-
PDF
- Split View
-
Views
-
Cite
Cite
Martina Beck, Ines Wyrsch, James Strutt, Rinukshi Wimalasekera, Alex Webb, Thomas Boller, Silke Robatzek, Expression patterns of FLAGELLIN SENSING 2 map to bacterial entry sites in plant shoots and roots, Journal of Experimental Botany, Volume 65, Issue 22, December 2014, Pages 6487–6498, https://doi.org/10.1093/jxb/eru366
Close - Share Icon Share
Abstract
Pathogens can colonize all plant organs and tissues. To prevent this, each cell must be capable of autonomously triggering defence. Therefore, it is generally assumed that primary sensors of the immune system are constitutively present. One major primary sensor against bacterial infection is the FLAGELLIN SENSING 2 (FLS2) pattern recognition receptor (PRR). To gain insights into its expression pattern, the FLS2 promoter activity in β-glucuronidase (GUS) reporter lines was monitored. The data show that pFLS2::GUS activity is highest in cells and tissues vulnerable to bacterial entry and colonization, such as stomata, hydathodes, and lateral roots. GUS activity is also high in the vasculature and, by monitoring Ca2+ responses in the vasculature, it was found that this tissue contributes to flg22-induced Ca2+ burst. The FLS2 promoter is also regulated in a tissue- and cell type-specific manner and is responsive to hormones, damage, and biotic stresses. This results in stimulus-dependent expansion of the FLS2 expression domain. In summary, a tissue- and cell type-specific map of FLS2 expression has been created correlating with prominent entry sites and target tissues of plant bacterial pathogens.
Introduction
Plant pathogens use a variety of different strategies to invade their hosts, which are tightly associated with the lifestyle of the pathogen as well as with plant development (Faulkner and Robatzek, 2012). The general aim of a pathogen is to invade and access plant tissues where it can find nutrients for its own development. Bacterial phytopathogens typically try to reach the apoplastic space between cells where they can multiply and reprogramme host metabolism by the injection of bacterial effectors into the extra- and intracellular space. During a susceptible interaction, as observed between Arabidopsis thaliana and Pseudomonas syringae, the Gram-negative bacterium enters the host tissue (typically leaves) via natural openings (stomata) or wound sites, from where it propagates in the apoplastic spaces, causing water-soaked, chlorotic (and later also necrotic) lesions (Preston, 2000).
Lacking a circulatory system and specialized immune cells, plants depend upon the ability of every cell to recognize potentially pathogenic microbes and initiate immunity. For this, plants exploit cell surface-localized pattern recognition receptors (PRRs), which allow the detection of conserved microbial molecules, so-called pathogen-associated molecular patterns (PAMPs) (Boller and Felix, 2009). In the case of immunity against bacterial pathogens, a major PRR is the receptor kinase FLAGELLIN SENSING 2 (FLS2) which recognizes bacterial flagellin through its conserved elicitor-active epitope flg22 (Gomez-Gomez et al., 1999). Studies show that flg22 triggers defence responses in whole seedlings, leaves, and roots (Zipfel et al., 2004; Millet et al., 2010; Jacobs et al., 2011). This suggests that the receptor is expressed in these tissues, which is consistent with findings of mRNA expression studies and FLS2–green fluorescent protein (GFP) imaging (Gomez-Gomez and Boller, 2000; Robatzek et al., 2006). These observations generally imply that defence components such as FLS2 might be constitutively expressed, but this might lead to an unwanted activation of defence responses which can negatively impact plant processes such as growth. A typical response, which can be observed for plants that are exposed long term to flagellin, is the reduction in plant growth, due to a defined trade-off between immune and hormonal signalling (Gomez-Gomez et al., 1999; Navarro et al., 2006, 2008; Lozano-Duran et al., 2013).
Publicly available gene expression data (Arabidopsis eFP browser; Faulkner and Robatzek, 2012) revealed that FLS2 is not expressed at similar levels throughout the plant. For example, FLS2 does not have measurable expression in root cells, despite flg22 triggering some defence responses in this organ (Millet et al., 2010; Jacobs et al., 2011). In leaves, FLS2 exhibits a more specific cellular function since flg22 perception seems to play a predominant role in stomatal immunity (Zipfel et al., 2004; Zeng and He, 2010). Recent studies showed that FLS2 transcriptional activation depends on ethylene signalling involving binding of the transcription factors ETHYLENE-INSENSITIVE 3 (EIN3) and ETHYLENE-INSENSITIVE3-LIKE 1 EIL1 (Boutrot et al., 2010; Mersmann et al., 2010), and is positively regulated by its own ligand and other PAMPs (Zipfel et al., 2004, 2006). These observations indicate that FLS2 expression is under spatio-temporal control, but the extent to which the transcription of FLS2 is regulated remains unknown.
