting. The remaining samples were on-bead digested with trypsin (24). Samples were adjusted to 3% formic acid, frozen at –80°C until analyzed by LC–MS as described previously (25). For details, see Supplementary Material and Methods.
For ssAP-MS/MS analysis, MS data generated by Q-Exactive were analyzed using the ProHits software package (26) and then searched using Mascot (version) and Comet (v2014.02 rev. 2) search engines. The searches were performed against the RefSeq database release 57 including a decoy set. Two missed cleavages were allowed in the search parameters for +2 to +4 precursor ions with a 10 ppm error tolerance, and a 0.6 Da error tolerance on fragmented ions. The output from each search engine were analyzed through the Trans-Proteomic Pipeline (27) (v4.7 POLAR VORTEX rev 1) by means of the iProphet pipeline using a 5% FDR (28). For details on data filters, normalization, and identification of Nol12-associated proteins, see Supplementary Material and Methods.
Northern blotting
Total RNA from scramble and NOL12 siRNA transfected cells was extracted with TRIzol (Invitrogen) according to the manufacturer's protocol. For RNA analysis, 3 or 8 μg of total RNA extracted from HCT116 WT and HCT116 p53−/− cells were separated on either 1% agarose-formaldehyde gels in Tricine–Triethanolamine or 8% acrylamide–urea gels (29), and transferred to a nylon membrane (see Supplementary Material and Methods). Probes used for hybridization are shown in Supplementary Table S9. Probes identified with terminal amines were fluorescently labeled with DyLight™ 800 NHS Ester as previously described (30).
eCLIP analysis
NOL12 eCLIP data (accession ENCSR820DQJ), including raw reads as well as standard genomic mapping of reads, was obtained from the ENCODE consortia (https://www.encodeproject.org/). Binding to ribosomal rRNA was quantified using a family-aware repeat element mapping pipeline (Van Nostrand, E.L., et al., in preparation). To summarize relative enrichment between IP and input, relative information was defined as the Kullback-Leibler divergence (relative entropy): |${p_i} \times lo{g_2}( {\frac{{{p_i}}}{{{q_i}}}} )$|, where pi is the fraction of total reads in IP that map to a queried element i (peak, gene, or repetitive element), and qi is the fraction of total reads in input for the same element.
Cell cycle profiling and quantification of apoptosis
To profile cell cycle distribution, siRNA- and/or drug-treated cells were harvested by trypsinization at the indicated time points, washed twice with PBS and fixed with 70% ethanol at -20°C for >1 h. Fixed cells were rehydrated 2× in PBS then permeabilized, RNAse-treated and stained for DNA content with a modified Krishan Buffer (20 μg/ml propidium iodide, 0.1% Triton X-100, 0.2 mg/ml RNAse A in PBS) for 30 min at RT. Apoptosis was measured using the Annexin V-FITC apoptosis detection kit (Sigma-Aldrich) according to the manufacturer's instructions. Briefly, siRNA- and/or drug-treated cells were harvested at the indicated time points and resuspended at 5 × 105 cells/ml. Cells were treated with Annexin V-FITC (250 ng/ml) and propidium iodide (500 ng/ml). In both cases, staining was quantified by flow cytometry and results were analyzed using the FlowJo software.
Cellular Fractionation
HCT116 cells (∼3 × 106 per condition) treated or not with Actinomycin D (1 μg/ml, 3 h) were collected by scraping followed by centrifugation at 500 × g for 5 min. To collect the cytoplasmic and nucleoplasmic fractions, the Nuclear Extract Kit from Active Motif® was used according to the manufacturer's instructions, except that the nuclear fractions were split in two and either treated or not with 100 μg/ml RNAse A for 30 min at 4°C in Complete Lysis Buffer (Active Motif). To prepare the chromatin fraction, pelleted untreated/RNAse A-treated chromatin yielded by the Nuclear Extract Kit above was washed, resuspended, and disrupted by sonication (30% amplitude, 3 cycles of 10’ on followed by 30’ on ice) prior to addition of SDS-PAGE loading buffer. 0.2 volumes of the total volume of each fraction were analyzed by western blotting. For details, see Supplementary Material and Methods.
Hypersensitivity and recovery assays
9 × 105 HCT116 WT cells were transfected with 10 nM siSCR or siNOL12#5 using Lipofectamine RNAiMAX (ThermoFisher) and cultured in six-well dishes for 24 h, when media was replaced with fresh DMEM + 10% FBS containing the indicated concentration(s) of H2O2 for 30 min. After treatment with either 0.1 mM H2O2 or 50 μM etoposide for 30 min, cells were allowed to recover in regular DMEM + 10% FBS for the indicated times at 37°C. Cells were analyzed by western blotting as described elsewhere.
Super-Resolution structured illumination microscopy (SR-SIM)
HCT116 WT cells were grown on poly-L-lysine-coated glass coverslips, fixed, permeabilized and incubated for immunofluorescence (IF) with antibodies against the indicated proteins (Supplementary Table S7). SR-SIM was performed on a Zeiss Axio Observer.Z1 coupled to a Zeiss Elyra PS.1 using a Plan-APOCHROMAT 63×/1.4 Oil DIC M27 objective lens (NA = 1.4). Three angles of the excitation grid, each with five phases, were acquired for each channel and z-stack slice using an Andor iXon3EM+ 885 EMCCD camera. SIM images were processed using the Zen SP1 Black 2011 software and analyzed using the Fiji package of ImageJ. For more details, see Supplementary Material and Methods.
RESULTS
Nol12 associates with a diverse interactome
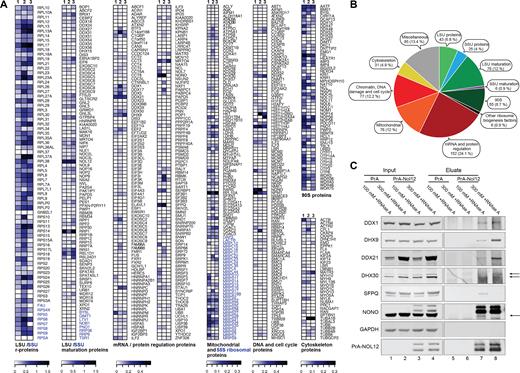
In order to more thoroughly investigate Nol12’s cellular niches, we determined its interactome using Protein A-tagged Nol12 (PrA-Nol12) expressed in HEK293T cells by single-step affinity purification (ssAP) coupled to semi-quantitative mass spectrometry (MS), and analyzed it against untagged and PrA-only expressing cell lines (Supplementary Figure S1A and B) (31). The highest total spectrum count values found in the control samples (Untagged, PrA; Supplementary Table S1) were subtracted from that of a PrA-Nol12 sample under the respective condition (100 or 300 mM NaCl ± RNAse A). A peptide was considered ‘real’ in a given PrA-Nol12 sample (100 or 300 mM NaCl ± RNAse A) if the spectrum count value in each replicate was ≥2-fold greater than that recorded in the control samples (untagged and PrA alone) under the same purification conditions (Supplementary Tables S1 and S2). The total number of unique peptide spectra recorded for the preys was determined, and preys were defined as Nol12-associated proteins if at least three enriched peptides were identified in both replicates. For each prey protein identified, the number of unique peptide spectra was normalized against those of the bait protein in order to obtain the number of unique peptide spectra per prey, which was used as a metric for the analysis of Nol12-associated proteins. We identified a total of 759 proteins, of which 496 were defined as enriched (≥3 peptides in all replicates; Supplementary Table S2). Functional classification of the identified proteins showed a complex interactome and suggested a highly diverse functionality for Nol12 in human cells (Supplementary Tables S3 and S5).
In line with a putative nucleolar and ribosome biogenesis-associated function (32), under low salt conditions PrA-Nol12-associated preys included 75 ribosome maturation factors required for large ribosomal subunit (LSU) maturation and 54 known 90S pre-ribosomes components, while only 8 proteins specific for small ribosomal subunit (SSU) maturation were identified (Figure 1A, column 1). In addition, 42 LSU and 27 SSU ribosomal proteins (RPs) were found (Figure 1A), suggesting a role for Nol12 during LSU maturation events. PrA-Nol12 also co-isolated a significant number of proteins from several non-ribosomal processes, including 51 proteins involved in genome maintenance and integrity (Figure 1A and B: ‘Chromatin associated’; Supplementary Table S3) such as Dhx9, PRKDC and MYBBP1A; furthermore, 94 nuclear mRNA maturation and turnover factors, including splicing and mRNA maturation factors (e.g. snRNP200, Srsf1, Thoc2, Mrto4), paraspeckle proteins (e.g. NONO, SFPQ), cytoplasmic processing (P) bodies components (e.g. Stau1, hnRnpA3, Fmr1, LaRP1) (Figure 1A and B: ‘mRNA metabolism’; Supplementary Table S3) (33) as well as 69 proteins with functions in mitochondria, including many involved in mitochondrial ribosome biogenesis (Figure 1A and B: ‘Mitochondria’; Supplementary Table S3).