Here, it is demonstrated that the FLS2 promoter is active in a cell type- and tissue-specific manner and is up-regulated in response to hormones and stress. Using transgenic Arabidopsis plants producing β-glucuronidase (GUS) under the control of the FLS2 promoter, pFLS2::GUS activity was detected in all organs, with the highest levels found in hydathodes, stomata, and the vasculature, representing prominent entry sites and target tissues of bacteria in plants. Tissue-specific Ca2+ measurement shows that the vasculature is responsive to flg22. Detailed imaging revealed, furthermore, that FLS2 is present in roots but restricted to outgrowing lateral roots (LRs) and the inner central cylinder, suggesting a specific role for FLS2 in these tissues. Hormones, wounding, and abiotic and biotic stress can differentially activate pFLS2::GUS in specific tissue layers. Altogether, this study provides a detailed expression map of a major plant immune receptor and reveals spatio-temporal control of the PRR promoter activity for optimal plant defences under pathogen attack.
Materials and methods
Plant materials and growth conditions
The A. thaliana transgenic plants used in this study (accession Columbia-0, if not otherwise indicated) were fls2 (Zipfel et al., 2004) and DR5::GFP (Benkova et al., 2003); courtesy of J. Friml. For microscopy, stress treatments, and developmental studies, seedlings were grown for 6–8 d on sterile 1× Murashige and Skoog (MS) plates supplemented with 1% sucrose and 0.8% phytoagar (w/v) under 16h light at 22 °C. For Ca2+ measurement, Col-0 35S:AEQ and the GAL4-mediated vascular enhancer trap line KC274 were used (Marti et al., 2013). Seeds were surfaced sterilized and sown on 0.5× MS medium with 0.8% agar (w/v). Seedlings were grown in long days at 19 °C at light intensity 50 μmol m–2 s–1 (Sanyo MLR30 growth cabinet) for 12 d. For non-sterile conditions used in developmental studies, plants were grown for 2–8 weeks on soil under controlled environments (12h light, 22 °C, and 60% humidity).
Gene constructs and plant transformation
The promoter of FLS2 (988bp) was used from pFLS2::FLS2–GFP (Robatzek et al., 2006) and fused to the GUS gene, which was isolated from pGUS Topo via BamHI and HindIII restriction sites and inserted into pFLS2::pCAMBIA2300, resulting in pFLS2::GUS-pCAMBIA2300. Col-0 plants were transformed via Agrobacterium-mediated transformation with the floral dipping method (Clough and Bent, 1998). Transformants were selected for kanamycin resistance. The experiments were repeated in two independent transgenic lines of the T3 generation.
GUS staining
All samples were processed according to the method described by De Block and Debrouwer (1992), with 1mM 5-bromo-4-chloro-3- indolyl-d-glucuronide (X-Gluc) in staining buffer [0.1M NaH2PO4, 0.1M Na2HPO4, 10mM EDTA, 2mM FeK3(CN)6, 2mM FeK4(CN)6·3 H2O, pH 7.0, 0.1% (v/v) Triton X-100] at room temperature for 2–18h. Samples were fixed and destained with ethanol/acetic acid (50% v/v). Specimens were examined and documented using a Leica M165 FC stereomicroscope.
Embedding and sectioning
Tissue was fixed in 2.5% glutaraldehyde or 4% paraformaldehyde, followed by a ethanol series of 30, 50, 70, 90, and 100%, for 30min each. Pre-infiltration of the tissue was done for 2h with 50:50 (v/v) ethanol:Technovit®7100 (Heraeus-Kulzer, Germany) base liquid. The preparation solution (Technovit®7100; see the supplier’s embedding protocol) was infiltrated and tissue samples were left for polymerization. Samples were sectioned to 10 μm thickness by using an Ultracut E ultramicrotome (Reichert-Jung, Germany).
Microscopy
Standard confocal laser microscopy was performed using a Leica SP5laser point scanning microscope. GFP/propidium iodide was excited using the 488nm argon laser, and fluorescence emissions were captured between 500nm and 550nm for GFP and between 580nm and 640nm for propidium iodide. Seedlings were incubated for 20min in 10 μg ml–1 propidium iodide solution.
Stress treatments
The chemicals were diluted in half-strength MS medium to their respective working solutions: 10 μM flg22 (10mM in dH2O), 50 μM salicylic acid [SA; 100mM in dimethylsulphoxide (DMSO)], 1mM H2O2 (1.5M), 10 μM 1-aminocyclopropane-1-carboxylic acid (ACC; 10mM in dH2O), and 10 μM indole acetic acid (IAA; 100mM in dH2O). Half-strength MS medium was used as mock treatment. For each treatment, seedlings (8–10 d after germination) were transferred from agar plates and incubated in the respective solutions for 48h under 16h light at 22 °C, followed by GUS staining. For bacterial stress and wound treatments, detached leaves of 3-to 4-week-old soil-grown plants were used. Detached leaves were submerged in 10mM MgCl2 (mock) or with Pseudomonas syringae pv. tomato DC3000 (Pto DC3000; OD 0.1) in 10mM MgCl2 solution, with slight shaking for 24h at room temperature. Wound stress was inflicted by a sharp needle on 10 detached leaves mounted on half-strength MS agar and left on plates for 4–6h at room temperature before staining. All stress treatments were performed on at least 10 seedlings or 10 leaves of the two independent T3 transgenic lines at the same developmental stage. Images show representative results of three biological repetitions.
Ca2+ measurements
Seedlings grown for 12 d were supplied with half-strength MS liquid medium supplemented with 20 μM coelenterazine (Nanolight) and incubated overnight in the dark at room temperature. Luminescence measurements were performed using a FLUOstar OPTIMA plate reader (BMG LABTECH). Luminescence from single wells was measured over 35 s, and flg22 (EZBiolab) dissolved in half-strength MS was injected to a final concentration of 100nM and measured at 15 s intervals for 1200 s. Mock treatment (water, 35 s) was performed under the same conditions. At the end of the experiment, the remaining aequorin (AEQ) pool was discharged by treatment with a final concentration of 1M CaCl2 in 10% (v/v) ethanol. Luminescence values were converted to estimates of intracellular Ca2+ ([Ca2+]i) according to Fricker et al. (1999).
LR growth analysis
Col-0 and fls2 were germinated on 1× MSN plates and transferred 3 d after germination in liquid 1× MS medium with or without 1 μM flg22. After 6 d, the root length and number of LRs were determined.
Immunoblot and ConA precipitation
A 100mg aliquot of root tissue of seedlings (Col-0) grown for 2 weeks vertically on 1× MS plates was homogenized in 0.2ml of cold IP buffer [50mM TRIS-HCl pH 8, 150mM NaCl, 1% (v/v) Nonidet P40, and protease inhibitor cocktail] and incubated for 1h at 4 °C followed by a centrifugation step (10 000 g for 10min, three times). The supernatant was incubated for 1h at 4 °C with concanavalin A (ConA)–Sepharose beads (Amersham Biosciences) to enrich samples for glycosylated proteins. This was used because FLS2 is highly glycosylated (Häweker et al., 2010) and weakly detectable in root total extracts. The beads were collected and washed three times with ice-cold IP buffer. After denaturation in SDS–PAGE sample loading [0.35M TRIS-HCl pH 6.8, 30% (v/v) glycerol, 10% (v/v) SDS, 0.6M dithiothreitol, and 0.012% (w/v) bromophenol blue], proteins retained on the beads were eluted by SDS–PAGE sample loading buffer and separated by 7% SDS–PAGE. FLS2 was detected by immunoblot analyses with anti-FLS2 antibodies (Chinchilla et al., 2006).
MAPK activation in roots
Isolated roots of 2-week-old plants (n=12) were placed in dH2O for 16h. Flg22 at 1 μM was added for 10min and tissue (50mg per sample) was shock frozen. To the ground material, 50 μl of SDS–PAGE sample loading [0.35M TRIS-HCl pH 6.8, 30% (v/v) glycerol, 10% (v/v) SDS, 0.6M dithiothreitol, and 0.012% (w/v) bromphenol blue] was added. Total proteins were separated by electrophoresis in a 12% SDS–polyacrylamide gel and electrophoretically transferred to a polyvinylidene fluoride membrane according to the manufacturer’s instructions (Bio-Rad). Transferred proteins were detected with Ponceau-S. Polyclonal primary antibodies against phospho-p44/42 mitogen-activated protein kinase (MAPK; Cell Signaling Technologies) were used, with alkaline phosphatase-conjugated anti-rabbit as secondary antibodies. Signal detection was performed using CDPstar (Roche).
Microarray
Landsberg erecta (ecotype Ler) seedlings and fls2-17 (Zipfel et al., 2004) were grown in liquid culture under constant shaking in 1× MS medium for 21 d. Plants were mock or flg22 (10 μM, 30min) treated, and roots were harvested and stored at –80 ºC for sample preparation. Experimental conditions for RNA extraction, microarray hybridizations, and statistical analyses were performed as in Zipfel et al. (2004).