Nol12 associates with proteins involved in various cellular processes. PrA-Nol12-associated complexes were affinity purified from HEK293T cells. Peptides enriched more than 2-fold over negative controls (untagged and PrA-only cell lines) and present in ≥3 peptides in all replicates, were identified and mapped to the human proteome. (A) Heatmap analysis of semi-quantitative AP-MS-identified preys associated with PrA-Nol12 under different conditions (see Supplementary Table S3 for classification, and Supplementary Table S2 as well as Materials and Methods for quantification). Quantified results from pullouts using 100 mM NaCl without RNase A treatment are shown in columns 1, for 100 and 300 mM NaCl with RNase A treatment in columns 2 and 3, respectively. (B) Pie chart compiling the numbers or preys as well as their overall percentages that were co-purified with Nol12 using 100 mM NaCl without RNase A treatment (see Supplementary Figure S1A for 100 mM and 300 mM NaCl with RNase A). (C) Western analysis of selected AP-MS preys co-isolated with PrA-Nol12 from HEK293 cells. Complexes were affinity-purified from cell lysates with IgG-coupled Dynabeads under conditions identical to those used for AP-MS (A). For each condition, complexes were resolved by SDS-PAGE and the indicated proteins were detected by western blotting using antibodies listed in Supplementary Table S7. Arrows indicate the protein of interest, cross-reacting bands are indicated with an *.
To determine the dependency of Nol12 associations on RNA as well as their stability, we investigated the Nol12 interactome in the presence of RNase A and/or increased salt. Components of the early (90S) pre-ribosomes were reduced by RNAse digestion, while the majority of LSU maturation factors and RPLs were considerably less sensitive to RNAse treatment (Figure 1A, column 2; Supplementary Figure S1C; Table S3), suggesting an RNA-dependency of Nol12 recruitment to 90S pre-ribosomes over 60S pre-ribosomes; however, an incomplete RNA digest due to the protection of pre-rRNA within LSU pre-ribosomal complexes cannot be ruled out. LSU maturation factors and RPLs were also considerably stabilized under increased salt conditions (Figure 1A, column 2; Supplementary Figure S1C; Table S3). Of the non-ribosome complexes, only a small number of co-isolated ‘mRNA/protein regulation proteins’ and ‘DNA and cell cycle proteins’ showed sensitivity to RNAse treatment (Figure 1A, column 2), suggesting that most Nol12 interactions in this group are not RNA-dependent; however, the majority of ‘mRNA/protein regulation proteins’ exhibited salt sensitivity compared to the ‘DNA and cell cycle protein’ group, suggesting labile or transient associations of Nol12 with interactors involved in mRNA metabolism.
To further confirm the presence of selected, high-ranked Nol12-interactors we performed co-affinity-purification coupled to western blot analysis. As shown in Figure 1C, specific association of proteins with different cellular functions co-isolating with Nol12, including ribosome biogenesis (Ddx21), mRNA metabolism (Ddx1), mitochondria (Dhx30), paraspeckles (SFPQ and NONO) and DNA maintenance (Dhx9), was confirmed using specific antibodies directed against prey proteins (Supplementary Table S7), supporting the specificity of the interactions shown by mass spectrometry.
Nol12 localizes to different subcellular compartments
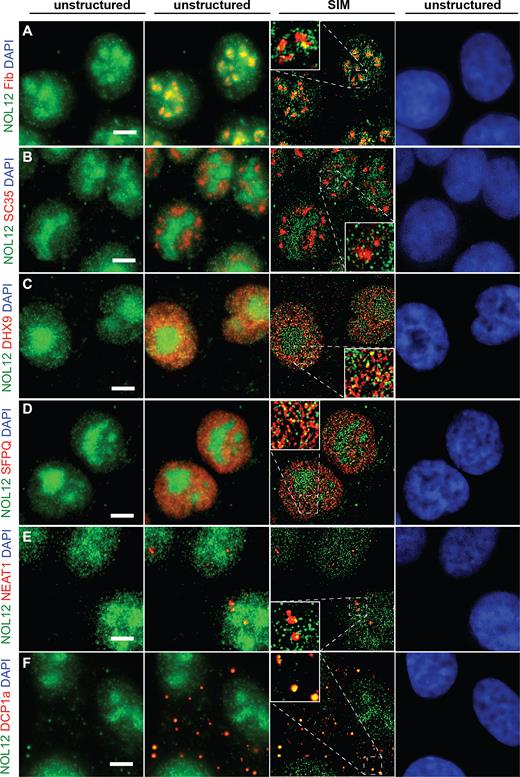
Since proteomic analysis of Nol12-associated complexes identified both nuclear and cytoplasmic proteins, we carried out fluorescent microscopy in HCT116 WT cells to further investigate the presence of Nol12 within specific subcellular compartments using an anti-Nol12 antibody (Sigma, Immunogen aa 1–201; see Supplementary Table S7) and structure illumination super-resolution microscopy. 3D volumes spanning the entire volume of HCT cells were acquired, and spatial overlap of fluorescent signal analyzed by investigating co-localization in individual z-planes (Materials and Methods). Consistent with a nucleolar function, a large fraction of Nol12 localized to nucleoli, with a small amount in the dense fibrillar component (DFC), marked by co-staining for the early ribosomal biogenesis factor Fibrillarin (Figure 2A) (34). The majority of nucleolar Nol12 was found within the granular component (GC) co-localizing with NPM1 (6), consistent with a 90S pre-ribosomes association and a role in early LSU maturation (Supplementary Figure S2A).
Nol12 localizes to different subcellular structures. Localization of endogenous Nol12 in HCT116 WT cells. HCT116 cells were grown on poly-l-lysine-coated glass coverslips, fixed, permeabilized, and incubated for immunofluorescence (IF) (A–C, D, F), or immunofluorescence-fluorescence in situ hybridization (IF-FISH) (E) with antibodies against the indicated proteins (Supplementary Table S7) and/or fluorescently-labeled probes against NEAT1_1/2. Endogenous Nol12 localization was compared to that of the nucleolar marker Fibrillarin (A), the splicing factor SC35 (B), the nuclear DNA/RNA helicase Dhx9 (C), the paraspeckle component SFPQ (D), the lncRNA NEAT1_1/2 (E), and GW/P-body component Dcp1a (F). DNA was visualized by DAPI staining. Cells were imaged using Structured Illumination Microscopy (SIM) and raw (unstructured) and SIM images were computed and processed using Zen SP1 Black Software and the Fiji package of ImageJ. Scale bar = 5 μm.
In addition to its nucleolar localization, Nol12 was also present within numerous distinct foci throughout the nucleoplasm (Figure 2). Nol12 was excluded from the splicing speckles, labeled here by SC35 (Figure 2B), as well as polycomb complexes (Supplementary Figure S2B) or PML bodies (Supplementary Figure S2C) (35). However, concurrent with our AP-MS data, Nol12 foci showed partial, yet distinct, overlap with both Dhx9, a multifunctional nuclear helicase that resolves R- and D-loops (Figure 2C) (36), as well as with the paraspeckle protein SFPQ (Figure 2D) (37,38). Nol12’s presence in paraspeckles was further confirmed by its co-localization with the long non-coding RNA NEAT1, another known component of paraspeckles (Figure 2E) (39–41).
Nol12-associated complexes identified by MS also included cytoplasmic components, in particular, GW/P-body proteins (Figure 1A and B). We observed several bright, discrete cytoplasmic foci in HCT116 WT cells that overlapped with co-staining for Dcp1a (Figure 2F), a key component of the mRNA-containing GW/P-bodies (42), suggesting a function for Nol12 in mRNA metabolism. However, no co-localization was observed with the stress granule marker Tia1 upon arsenite treatment (Supplementary Figure S2D). The specificity of Nol12 localization was confirmed using a second anti-Nol12 antibody raised against a different epitope within the protein (Bethyl Laboratories, Immunogen aa163–213; data not shown).
To further confirm Nol12’s presence in both the nucleus and cytoplasm, we performed cell fractionation of HCT 116 WT cells and compared the fractionation pattern to those of known cytoplasmic (GAPDH), nuclear (CDK9), and chromatin-associated proteins (H3). Nol12 was found both in the cyto- and nucleoplasmic fractions, and a small amount also in the chromatin fraction (Supplementary Figure S2E, lanes 1, 2 and 3). Pre-treatment with RNAse showed that the majority of nucleoplasmic Nol12 was RNA dependent, while the chromatin associated fraction was not (Supplementary Figure S2E, lanes 4 and 5). Due to high numbers of DNA repair proteins found associated with PrA-Nol12 by AP-MS, we also determined the subcellular distribution under DNA stress conditions. Cells were pretreated with 1 μg/ml Actinomycin D (Act D), which intercalates into DNA inducing double-strand breaks and nucleolar disruption, for 3 h in the presence or absence of RNAse (43). Analysis of Nol12 in different fractions revealed not only an increase in cytoplasmic Nol12 but also the appearance of an RNA-independent protein pool in the nucleoplasm suggesting that Nol12 may dissociate from RNA under DNA damage conditions (Supplementary Figure S2E, lanes 6 and 9). As the increase in nucleoplasmic RNA-independent Nol12 coincides with the disruption of nucleolus, it could represent released nucleolar in addition to nucleoplasmic Nol12, not associated with RNA. Moreover, the Nol12 bands of differing migration detected in the cytoplasmic and nucleoplasmic fractions suggests that Nol12 may be posttranslationally modified in different cell compartments. No change in our control proteins was observed under these conditions. Taken together, our data shows that Nol12 is localized to the nucleolus, nucleus and cytoplasm where it may fulfill different functions.