Results
FLS2 is highly expressed in stomata, hydathodes, and wound sites in leaves
To investigate the promoter activity of FLS2 at the tissue level, transgenic A. thaliana lines containing the putative promoter sequences of the FLS2 gene fused to GUS were generated. An ~900bp genomic sequence upstream of the start codon of FLS2 was used (Supplementary Fig. S1 available at JXB online), which was sufficient to complement fully an fls2 mutant expressing the FLS2–GFP fusion protein (Zipfel et al., 2004). In silico motif analysis of the promoter sequence 900bp upstream of At5g46330 revealed the presence of a TATA box motif and several cis-elements such as W-boxes, known binding sites of WRKY transcription factors (Supplementary Fig. S1). Two binding sites in the region were previously shown to be occupied by EIN3 and EIL1, transcription factors of the ethylene pathway mediating FLS2 expression (Boutrot et al., 2010).
By monitoring GUS accumulation in the pFLS2::GUS lines during plant development, it could be confirmed that the FLS2 promoter exhibited expression in all organs examined (Supplementary Fig. S2 at JXB online). In 2-day-old seedlings, a clear blue staining could be detected in the developing cotyledons and root. In older seedlings, a prominent staining occurred additionally in the vascular tissue of cotyledons and the hypocotyl (Supplementary Fig. S2). At later stages of plant development, stipules, small leaf-like appendage at the bases of leaves, as well as floral and reproductive organs including petals, stamen, and the dehiscence zone in mature, conferred a clearly visible pFLS2::GUS expression (Supplementary Fig. S2).
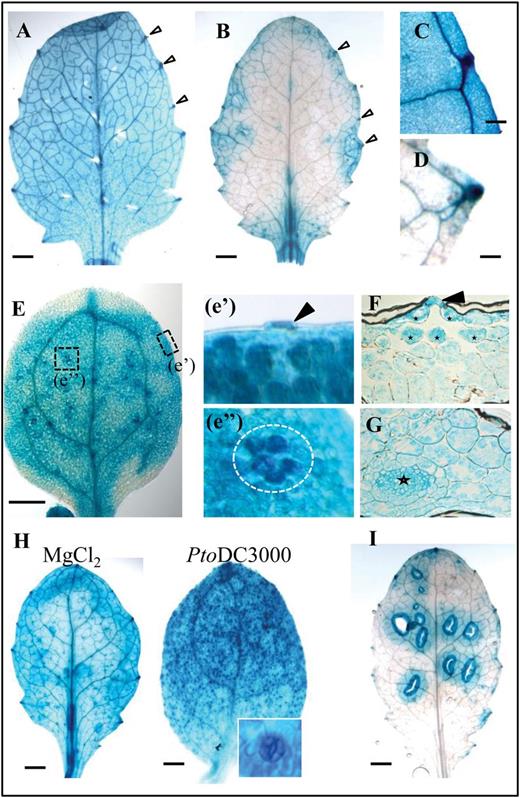
As FLS2-mediated immunity is predominantly studied in the Arabidopsis interaction with the leaf-infecting pathogen Pto DC3000, the basal pFLS2::GUS expression in different leaf developmental stages was studied (Supplementary Fig. S3A at JXB online). In cotyledons and the first pair of true leaves, the promoter expression showed a homogenous pattern throughout the leaf tissue, with higher expression levels in the vascular tissue and hydathodes (Fig. 1A, C, E). In younger leaves, GUS staining exhibited a more patchy distribution throughout the leaves (Fig. 1B; Supplementay Fig. S3B), but continuously showed a strong staining in hydathodes (Fig. 1D). At the cellular level, pFLS2::GUS expression was significantly visible in the mesophyll and phloem, as well as in epidermal cells, such as in the guard cells of the stomata (Fig. 1E, G). Notably, the mesophyll cells underneath the stomatal openings, forming the substomatal cavity, had clear promoter activity as revealed by cross-sectioning of leaf tissues (Fig. 1F).
FLS2 is differentially activated in leaves. Representative images of pFLS2::GUS expression. (A) First pair of true leaves. (B) Second pair of true leaves. Arrows show strong expression in hydathodes from (C) cotyledons and (D) the second pair of true leaves. (E) Promoter activity in cotyledons; dashed boxes show expression (e’) in stomata (arrow) and (e’’) a group of mesophyll cells (circle). (F) Cross-section of cotyledons shows guard cell expression (arrow) and high GUS staining in mesophyll cells surrounding the stomatal cavity (asterisks); (G) shows high expression in leaf veins (asterisk) and mesophyll. (H) Pto DC3000 increases promoter activity in stomata from the first pair of true leaves compared with mock (MgCl2) treatment. The inset shows an enlarged stoma. (I) Wound-induced GUS staining in the second pair of true leaves. (A, B, E, H, I) bar=1mm, (C, D) bar=0.1mm.