Loss of Nol12 leads to an increase in cellular stress and p53-independent cell cycle arrest and apoptosis
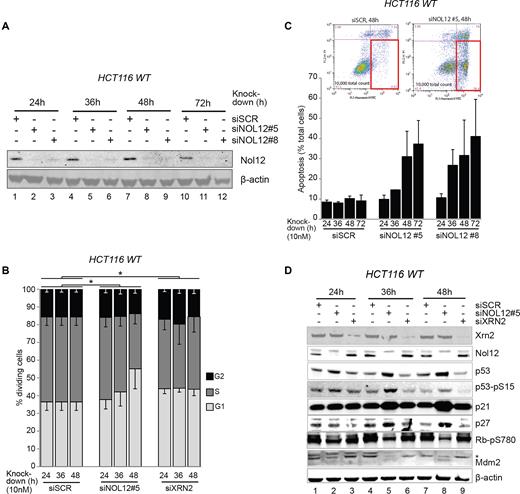
To investigate the cellular function of Nol12, we first determined the overall effect of Nol12 depletion on cell proliferation and survival in HCT116 WT cells, using small interfering RNAs (siRNAs; siNOL12) against NOL12 mRNA (Figure 3A, siNOL12#5 and #8) (20,22). Compared to cells transfected with a scrambled control siRNA (siSCR), knockdown of NOL12 resulted in a robust G1/S arrest between 36–48 h (Figure 3B), and induced a strong apoptotic response within 36–48 h of siRNA transfection, as evidenced by both increased annexin V binding without concurrent propidium iodide infiltration (Figure 3C), and by proteolytic activation of caspase 3 (Supplementary Figure S3A). By comparison, knockdown of the multifunctional protein Xrn2 induced only a subtle S-phase delay within 24 h of siRNA treatment (Figure 3B), and no apoptotic response (Figure 3C) (32). Analysis of HCT WT whole cell lysates by western blotting 24, 36 and 48 h after transfection revealed that depletion of Nol12, but not Xrn2, resulted in progressive accumulation of the key cell cycle- and apoptosis-regulatory protein p53, as well as p21WAF1/CIP1, p27KIP1, and hypo-phosphorylation of Mdm2 (Figure 3D, lanes 5 and 8 versus lanes 6 and 9). These changes coincided with the downregulation of S-phase-promoting phosphorylation of Rb and increased phosphorylation of p53 on Ser15, a stabilizing modification by PIKK-family kinases (ATM, ATR and DNA-PK) usually observed in response to DNA damage (Figure 3D, lanes 5 and 8) (44), suggesting that loss of Nol12 elicits significant cellular stress, activating both cell cycle arrest and apoptosis response pathways.
Nol12 knockdown causes G1/S arrest and apoptosis. (A) HCT116 WT cells were transfected with 10nM of silencing RNAs, siSCR (control), siNOL12#5 or siNOL12#8, for the times indicated. Whole cell extracts (WCEs) of siRNA-treated cells were resolved on 4–12% Bis–Tris gradient gels, transferred to PVDF membrane and blotted with the indicated primary antibodies. Knock-down efficiency of NOL12-targeting siRNAs was tested by aprobing membrane with an anti-Nol12 antibody (Sigma). Beta-actin was used as a loading control. (B) HCT116 WT cells were transfected with siSCR, siXRN2, or siNOL12#5 (10 nM) for the times indicated. Propidium iodide (PI)-stained DNA content was quantified by FACS, and G1 (2n), S (2n-4n) and G2 (4n) populations determined. Data represent mean ± S.D. for five independent experiments. *P < 0.05 by Student's two-tailed t-test on area under the curve (AUC) of G1 or AV+ PI− populations as appropriate. (C) HCT116 WT cells were transfected with siSCR, siNOL12#5, or siNOL12#8 (10 nM) for the times indicated. Annexin V-FITC-positive, PI-negative apoptotic cells (gated, inset) were quantified by FACS. Data represents mean±s.d. for two independent experiments. (D) HCT116 WT cells were transfected with siSCR, siXRN2, or siNOL12#5 (10 nM) for the times indicated. Whole cell extracts (WCEs) of siRNA-treated cells were resolved on 4–12% Bis–Tris gradient gels, transferred to PVDF membrane and blotted with the indicated primary antibodies (Supplementary Table S7). Representative images of biological replicates are shown. Asterisk denotes the double phosphorylated form Mdm2-pS166/pS186/S188.
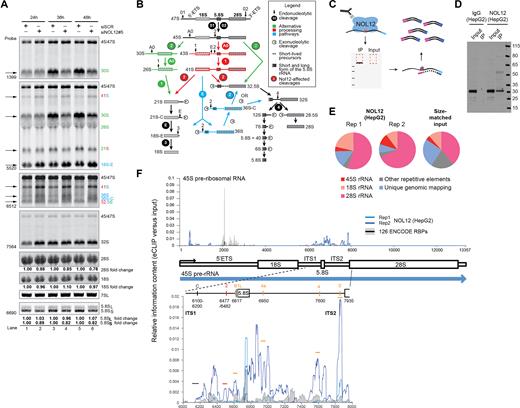
Nol12 was previously suggested to be a putative ribosome biogenesis factor with a role in ITS1 processing based on its S.cerevisiae homolog Rrp17p and studies in HeLa cells, where in the latter ribosome processing events were surveyed 60 h after siNOL12 transfection (22). To determine whether the function of Nol12 in ribosome biogenesis could be the cause for the G1/S cell cycle arrest and apoptosis observed 36–48 h after Nol12 knockdown, cells were transfected with siNOL12 (Figure 4A, lanes 2, 4 and 6) or a siSCR control (Figure 4A, lanes 1, 3 and 5), and pre-rRNA processing was analyzed by Northern blotting 24, 36 and 48 h after transfection. Knockdown of NOL12 in HCT WT cells resulted in rRNA processing defects affecting precursors produced by cleavage at site 2 within ITS1 (Figure 4A): the large subunit precursor 32.5S, and the small subunit pre-rRNAs 30S, 26S and 21S were noticeable decreased after 36 h compared to the SCR control (Figure 4A). Conversely, levels of site E (also termed site 2a, (43)) cleavage products, namely 36S pre-rRNAs, its subsequent 5′end processed form 36S-C as well as the small subunit precursor 18S-E were increased as early as 24 h post NOL12 siRNA treatment (Figure 4A), suggesting an increased number of cleavage events at site E/2a in the absence of Nol12. Mature 28S, 18S and 5.8S rRNA levels were overall only modestly decreased, with LSU rRNAs (28S, 5.8S) more affected than SSU ones (18S). In higher eukaryotes, processing of Pol I-transcribed pre-rRNA occurs along three parallel pathways, presumably to ensure biogenesis of ribosomes (Figure 4B) (10). The major pathways require an endonucleolytic cleavage at site 2 to separate SSU and LSU pre-rRNAs (Figure 4B; green and red, respectively), while an alternative, minor pathway produces downstream precursors via endonucleolytic cleavage at site E (Figure 4B; blue) (10). The decrease of ‘Major Pathway’ precursors in response to Nol12 depletion together with a concomitant increase in products of site E cleavage along the ‘Minor Pathway’ (Figure 4A; 36S, 36S-C and 18S-E, arrows) suggests a role for Nol12 in cleavage events at site 2, and a rerouting of ribosome processing events from cleavage at site 2 to site E to sustain ribosome production in the absence of Nol12.