The substomatal expression pattern is correlated to cells exposed to early invasion of bacteria, which enter the apoplastic space underneath stomata. To visualize the entry of bacteria in Arabidopsis leaves, Col-0 plants were incubated with a GFP-transformed Pto DC3000 strain (Supplementary Fig. S4A at JXB online). The GFP-labelled bacteria were clearly visible in epidermal cells and within the openings of stomata (Supplementary Fig. S4A). Bacterial accumulation was often detectable in the intercellular space of mesophyll cells directly underneath stomata (Supplementary Fig. S4A). Next it was tested whether the presence of bacteria on the leaf surface would have an influence on the FLS2 promoter activity. Overnight incubation of 14- to 18-day-old plants with Pto DC3000 led to a strong visible GUS staining in stomatal guard cells in leaves and the hypocotyl (Fig. 1H; Supplementary Fig. S3D).
Bacteria also take advantage of wound sites and cracks in the epidermis to enter plant tissues, and therefore the influence of wounding on the FLS2 promoter activity was investigated. In general, in young leaves the pFLS2::GUS activity was very low in the absence stimuli (Fig. 1B). By contrast, wounding of leaves led to up-regulation of the promoter around the wound sites (Fig. 1I; Supplementary Fig. S3C), which was not obvious in cotyledons and first true leaves (Supplementary Fig. S3C). All these findings reveal that high levels of FLS2 expression in leaves occur in cells and tissues that represent natural entry sites of bacteria, or can become entry sites due to wounding.
FLS2 shows specific expression patterns and flg22 responses in roots
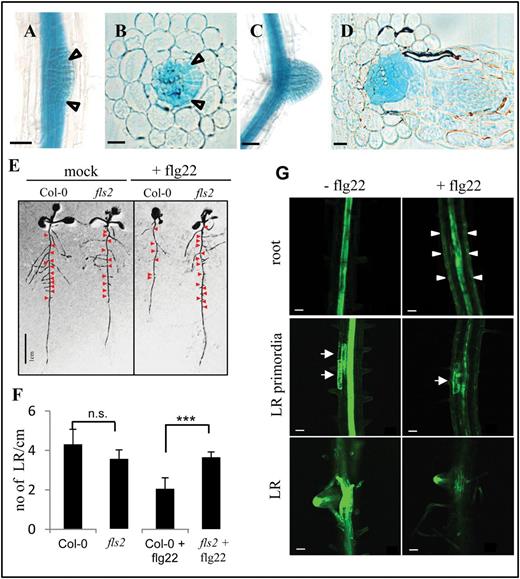
In roots, the pFLS2::GUS lines showed a basal expression in the root vascular cylinder starting at the root differentiation zone; no GUS expression could be observed in the root meristematic zone (Fig. 2A, B). Under sterile conditions, the highest expression was restricted to the inner cellular layers of the root, the vascular cylinder (Fig. 2B). In root cross-sections, a pronounced accumulation of GUS precipitate was observed in cells inside the endodermis (Fig. 2C) and this expression maximum correlated with a high accumulation of the pFLS2::FLS2–GFP fusion protein in the stele as revealed by co-staining the roots with the apoplastic tracer propidium iodide, uptake of which is blocked at the endodermis (Alassimone et al., 2010) (Fig. 2D). These observations are consistent with the accumulation of the native FLS2 protein in roots as revealed by immunoblot analysis (Fig. 2G). This basal expression pattern of FLS2 in roots may protect the plant from bacterial infections of the vasculature and ultimately colonization throughout all tissues.
Roots exhibit specific FLS2 expression patterns and tissue-specific responsiveness to flg22. In sterile-grown roots (8 d after germination) of pFLS2::GUS, the promoter activity is not present in root tips (A), but shows a high expression in the root stele (B) as revealed by root cross-section (C); bar=10 μm. (D) Confocal micrographs of pFLS2::FLS2–GFP show accumulation of GFP signal in the inner part of the stele (arrowheads point to inhibited uptake of propidium iodide at the endodermis; bar=10 μm. (E) Digital cross-section with plasma membrane localization of FLS2–GFP at cortical cells (arrowheads) and in the root cylinder (arrow). Autofluorescence of xylem is marked with asterisks. (F) Changes in [Ca2+]i values in mock-treated control (water, 35 s) or in response to flg22 (100nm, 35 s) in 35S::AEQ seedlings and the vasculature enhancer trap line KC274. Luminescence was measured over 1200 s. Data are presented as means ±SD, n=4 (mock), n=6 (flg22). (G) Immunoblot of detected FLS2 protein in roots and shoots. Samples were enriched for glycosylated proteins using ConA. (H) Immunoblot detection of phosphorylated MAPK present in Col-0 after 1 μM flg22 (10min) treatment but not in fls2. (I) Gene ontology of enriched genes specifically up-regulated in Ler roots after flg22 treatment (10 μM, 30min).