Nol12 is enriched at pre-rRNA and required for efficient rRNA processing. (A) Total RNA extracted at different time points from HCT116 WT cells transfected with 10nM siNOL12#5 or siSCR was analyzed by northern blotting using probes hybridizing to either 5′-ETS, ITS1 or ITS2 regions of the pre-rRNA to detect processing intermediates. RNA loading was monitored by methylene blue staining of mature 18S and 28S rRNAs (large RNAs) or northern blotting for the 7SL RNA (small RNAs). Changes in levels of mature rRNAs (5.8S, 18S and 28S) were quantified and normalized against 47S pre-rRNA for each lane. A schematic of rRNA maturation pathways in human cells is shown in (B). (C) Schematic of eCLIP. Nol12 is crosslinked to bound RNA, and Nol12 and associated RNA are subjected to immunoprecipitation (IP). After 3′ adapter ligation, both IP and input samples are subjected to PAGE electrophoresis and transferred to nitrocellulose membranes, after which the region from Nol12 (estimated size 25 kDa) to ∼75 kDa above is isolated and RNA is extracted. After reverse transcription, a 3′ cDNA adapter is ligated, and samples are PCR amplified to obtain final library. (D) IP-western image for Nol12 in HepG2, as well as IP performed with isotype control IgG. (E) Pie charts indicate the fraction of reads in Nol12 eCLIP replicates, or size-matched input, associated with ribosomal RNA (red-pink), other repetitive elements (grey), or uniquely mapping to the genome (blue). (F) Nol12 enrichment at rRNA. Lines indicate relative information in IP versus input at each position along (top) the 45S pre-ribosomal RNA or (bottom) the Internal Transcribed Spacers 1 and 2 (ITS1 and ITS2). Blue lines indicate biological replicates of Nol12 in HepG2, whereas black line indicates the mean observed across 181 eCLIP experiments in K562 and HepG2 cell lines (profiling 126 total RBPs) generated by the ENCODE consortia (with standard deviation indicated in gray).
To validate the association between Nol12 and rRNA, we obtained Nol12 eCLIP profiling data performed in HepG2 cells by the ENCODE project (Figure 4C and D). As standard data processing discards reads that map to repetitive elements, we re-mapped sequencing reads to ribosomal RNA using a family-aware repeat mapping pipeline (Van Nostrand, E.L., et al., in preparation). This analysis indicated that 81.2% (rep1) and 89.4% (rep2) of reads in Nol12 eCLIP map to all rRNA transcripts, including the 45S precursor, with 55.2% and 68.3% mapping to just the 28S rRNA, significantly more than observed in the size-matched paired input (54.5% for all rRNA or 40.0% for 28S specifically) (Figure 4E; Supplementary Figure S4A; Table S6). We observed enrichment of Nol12 at multiple positions throughout the 45S pre-rRNA, particularly in the ITS1 and ITS2 regions, which were enriched both relative to the size-matched paired input as well as versus all 181 publicly available eCLIP datasets in HepG2 and K562 (covering 126 RBPs), suggesting these enriched loci are specific to Nol12. Within ITS1, Nol12 eCLIP mapped to sequences around site 2, supporting a role of Nol12 in efficient site 2 processing, and B1L (Figure 4F). Nol12 enrichment was also observed within the ITS2, around sites 4a, 4, and 3′, and Sloan and colleagues observed a decrease in 12S pre-rRNA, a product of ITS2 processing, upon loss of Nol12 (43); however, any role for Nol12 in ITS2 processing remains to be determined.
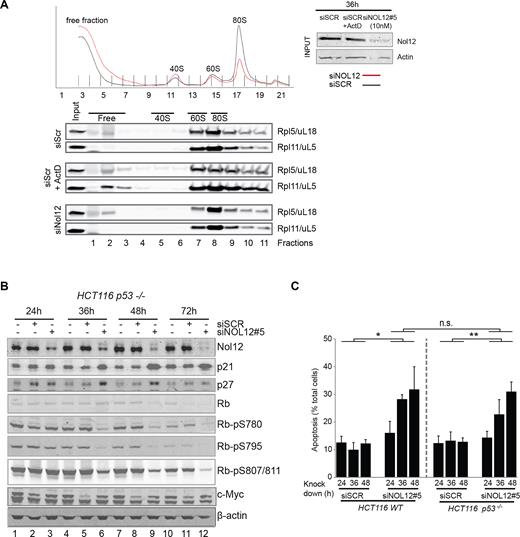
As levels of mature rRNAs are not significantly affected in the absence of Nol12, it is unlikely that the protein's role in ribosome biogenesis is the direct cause of cell death. However, loss of some nucleolar proteins, ribosome biogenesis factors or ribosomal proteins have been shown to trigger a nucleolar stress response via a release of ribosomal proteins Rpl5/uL18 and Rpl11/uL5 from pre-ribosomes into the nucleoplasm, where they bind and downregulate the p53-ubiquitin E3 ligase Mdm2, leading to subsequent p53 accumulation, cell cycle arrest and apoptosis (12). Moreover, the Nol12-mouse homologue, Nop25, was suggested to function in the maintenance of nucleolar architecture (19,20). To determine whether Nol12 depletion leads to a nucleolar stress response, we used sucrose gradient centrifugation to determine the release of Rpl11/uL5 and Rpl5/uL18 from pre-ribosomes and an increase in their free fractions (45). No additional increase in free Rpl11/uL5 or Rpl5/uL18 (Figure 5A) was observed upon depletion of Nol12 compared to the positive control, where an increase of Rpl11/uL5 and Rpl5/uL18 in the free fraction could be observed upon Actinomycin D treatment and induction of nucleolar stress. We furthermore determined whether NOL12 kd-mediated G1/S arrest and apoptosis were dependent on p53 stabilization. To that end, Nol12 was depleted in HCT116 cells lacking functional p53 (HCT116 p53−/−(46)). Both apoptosis (Figure 5C) and G1/S arrest (Supplementary Figure S5B) were identical to p53+/+ cells upon loss of Nol12, while no cell cycle delay was detected upon XRN2 kd. While p53-dependent pathways represent the major effector mechanisms of nucleolar stress, p21WAF1/CIP1, p27KIP1 and c-myc were previously shown to activate nucleolar stress-dependent cell cycle arrest in a p53-independent manner (12). Indeed in HCT116 p53−/− cells, p21WAF1/CIP1 and p27KIP1 were both upregulated following NOL12 kd, while c-myc, another target of Rpl11/uL5, was unaltered (Figure 5B, lanes 3, 6, 9, and 12) (12). In addition, co-depletion of Nol12, p53 and p21WAF1/CIP1 or p27KIP1 in HCT WT cells (Supplementary Figure S5C), or of Nol12 and either p21WAF1/CIP1 or p27KIP1 in HCT p53−/− cells failed to rescue the G1/S arrest (Supplementary Figure S5D), and pre-rRNA maturation defects upon NOL12 kd in HCT p53−/− cells were identical to those observed in HCT WT cells (Supplementary Figure S5E, lanes 2 and 4).
NOL12 kd-induced apoptosis is independent of p53 and nucleolar stress. (A) RPL5/uL18 and RPL11/uL5 do not freely accumulate in the nucleoplasm upon Nol12 depletion as part of a nucleolar stress response. Cell lysates from HCT cells transfected for 36h with 10nM of scramble, Nol12 #5 siRNA, or scramble and treated with 0.05μg/ml Actinomycin D for 3 h, were fractionated on a sucrose gradient. Fractions were analyzed by western blotting for the presence of both Rpl5/uL18 and Rpl11/uL5 (antibodies listed in Supplementary Table S7). The efficiency of Nol12 knockdown was assessed by western blotting as shown in the input panels. (B) HCT116 p53−/− cells were transfected with siSCR or siNOL12#5 (10 nM) for the times indicated. Whole cell extracts (WCEs) of siRNA-treated cells were resolved on 4–12% Bis–Tris gradient gels, transferred to PVDF membrane and blotted with the indicated primary antibodies (Supplementary Table S7). Representative images of biological replicates are shown. (C) HCT 116 WT or p53−/− cells were treated with siSCR or siNOL12#5 (10 nM) for the times indicated, and apoptotic cells quantified. Annexin V-FITC-positive, PI-negative apoptotic cells (gated, inset) were quantified by FACS. Data represents mean ± S.D. for two independent experiments. *P < 0.5, **P < 0.01, n.s., non-significant by Student's two-tailed t-test on the area under the curve (AUC) of G1 or AV+ PI− populations as appropriate.
Taken together our data suggests that the NOL12 kd-induced cell cycle arrest and apoptosis are not the result of a nucleolar stress response, and, furthermore, are not dependent on p53, c-myc, p21 or p27, and are thus qualitatively distinct from a nucleolar stress response suggesting that the G1/S delay and apoptosis observed upon NOL12 kd can occur even in the absence of p53.
Nol12-induced apoptosis is dependent on events downstream of the DNA damage-sensing kinase ATR
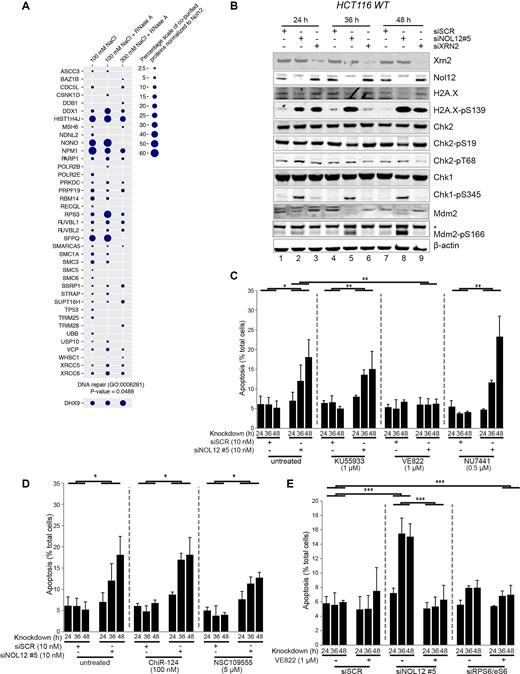
The phosphorylation of p53 on Ser15, an event associated with the activity of the DNA damage-sensing PIKK-family kinases ATM, ATR and DNA-PK following Nol12 depletion (Figure 3D) and identification of 40 proteins associated with DNA repair pathways (GO: 0006281) associated with PrA-Nol12 by AP-MS (Figure 6A), led us to examine the activation of DNA damage response (DDR) pathways upon Nol12 depletion. Western analysis of whole cell lysates from NOL12 kd cells revealed the induction of signaling cascades typically associated with a DDR (Figure 6B). Phosphorylation of the variant histone H2A.X on Ser139 (γH2A.X), an early event in DNA damage recognition, was increased 24 h after siNOL12 transfection (Figure 6B, lane 2), an effect also seen upon kd of XRN2 although to a lesser extent (Figure 6B, lane 3). NOL12 kd also resulted in a robust and rapid phosphorylation of Chk1 on Ser345 after only 24 h, indicating activation of the ATR kinase (Figure 6B, lane 2), while only a mild increase in phosphorylation of Chk2 at Thr68 and a decrease of Chk2pSer19, targets of DNA-PK and ATM kinases, respectively, were observed, suggesting that loss of Nol12 triggers activation of the ATR-Chk1 arm of the DDR cascade, and, to a lesser extent, ATR-Chk2 (47). In addition, we observed phosphorylation of Mdm2 on Ser166 (Figure 6B, lanes 2, 5 and 8) in NOL12 kd but not XRN2 kd cells, with a concomitant downregulation of Mdm2. Mdm2pS166 is an activating phosphorylation mediated by p-Akt upon oxidative stress that propagates p53 degradation, but is blocked by p53-phosphorylation on Ser15/Ser20. Upon increased oxidative stress and Mdm2pS166 phosphorylation, p53pS15 is degraded, an effect also observed upon NOl12 kd in HCT WT cells after 48 h (Figure 3D, lane 8), suggesting that activation of p53pS15 and activation of Mdm2pS166 may be caused by different upstream events and effectors, leading to the activation of different downstream pathways (44,48); moreover, the data also suggests that the Mdm2pS166-triggering event may be the one resulting in apoptosis of Nol12-depleted cells.
Nol12 associates with DDR factors and its loss causes activation of DNA damage response pathways via ATR signaling. (A) PrA-Nol12 associated proteins identified by MS analysis with a ‘DNA repair’ gene ontology annotation (GO:0006281) and the DNA repair implicated DNA/RNA helicase Dhx9. Circles represent semi-quantitative analysis based on peptide numbers identified. (B) HCT116 WT cells were transfected with siSCR, siXRN2, or siNOL12#5 (10nM) for the times indicated. Whole cell extracts (WCEs) of siRNA-treated cells were resolved on 4–12% Bis–Tris gradient gels, transferred to PVDF membrane and blotted with the indicated primary antibodies (Supplementary Table S7). Representative images of biological replicates are shown. Asterisk denotes the double phosphorylated form Mdm2-pS166/pS186/S188. (C) HCT116 WT cells were transfected with siSCR or siNOL12#5 (10 nM) as indicated and grown in the presence of KU55933 (1 μM; ATM inhibitor), VE822 (1 μM; ATR inhibitor) or NU7441 (0.5 μM; PRKDC inhibitor) for the indicated times prior to quantification of apoptotic cells by FACS as in Figure 3C. Data represents mean±s.d. for four independent experiments. * P < 0.5, ** P<0.01 by Student's two-tailed t-test (D) HCT116 WT cells were transfected with siSCR or siNOL12#5 (10nM) as indicated and grown in the presence of ChiR-124 (100 nM; Chk1 inhibitor) or NSC109555 (5 μM; Chk2 inhibitor) for the indicated times prior to quantification of apoptotic cells as in Figure 3C. Data represents mean ± S.D. for four independent experiments. *P < 0.5 by Student's two-tailed t-test. (D) HCT116 WT cells were transfected with siSCR, siNOL12#5 or siRPS6/eS6 (10 nM) as indicated and grown in the presence of VE822 (1 μM; ATR inhibitor) for the indicated times prior to quantification of apoptotic cells as in Figure 3C. Data represents mean ± S.D. for four independent experiments. *P < 0.5, **P < 0.01, ***P < 0.005 by Student's two-tailed t-test.
ATM, DNA-PK and ATR are activated by different DNA damage signals. ATM and DNA-PK recognize double-strand breaks (DSBs), while ATR recognizes regions of single-strand DNA that arise from a range of insults including single-strand breaks (SSBs), DNA oxidation, and stalled replication forks; however, some cross-talk between the pathways exists (49,50). In order to investigate the relationship between ATM, ATR, and DNA-PK and Nol12 function, HCT WT cells transfected with siSCR or siNOL12 were grown in the presence of specific inhibitors of ATM (KU55933), ATR (VE822) and DNA-PK (NU7441) (Figure 6C) (51–53). Cells were assayed for apoptosis 24, 36 and 48 h post-transfection, as previously described. While neither KU55933 nor NU7441 affected the induction of apoptosis following NOL12 kd, treatment with VE822 reduced the apoptotic response to that of background (i.e. siSCR + VE822; Figure 6c). Dose-response analyses revealed that VE822 inhibited NOL12 kd-dependent apoptosis with an IC50 of 92.2nM (R2 = 0.982), 4.9-fold higher than the published IC50, while KU55933, for comparison, inhibited apoptosis by only 38% at the highest concentration tested (20μM, 1550-fold over the published IC50). Next, we tested the relation of NOL12 kd-induced apoptosis to Chk1 and Chk2, the kinases downstream of ATM and ATR. Surprisingly, treatment with the Chk1-inhibitor ChiR124 did not reduce the apoptotic response upon NOL12 kd (Figure 6D, middle), while a mild reduction upon treatment with the Chk2-inhibitor NSC109555 was observed (Figure 6D, right) (54,55). These results suggest that, while NOL12 kd-induced apoptosis requires signaling via ATR, it may do so independently of Chk1, despite leading to its phosphorylation; instead, apoptosis upon loss of Nol12 may be dependent on signaling via Chk2. Importantly, knockdown of the ribosomal protein gene, RPS6/eS6, did not result in an apoptotic response and was insensitive to VE822 treatment, confirming that the NOL12 kd-related apoptotic response is unlikely to be the result of nucleolar or ribosomal stress (Figure 6E). Overall, these data suggest that Nol12 may function in an ATR-induced DNA damage response.
Given that the cell cycle arrest arising from NOL12 kd was resistant to inhibition of nucleolar stress pathways, we tested whether it too was dependent on DNA damage signaling. To this end, we carried out cell cycle profiling experiments in the presence of caffeine, a PIKK kinase inhibitor that targets both ATM and ATR, and the DNA-PK inhibitor NU7441 (56). Consistent with observations described above, the <2n cell cycle population (consisting of apoptotic/necrotic cells and cell debris) was significantly decreased in the presence of caffeine but not NU7441 (Supplementary Figure S6A), as was the total apoptotic cell count (Supplementary Figure S6B). However, the induction of G1/S arrest was unchanged in the presence of either caffeine or NU7441 (Supplementary Figure S6A), suggesting that the cell cycle arrest and apoptotic phenotypes of the NOL12 kd arise by different mechanisms and, possibly, different upstream events.