One of the earliest responses to PAMPs is a transient and rapid (within seconds) increase of free [Ca2+]i, which subsequently (within minutes) declines to steady-state [Ca2+]i levels (Blume et al., 2000; Ranf et al., 2008). This [Ca2+]i increase was shown to be crucial for many downstream responses. To test whether the vasculature tissue is sensitive to flg22 stimulation, the GAL4-mediated vascular enhancer trap line KC274 expressing AEQ specifically in the vasculature (Marti et al., 2013) was exploited. Treatment with flg22 induced a rapid increase in [Ca2+]i in both the vasculature-specific KC274;UAS AEQ line and in the line in which aequorin was expressed constitutively under the control of the Cauliflower mosaic virus (CaMV) 35S promoter (Fig. 2F). The magnitude of the reported flg22-induced increase in [Ca2+]i was greater when AEQ was targeted specifically to the vasculature tissue in KC274 (Fig. 2F), suggesting that FLS2 in the vasculature mediates a typical early flg22 response and indicates that this tissue contributes to the source of the PAMP-induced [Ca2+]i burst in plants.
To gain further insights into the functional relevance of the presence of FLS2 in roots, the phosphorylation of MAPKs upon flg22 elicitation was studied. Immunoblot analysis revealed a specific flg22-induced activation of MAPK in root tissue of Col-0 but not fls2, demonstrating that FLS2 in roots activates similar signalling responses to those shown for leaf tissues (Fig. 2H).
The root’s response to flg22 elicitation was further explored on a more global scale, and whole-transcriptome expression analysis was performed. Sterile grown seedlings (Ler) were mock and flg22 treated, and roots were harvested after 30min. ATH1 microarray expression analysis revealed flg22-regulated genes overlapping with those identified from whole-seedling expression analysis (Zipfel et al., 2004), but also identified ~75 genes specifically up-regulated in roots (Fig. 2I). Fifty-three of these genes showed a >2.5-fold induction after flg22 treatment (Supplementary Table S1 at JXB online). Sixty-five of these genes have their highest expression values during root development (eFP Browser http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), which confirmed the enrichment for root-specific processes (Supplementary Table S1). After flg22 elicitation, in the roots transcriptional changes were seen of genes with roles in hormone and stress signalling, such as auxin- and ethylene-mediated pathways (AT1G59500, AT5G65600, AT1G72360, and AT5G46080), root and LR development (AT4G31500 and AT5G13080), or signalling and defence pathways (AT2G17060 and AT3G21650) (Table 1). Taken together, the data show that not only FLS2 promoter activity, but also the functional protein, is present in roots. Root-specific activation of FLS2 reveals a subset of genes which are specifically enriched after flg22 treatment, indicating additional functions of this receptor in roots.
flg22-induced genes in roots: candidates with maximum expression in roots
| . | Gene . | flg22 fold induction . | Maximum expression level . | Annotation . | Biological process . |
|---|---|---|---|---|---|
| Hormone and stress signalling | AT1G59500 | 6.67 | 3915.41a | GH3.4; indole-3-acetic acid amido synthetase | Auxin homeostasis, response to auxin stimulus |
| AT5G65600 | 5.3 | 1132.13b | Legume lectin family protein/ protein kinase family protein | Protein phosphorylation, response to ethylene stimulus | |
| AT1G08050 | 4.46 | 2268.52b | Zinc finger (C3HC4-type RING finger) family protein | MAPK cascade, abscisic acid-mediated signalling pathway, cell communication | |
| AT5G11920 | 4.2 | 8344.6b | AtcwINV6 (6-&1-fructan exohydrolase) | Carbohydrate metabolic process, regulation of hydrogen peroxide metabolic process | |
| AT1G15670 | 4.05 | 14417.2b | Kelch repeat-containing F-box family protein | Negative regulation of cytokinin-mediated signalling pathway | |
| AT5G67340 | 3.94 | 3905.65b | Armadillo/beta-catenin repeat family protein | Endoplasmic reticulum–nucleus signalling pathway, MAPK cascade, negative regulation of defence response | |
| AT1G72360 | 3.76 | 11466.3b | Ethylene-responsive element- binding protein | Cellular response to ethylene stimulus, regulation of transcription | |
| AT3G28580 | 3.76 | 8339.31b | AAA-type ATPase family protein | Response to abscisic acid stimulus, response to ethylene stimulus | |
| AT5G46080 | 3.62 | 890.14b | Protein kinase family protein | Ethylene biosynthetic process, protein phosphorylation | |
| AT5G01550 | 3.08 | 1208.89b | LECRKA4.2 (LECTIN RECEPTOR KINASE A4.1) | Abscisic acid-mediated signalling pathway, protein phosphorylation, response to chitin | |
| AT3G13100 | 2.66 | 2294.74b | ATMRP7; ATPase | Response to other organisms, salicylic acid biosynthetic process | |
| Root development | |||||
| AT4G31500 | 3.44 | 17621.3b | CYP83B1 (CYTOCHROME P450 MONOOXYGENASE) | Adventitious root development, callose deposition in cell wall during defence response | |
| AT1G67980 | 3.42 | 1164.11b | CCoAMT; caffeoyl-CoA O-methyltransferase | Lignin biosynthetic process | |
| AT3G45960 | 2.83 | 1535.55b | ATEXLA3 (Arabidopsis thaliana expansin-like a3) | Plant-type cell wall loosening, plant-type cell wall organization | |
| AT5G13080 | 2.58 | 3789.3b | WRKY75; transcription factor | Cellular response to phosphate starvation, lateral root development, response to ethylene stimulus | |
| Signalling/defence | |||||
| AT2G17060 | 3.79 | 561.85b | Disease resistance protein (TIR- NBS-LRR class) | Defence response, signal transduction | |
| AT4G28350 | 3.66 | 1223.16b | Lectin protein kinase family protein | Defence response to fungus, protein phosphorylation, response to chitin | |
| AT1G64400 | 3.09 | 2202.11b | Long-chain-fatty-acid–CoA ligase | Defence response to insect, fatty acid biosynthetic process | |
| AT3G21650 | 2.74 | 900.6b | Serine/threonine protein phosphatase 2A (PP2A) | Signal transduction | |
| . | Gene . | flg22 fold induction . | Maximum expression level . | Annotation . | Biological process . |
|---|---|---|---|---|---|
| Hormone and stress signalling | AT1G59500 | 6.67 | 3915.41a | GH3.4; indole-3-acetic acid amido synthetase | Auxin homeostasis, response to auxin stimulus |
| AT5G65600 | 5.3 | 1132.13b | Legume lectin family protein/ protein kinase family protein | Protein phosphorylation, response to ethylene stimulus | |
| AT1G08050 | 4.46 | 2268.52b | Zinc finger (C3HC4-type RING finger) family protein | MAPK cascade, abscisic acid-mediated signalling pathway, cell communication | |
| AT5G11920 | 4.2 | 8344.6b | AtcwINV6 (6-&1-fructan exohydrolase) | Carbohydrate metabolic process, regulation of hydrogen peroxide metabolic process | |
| AT1G15670 | 4.05 | 14417.2b | Kelch repeat-containing F-box family protein | Negative regulation of cytokinin-mediated signalling pathway | |
| AT5G67340 | 3.94 | 3905.65b | Armadillo/beta-catenin repeat family protein | Endoplasmic reticulum–nucleus signalling pathway, MAPK cascade, negative regulation of defence response | |
| AT1G72360 | 3.76 | 11466.3b | Ethylene-responsive element- binding protein | Cellular response to ethylene stimulus, regulation of transcription | |
| AT3G28580 | 3.76 | 8339.31b | AAA-type ATPase family protein | Response to abscisic acid stimulus, response to ethylene stimulus | |
| AT5G46080 | 3.62 | 890.14b | Protein kinase family protein | Ethylene biosynthetic process, protein phosphorylation | |
| AT5G01550 | 3.08 | 1208.89b | LECRKA4.2 (LECTIN RECEPTOR KINASE A4.1) | Abscisic acid-mediated signalling pathway, protein phosphorylation, response to chitin | |
| AT3G13100 | 2.66 | 2294.74b | ATMRP7; ATPase | Response to other organisms, salicylic acid biosynthetic process | |
| Root development | |||||
| AT4G31500 | 3.44 | 17621.3b | CYP83B1 (CYTOCHROME P450 MONOOXYGENASE) | Adventitious root development, callose deposition in cell wall during defence response | |
| AT1G67980 | 3.42 | 1164.11b | CCoAMT; caffeoyl-CoA O-methyltransferase | Lignin biosynthetic process | |
| AT3G45960 | 2.83 | 1535.55b | ATEXLA3 (Arabidopsis thaliana expansin-like a3) | Plant-type cell wall loosening, plant-type cell wall organization | |
| AT5G13080 | 2.58 | 3789.3b | WRKY75; transcription factor | Cellular response to phosphate starvation, lateral root development, response to ethylene stimulus | |
| Signalling/defence | |||||
| AT2G17060 | 3.79 | 561.85b | Disease resistance protein (TIR- NBS-LRR class) | Defence response, signal transduction | |
| AT4G28350 | 3.66 | 1223.16b | Lectin protein kinase family protein | Defence response to fungus, protein phosphorylation, response to chitin | |
| AT1G64400 | 3.09 | 2202.11b | Long-chain-fatty-acid–CoA ligase | Defence response to insect, fatty acid biosynthetic process | |
| AT3G21650 | 2.74 | 900.6b | Serine/threonine protein phosphatase 2A (PP2A) | Signal transduction | |
a Lateral root.