Nol12 associates with sites of ATR-activating DNA insults
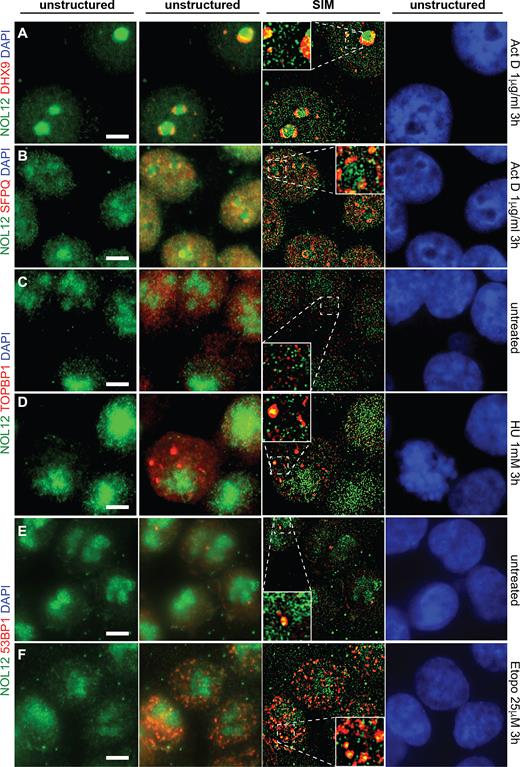
As PrA-Nol12 co-isolated numerous proteins implicated in DNA damage response and repair pathways, and NOL12-kd resulted in the activation of ATR-mediated DDR signaling, we next investigated whether Nol12 was localized to sites of DNA repair upon different insult (Figure 7). Association of Nol12 with Dhx9 and SFPQ was not RNA dependent and therefore suggested a potential DNA-related association with these proteins. Moreover, both Dhx9 and SFPQ have previously been implicated in DNA damage, when both are recruited to sites of damage and into nucleolar caps (36). Upon Act D treatment at concentrations known to affect overall genomic stability (1 μg/ml, 3 h), co-localization of Nol12 with Dhx9 was increased in both nucleolar caps as well as individual foci suggesting, in line with our proteomic data, a potential co-function for Nol12 in genome instability management (Figure 7A) (36). Under the same conditions, Nol12 partially co-localized with SFPQ to nucleolar caps (Figure 7B) (7,57). However, unlike SFPQ and Dhx9, Nol12 did not retreat into nucleolar caps, but instead formed a peripheral nucleolar ring while still maintaining its presence within the nucleolus, nucleoplasmic foci, and the cytoplasm. Co-localization of Nol12 with the nucleolar protein Fibrillarin was not significantly altered upon Act D treatment (Supplementary Figure S7A), whereas co-localization with NPM1 was diminished (Supplementary Figure S7B), suggesting divergent function for Nol12 and NPM1 under DNA stress conditions.
Nol12 localizes to sites of DNA insults. HCT116 WT cells were grown on poly-l-lysine-coated glass coverslips, fixed, permeabilized and incubated for immunofluorescence (IF) with antibodies against the indicated proteins (Supplementary Table S7). Endogenous Nol12 localization was compared to that of the RNA/DNA DEAD-box helicase Dhx9 (A) and the paraspeckle component SFPQ (B) in actinomycin D-treated (1 μg/ml 3 h) cells. Co-localization of endogenous Nol12 was also determined with the replication fork marker TOPBP1 in untreated (C) and HU-treated (1 mM 3 h) cells (D), and the DNA repair protein 53BP1 in untreated (E) and etoposide-treated (25 μM 3 h) cells (F). DNA was visualized by DAPI staining. Images are representative of three independent experiments. Cells were imaged using Structured Illumination Microscopy (SIM) and raw (unstructured) and SIM images were computed and processed using Zen SP1 Black Software and the Fiji package of ImageJ. Scale bar = 5 μm.
Since NOL12 kd-induced apoptosis was reduced upon inhibition of ATR (Figure 6C; VE822), we further investigated whether Nol12 re-localized to sites of ATR activating insults, such as stalled replication forks, by determining the protein's co-localization with TOPBP1, a checkpoint protein that is localized with ATR at sites of DNA replication stress (58). In untreated cells, Nol12 and TOPBP1 showed no spatial overlap (Figure 6C); however, upon treatment with the replication fork stalling agent hydroxyurea (HU), Nol12 and TOPBP1 were found to co-localize within large foci (Figure 7D ; 1 mM HU, 3 h), suggesting that Nol12 is localized to sites of DNA replication stress. We further examined if Nol12 co-localized with 53BP1, a protein recruited predominantly to DNA double-strand breaks, upon treatment of cells with the topoisomerase II inhibitor etoposide, known to produce both DNA double and single strand breaks (59–62). While in untreated cells little 53BP1 was detected (Figure 7E), Nol12 and 53BP1 were found to co-localize within nucleoplasmic foci upon etoposide treatment (Figure 7F; 25 μM Etopo, 3 h) (63). Taken together, our data suggests that Nol12 localizes to sites of replication stress, which activate ATR-mediated DNA repair pathways, where it co-localizes with TOPB1 and 53BP1.
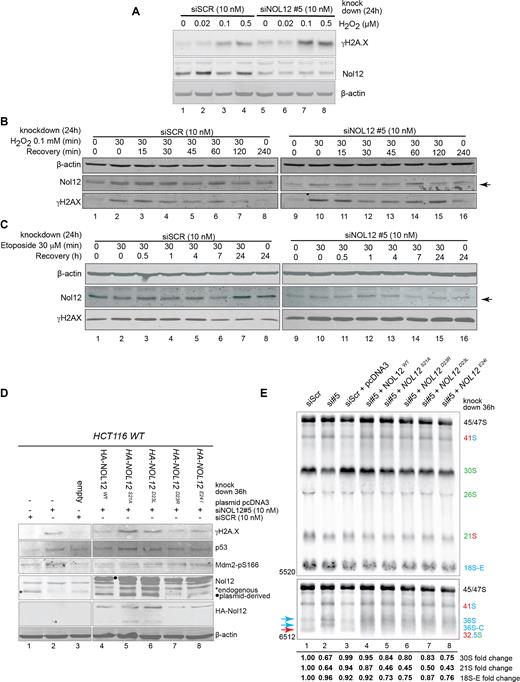
Following these observations, we next assayed whether loss of Nol12 sensitized cells to DNA insults, resulting in activation of an ATR-mediated DNA repair. Therefore, we treated HCT116 WT cells, transfected for 24 h with either siSCR or siNOL12, with increasing amounts of hydrogen peroxide (H2O2) to induce oxidative damage and ATR-mediated DNA damage response, as previously shown in Driessens et al. (64). As shown in Figure 8A, H2O2 treatment led to increased levels of γH2A.X in the absence of Nol12 (Figure 8, lanes 7 and 8) compared to control knock-down cells (Figure 7, lanes 3 and 4). Moreover, recovery from both peroxide-induced (Figure 8B) and etoposide-induced (50 μM, 30 min; Figure 8C) (65) DNA insults was inefficient in Nol12-kd cells, suggesting that Nol12-depleted cells are not only more sensitive to DNA stress, but the absence of the protein may interfere with resolving the insult, potentially leading to apoptosis.
Loss of Nol12 prevents cell recovery from DNA damage and its conserved domain is required for its function in DNA stress and ribosome processing. (A) HCT116 WT cells grown in 12-well dishes were transfected with siSCR or siNOL12#5 (10 nM) for 24 h before being treated with the indicated concentrations of H2O2 in DMEM + 10% FBS for 30 min prior to 30 min recovery in unsupplemented DMEM + 10% FBS. Cells were harvested and 55 μg of protein from each condition was resolved by SDS-PAGE and the indicated proteins analyzed by western blotting. (B) HCT116 WT cells grown in 12-well dishes were transfected with siSCR or siNOL12#5 (10 nM) for 24 h before being treated or not with 0.1 mM H2O2 in DMEM + 10% FBS for 30 min then allowed to recover in unsupplemented DMEM + 10% FBS for the indicated time periods prior to harvesting and protein extraction. 55μg of protein from each condition was resolved by SDS-PAGE and the indicated proteins analyzed by western blotting. (C) HCT116 WT cells grown in 12-well dishes were transfected with siSCR or siNOL12#5 (10 nM) for 24 h before being treated or not with 50 μM etoposide in DMEM + 10% FBS for 30 min then allowed to recover in unsupplemented DMEM + 10% FBS for the indicated time periods prior to harvesting and protein extraction. 55 μg of protein from each condition was resolved by SDS-PAGE and the indicated proteins analyzed by western blotting. (D) HCT116 WT cells were transfected for 12h with either siSCR or siNOL12 (10 nM; lanes 1 and 2), or for 12 h with siSCR or siNOL12 (10 nM), followed by a 24 h transfection of empty pcDNA3 in the case of siSCR (lane 3), or with Nol12 WT, Nol12-S21A, Nol12-D23R, NOL12-D23L or NOL12-E24I in the case of siNOL12 (lanes 4–8). Whole cell extracts (WCEs) were then resolved on 4–12% Bis–Tris gradient gels, transferred to PVDF membrane and blotted with the indicated primary antibodies (Supplementary Table S7). * indicated endogenous Nol12, • plasmid-derived HA-tagged Nol12. (E) Northern blotting of rRNA using total RNA extracted from HCT116 WT cells transfected for 36h with either siSCR or siNOL12 (10nM; lanes 1 and 2), or for 12h with siSCR or siNOL12 (10nM), followed by a 24 h transfection of empty pcDNA3 in the case of siSCR (lane 3), or with Nol12 WT, Nol12-S21A, Nol12-D23R, NOL12-D23L, or NOL12-E24I in the case of siNOL12 (lanes 4–8). Pre-ribosomal RNAs were analyzed using probes hybridizing to the 5′ and 3′ ITS1 regions of the pre-rRNA to detect processing intermediates. Changes in levels of pre-rRNAs (30S, 21S and 18S-E) were quantified and normalized against 47S pre-rRNA for each lane.
Nol12’s conserved domain is required for its roles in RNA and DNA metabolism
The S. cerevisae homologue of Nol12, Rrp17, has previously been shown to contain a putative catalytic domain that is highly conserved (Supplementary Figure S8), and required for its function (21). To determine whether mutations within the conserved domain affect Nol12 function in vivo, we constructed four point mutants, targeting residues within the putative catalytic domain (Figure 8D). Aspartic acid (D) 23, in yeast D32, was previously shown to be important for Rrp17’s function (21), and was replaced with a negatively charged arginine or uncharged leucine; moreover, we exchanged its surrounding residues with uncharged amino acids (S to A and E to I). The point mutations were introduced into siRNA-insensitive plasmid-derived HA-Nol12 constructs, that were transfected into HCT 116 WT cells 12 h after treatment with siNOL12#5 to deplete endogenous NOL12. Expression of a wild-type Nol12 construct was able to rescue the Nol12-kd induced DNA damage phenotype as indicated by reduced γH2A.X levels (Figure 8D, lanes 2 and 4). Nol12 function on DNA stress was not affected in D23R and E24I mutants (Figure 8D, lanes 7 and 8); however, conversion of aspartic acid 23 to leucine and serine 21 to alanine disrupted Nol12 activity resulting in the activation of DNA damage response pathways (Figure 8D, lanes 5 and 6). Similarly, ribosomal RNA processing defects were rescued by a co-transfected wild-type Nol12 allele (Figure 8E, lanes 2 and 4; Supplementary Figure S8), while none of the mutants were able to complement the Nol12 kd induced phenotype (Figure 8E, lanes 5–8). Site E/2a (43) cleavage products, namely 36S pre-rRNAs, its subsequent 5′end processed form 36S-C were increased in all mutants over the SCR control, while the site 2 cleavage product, 32.5S, was decreased. The heterogeneous distribution of 32.5S, 36S, and 36S-C in cells expressing the wild-type Nol12 construct in the presence of siNOL12, may be due to a lower transfection efficiency of cells after Nol12 knockdown resulting in a mixed population (Figure 8E, lane 4). Overall, this data suggests the importance of the conserved putative nuclease domain in Nol12 function.
DISCUSSION
To counteract the breakdown of genome integrity, eukaryotic cells have developed a network of pathways to prevent and resolve DNA damage. Here, we identify the human protein Nol12 as a novel RBP required for both ribosome maturation and for the maintenance of genome integrity. Knockdown of NOL12 impaired cellular proliferation by inducing a G1/S cell cycle arrest and, furthermore, led to a rapid apoptotic response due to activation of the DDR pathway(s) proceeding via ATR kinase. Nol12 associated with chromatin-associated factors such as PARP, NPM1, as well as proteins implicated in DNA damage repair such as the paraspeckle components SFPQ and p54nrb/NONO, and the RNA/DNA DEAD-box helicases Dhx9 and DDX1, both involved in genome instability prevention. Moreover, upon DNA stress, Nol12 was localized to sites of replication stress and DNA insults in vivo together with TOPBP1 and 53BP1, respectively, suggesting a role for Nol12 in maintaining genome integrity. Nol12 was also associated with RNA metabolism pathways outside of ribosome biogenesis, namely in P-bodies, which is supported by both proteomic data and its co-localization with Dcp1a in vivo.
A role for Nol12 in the maintenance of genome integrity
While Nol12 is part of early 90S and pre-60S ribosomal subunits in vivo, where it is required for efficient separation of the large and small ribosomal subunit precursors via cleavage at site 2, the marked severity of the G1/S arrest, and the induction of apoptosis, suggest that it is Nol12’s role in genome maintenance and not ribosome biogenesis that is responsible for the rapid decrease in cell survival observed upon Nol12 kd. This is supported by the only moderate 20–25% reduction of levels in mature 28S and 5.8S rRNAs observed upon Nol12 kd. In Drosophila, knock-down of the Nol12 homologue Viriato resulted in loss of cell proliferation and apoptosis in a dMyc-dependent manner, and a nucleolar stress response (18,19). However, the apoptotic response upon NOL12 kd is outside of the typical nucleolar stress response that has been observed for many ribosome biogenesis factors and independent of both p53 and c-myc signaling and (22,32,66).
In higher eukaryotes, DNA damage sustained from DNA replication, oxidative stress, mitotic catastrophe and damaging agents can induce apoptosis if left unresolved (67). The increased sensitivity to several DNA-damaging agents, including etoposide and oxidative stress strongly indicate that Nol12-depleted cells have increased their sensitivity to, and/or lost their ability to resolve DNA stress. These factors are the likely direct cause for the observed rapid apoptotic response 36–48 h post-siRNA treatment. This notion is supported by the activation of the ATR-Chk1/2 DDR pathway—as evidenced by phosphorylation of Chk1pS345, Chk2pT68 and Mdm2pS166—and the abolition of the apoptotic response by pre-treatment with the ATR-specific inhibitor VE822 (Figure 7B, C and E) (47,48). ATR is a PI3K family kinase (alongside ATM and DNA-PK) that is activated by factors including TOPBP1, a BRCT domain containing protein, upon recruitment to sites of single-stranded (ss) DNA arising from DNA damage, replication stress or other structures [9]. Once ATR is active, it phosphorylates a number of substrates, including Chk1 at serine 345, to coordinate DNA repair and cell cycle progression (68,69). ATR-mediated apoptosis has previously been observed upon replicative stress via ribonucleotide reductase inhibition, interstrand cross-links, and oxidative or base damage of DNA, amongst others (49). It has recently been suggested that persistent formation of RNA:DNA hybrids (R-loops) by nascent transcript invasion of the DNA duplex can impede replication progression leading to replication fork stalls, DNA damage and rearrangements in a replication-dependent manner (70). Recently, several different RNA/DNA DEAD/H-box helicases—Ddx19, Ddx21 and Dhx9—were found to play important roles in facilitating the removal of R-loops as well as G-rich tetraplexes (71–74). A third DEAD/H-box helicase, Ddx1, was further reported to resolve RNA:DNA hybrids formed within end-resected regions of ssDNA at DNA DSBs (75). Three of these four of these helicases (Ddx1, Ddx21 and Dhx9) were found to copurify at high levels with Nol12 in an RNA-independent manner by affinity purification mass spectrometry (Figure 1A, C). Dhx9 additionally co-localized with Nol12 in vivo under normal (Figure 2C) and DNA damage conditions (Figure 7A), while Nol12 also co-localized with TOPBP1 to sites of replication stress (Figure 7D) and sites of DNA damage with 53BP1 (Figure 7F). While the precise role of Nol12 at these sites of replication stress and/or DNA damage requires considerable further study, the paradigm emerging from multiple studies of multifunctional RBPs suggest that Nol12 may participate directly in the regulation of DNA repair and/or DDR signaling, either through regulation of DDR-regulatory proteins or metabolism of one or more DNA damage-related structure(s) at the site of repair (76–78) Interestingly, recent data showed that 53BP1 can form nuclear foci as a result of replication stress which are largely confined to G1 (79); furthermore, TOPBP1 was shown to function with 53BP1 in the G1 DNA damage checkpoint (80). The fact that Nol12 co-localized with 53BP1 upon etoposide treatment, and the robust G1/S arrest observed upon loss of Nol12, strongly supports a role for the protein in maintenance of genome integrity in G1 and or during replication.
Nol12 and ribosome biogenesis
Redundant ribosome maturation pathways are considered vital to sustain ribosome levels and, in higher eukaryotes, this link is emphasized by the fact that loss of most ribosome biogenesis factors leads to a cell cycle arrest in G1/S, which is always preceded by the appearance of rRNA processing defects. However, while the G1/S arrest is part of a nucleolar stress response, it rarely leads to apoptosis in a majority of cell types and is largely dependent on signaling via p53 (12,32). Our characterization of Nol12’s role in pre-rRNA processing revealed the protein to be required for efficient processing of site 2 as all precursors directly downstream of this cleavage—30S and 21S, but mostly 32.5S—are significantly reduced in the absence of Nol12, while those upstream of the cleavage—most notably 41S—accumulate. A study by Sloan and colleagues has previously suggested a role for Nol12 in the processing of ITS1 around both site 2 and E/2a together with Xrn2, studying processing events 60 h after siRNA transfection, at which point significant levels of apoptosis can be observed in Nol12-depleted cells (22). Our results, which were observed as early as 24 h after Nol12 knockdown, mostly overlap with those by Sloan et al., confirming a role of Nol12 as a ribosome biogenesis factor essential for cleavage events at site 2. However, in contrast to Sloan et al., we observed an increase of 18SE pre-rRNA in addition to 36S and 36S-C, all products site E/2a cleavage (Figure 4A), indicating that Nol12 is not essential for cleavage at site E/2a, but rather that in the absence of Nol12 and a block of site 2 processing (as also observed by (43)), cleavage at site E/2a is increased as the pathway is rerouted to ensure ribosome production, which is supported by the moderate 20–25% reduction of mature large subunit rRNAs (28S and 5.8S). The observed variance could be due to the advanced stage of apoptosis of cells in Sloan et al., which is indicated by the overall drop in mature rRNAs (43), or differences in cell lines. eCLIP experiments also showed binding of Nol12 to ITS1 regions with particular enrichment around site 2, but not E/2a (Figure 4F), and a role for Nol12 at site 2 is further supported by the Nol12 pre-ribosomal interactome, in particular, the significant enrichment of Utp10, Rrp1, Rrp5, Ftsj3, Gtpbp4, and Rsl1D1 (Supplementary Table S3). The association with a high number of 90S factors suggests an early recruitment of Nol12 to pre-rRNA as Utp10 is a t-UTP factor that was suggested to be one of the earliest proteins to associate with the 47S precursor (81). The RNA binding proteins Rrp1 and Rrp5 were recently shown to be important for both cleavage at site 2 and, in the case of Rrp5, processing from site 2 to E by Rrp6 (22,82), while loss of Ftsj3 delayed processing at site 2, shifting events to the minor pathway via site E (83). It is conceivable that these proteins either aid in the recruitment of Nol12 to pre-ribosomes, or facilitate Nol12’s function at site 2, possibly through direct interaction. Nothing is known about the roles of Rsl1D1 and Gtpbp4 in pre-rRNA processing, yet their depletion results in a similar pattern of precursors than those observed upon loss of Nol12, suggesting a function in or around the same processing events (32).
Previously, Nol12 was suggested to be a putative 5′-3′ exonuclease based on its homology to the yeast pre-rRNA processing protein Rrp17 (42). From their data, Sloan et al. inferred that as site 2 cleavage in human cells was significantly inhibited by depletion of Nol12 as well as Xrn2, Rrp5 and Bop1, it is analogous to site A3 cleavage in yeast pre-rRNA processing, which is the entry site for Rrp17 exonucleolytic activity (22). Recent work by Goldfarb and Cech also reported a catalytic role for RNAse MRP in processing at site 2 cleavage similar to yeast (84). As complementary and/or overlapping functions of nucleases as part of redundant ribosomal processing pathways have been identified in S. cerevisiae (42), a role for Nol12 as putative nuclease at or around site 2 together with RNAse MRP and Xrn2 is also conceivable in human cells. Interestingly, eCLIP also revealed Nol12 enrichment across ITS2, particularly around sites 4 and 3′ suggesting an potential role for the protein in the maturation of 28S rRNA (21). Rrp17 was shown to be involved in ITS2 processing (42) and Sloan et al. observed a reduction in 12S upon Nol12 knockdown in HeLa cells (43); as such, a role for Nol12 within ITS2 could also be imagined, which will be examined in the future.
Nol12, a RBP with diverse cellular roles
In recent years, numerous RBPs have been found to possess pleiotropic functions in non-RNA pathways, in particular the DDR; many of these proteins thereby act as regulatory hubs that link different cellular pathways. The abundant nucleolar proteins NPM1 and NCL independently coordinate the transcription and maturation of both rRNAs and mRNAs, as well as acting as histone chaperones in multiple DNA repair pathways and regulating the availability of other DNA repair enzymes (6). The multifunctional RNA exonucleases Xrn2 and hRrp6 were recently shown to aid in the resolution of RNA:DNA hybrids and repair of DNA double-strand breaks, respectively, expanding their functions beyond the RNA realm (4,5); this function of Xrn2 is also the most likely cause for the accumulation of H2A.XpS139 observed in its absence. Numerous proteins that reside in the paraspeckle have been found to directly participate in the DDR, and particularly in the repair of double-strand breaks, though in some cases the mechanism remains elusive (85–89). Significantly, several previously-identified DNA repair proteins have subsequently been described to be RBPs; for example, human AP endonuclease 1 (APE1), a key enzyme in the base excision repair pathway, is responsible for both decay of c-myc mRNA and repair of oxidative damage to nucleolar rRNA (90,91).
The co-isolation of components from diverse cellular RNA/DNA homeostasis niches as well as its localization to different subcellular compartments supports the idea of Nol12 as another multifunctional RBP. Nol12 has also been identified in several recent high-throughput screens for mRNA-binding proteins (92,93) [38, 39], and, here, we found the protein was localized to GW/P-bodies, suggesting a putative role in mRNA metabolism. Moreover, Nol12’s association and co-localization with paraspeckle components suggests a potential function within these bodies, either in the stress-dependent nuclear retention of the putative adenosine-to-inosine edited Alu-repeat containing human mRNAs, or as a partner of one or more DNA damage-regulatory paraspeckle proteins such as FUS/TLS, SFPQ, NONO, RBM14 and RBMX (85–90,94). Interestingly, recent work has demonstrated binding of Dhx9 to inverted-repeat Alu elements and an increase in circular Alu-containing RNAs upon loss of Dhx9 (95). Given Nol12’s association with Dhx9, a role for the protein in these functions could additionally be considered.
Being a broad-specificity RNA-binding protein, Nol12’s multifunctionality is likely to be conveyed through different cofactors and/or posttranslational modifications (PTMs)—as has been reported for several multifunctional RBPs mentioned above (96,97), and we observed Nol12 bands differing in migration in nucleoplasmic and cytoplasmic fractions (Supplementary Figure S2E). Several phosphorylation sites within Nol12 have been identified by multiple high-throughput phosphorylation screens, most notably within a casein kinase II consensus sequence in both Nol12 (S134) and its mouse homologue Nop25 (S135) (98). Significantly, both the CKII subunit CSNK2A1 and the phosphatase PP1A are part of the Nol12 proteome (Supplementary Table S3), and CKII has been reported to modulate the catalytic activity and/or subcellular localization of several multifunctional RBPs including NPM1, NCL and APE1 (99–101). Numerous helicases were also found significantly enriched in Nol12 affinity purifications, including ribosome biogenesis-related helicases such as Ddx27, Ddx55 and Dhx15 (102–104), Ddx21, which is involved in both mRNA and rRNA transcription/processing and R-loop clearance (72,105), the GW/P-body proteins Ddx5 and Ddx17 (106), and the multifunctional RNA/DNA helicase Dhx9, implicated in the resolution of numerous aberrant DNA and RNA structures including triple-helices, G-quadruplexes, D-loops, most significantly, RNA:DNA hybrids (R-loops) (107). Dhx9 has also been shown to co-localize with H2A.XpS139 and DNA-PK in RNA-containing granules upon exposure of cells to low-dose actinomycin D and to protect Eμ-myc/Bcl-2 lymphoma cells against an ATR-dependent apoptotic response (108), suggesting it may be required for the resolution of stalled RNA polymerases and/or the resulting RNA:DNA hybrids. Indeed, the abundance of known and putative DNA repair/DDR-related DEAD/H-box helicases within the Nol12 interactome, and the wealth of evidence supporting the presence and significance of Nol12 at sites of replication stress and/or DNA repair, a direct and/or regulatory role for Nol12 in the resolution of unusual DNA structures or RNA:DNA hybrids in concert with Dhx9, or one or more other helicases, would provide an attractive and plausible explanation for the activation of an ATR-dependent apoptotic response following Nol12 depletion. The recent identification of RNA processing proteins in the DNA damage response by a number of screens, and the wealth of RBPs recently found to contribute to DDR signaling and DNA repair, speak to the extensive functional and regulatory connections between RNA metabolism and DNA repair pathways, and the significance of these connections to the pathogenesis of human disease (76,78,109). Overall, the putative multifunctionality of Nol12 defines it as a novel member of a family of proteins at the intersection of RNA metabolism and DNA maintenance and repair, and one that warrants further examination to define the mechanisms of its different functions in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Drs J. Archambault, N. Francis, E. Lecuyer, F. Robert, N. Watkins, B. Coulomb and F. Charron for gifts of reagents used in this study, Dr C. Denicourt for cell lines, D. Faubert of the IRCM Proteomics platform, and members of the Oeffinger and Zenklusen labs for critical reading.
FUNDING
Canadian Institutes of Health Research [MOP 106628-MO] to M.O.; Natural Sciences and Engineering Research Council of Canada [RGPIN 386315-MO] to M.O.; Fonds de recherche du Québec – Santé (PR2I) to M.O.; Canadian Foundation for Innovation to M.O. and D.Z.; Canadian Institute for Health Research [366682] to D.Z.; Fond de recherche du Quebec (Chercheur-boursier Junior 2) to D.Z.; NIH [HG007005] to G.W.Y.. Funding for open access charge: Canadian Institutes of Health Research [MOP 106628-MO].
Conflict of interest statement. None declared.
Present address: Karen E. Wei. Human Oncology and Pathogenesis Program, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
REFERENCES
Author notes
These authors contributed equally to this work as first authors.




Comments