b Root.
flg22-induced genes in roots: candidates with maximum expression in roots
| . | Gene . | flg22 fold induction . | Maximum expression level . | Annotation . | Biological process . |
|---|---|---|---|---|---|
| Hormone and stress signalling | AT1G59500 | 6.67 | 3915.41a | GH3.4; indole-3-acetic acid amido synthetase | Auxin homeostasis, response to auxin stimulus |
| AT5G65600 | 5.3 | 1132.13b | Legume lectin family protein/ protein kinase family protein | Protein phosphorylation, response to ethylene stimulus | |
| AT1G08050 | 4.46 | 2268.52b | Zinc finger (C3HC4-type RING finger) family protein | MAPK cascade, abscisic acid-mediated signalling pathway, cell communication | |
| AT5G11920 | 4.2 | 8344.6b | AtcwINV6 (6-&1-fructan exohydrolase) | Carbohydrate metabolic process, regulation of hydrogen peroxide metabolic process | |
| AT1G15670 | 4.05 | 14417.2b | Kelch repeat-containing F-box family protein | Negative regulation of cytokinin-mediated signalling pathway | |
| AT5G67340 | 3.94 | 3905.65b | Armadillo/beta-catenin repeat family protein | Endoplasmic reticulum–nucleus signalling pathway, MAPK cascade, negative regulation of defence response | |
| AT1G72360 | 3.76 | 11466.3b | Ethylene-responsive element- binding protein | Cellular response to ethylene stimulus, regulation of transcription | |
| AT3G28580 | 3.76 | 8339.31b | AAA-type ATPase family protein | Response to abscisic acid stimulus, response to ethylene stimulus | |
| AT5G46080 | 3.62 | 890.14b | Protein kinase family protein | Ethylene biosynthetic process, protein phosphorylation | |
| AT5G01550 | 3.08 | 1208.89b | LECRKA4.2 (LECTIN RECEPTOR KINASE A4.1) | Abscisic acid-mediated signalling pathway, protein phosphorylation, response to chitin | |
| AT3G13100 | 2.66 | 2294.74b | ATMRP7; ATPase | Response to other organisms, salicylic acid biosynthetic process | |
| Root development | |||||
| AT4G31500 | 3.44 | 17621.3b | CYP83B1 (CYTOCHROME P450 MONOOXYGENASE) | Adventitious root development, callose deposition in cell wall during defence response | |
| AT1G67980 | 3.42 | 1164.11b | CCoAMT; caffeoyl-CoA O-methyltransferase | Lignin biosynthetic process | |
| AT3G45960 | 2.83 | 1535.55b | ATEXLA3 (Arabidopsis thaliana expansin-like a3) | Plant-type cell wall loosening, plant-type cell wall organization | |
| AT5G13080 | 2.58 | 3789.3b | WRKY75; transcription factor | Cellular response to phosphate starvation, lateral root development, response to ethylene stimulus | |
| Signalling/defence | |||||
| AT2G17060 | 3.79 | 561.85b | Disease resistance protein (TIR- NBS-LRR class) | Defence response, signal transduction | |
| AT4G28350 | 3.66 | 1223.16b | Lectin protein kinase family protein | Defence response to fungus, protein phosphorylation, response to chitin | |
| AT1G64400 | 3.09 | 2202.11b | Long-chain-fatty-acid–CoA ligase | Defence response to insect, fatty acid biosynthetic process | |
| AT3G21650 | 2.74 | 900.6b | Serine/threonine protein phosphatase 2A (PP2A) | Signal transduction | |
| . | Gene . | flg22 fold induction . | Maximum expression level . | Annotation . | Biological process . |
|---|---|---|---|---|---|
| Hormone and stress signalling | AT1G59500 | 6.67 | 3915.41a | GH3.4; indole-3-acetic acid amido synthetase | Auxin homeostasis, response to auxin stimulus |
| AT5G65600 | 5.3 | 1132.13b | Legume lectin family protein/ protein kinase family protein | Protein phosphorylation, response to ethylene stimulus | |
| AT1G08050 | 4.46 | 2268.52b | Zinc finger (C3HC4-type RING finger) family protein | MAPK cascade, abscisic acid-mediated signalling pathway, cell communication | |
| AT5G11920 | 4.2 | 8344.6b | AtcwINV6 (6-&1-fructan exohydrolase) | Carbohydrate metabolic process, regulation of hydrogen peroxide metabolic process | |
| AT1G15670 | 4.05 | 14417.2b | Kelch repeat-containing F-box family protein | Negative regulation of cytokinin-mediated signalling pathway | |
| AT5G67340 | 3.94 | 3905.65b | Armadillo/beta-catenin repeat family protein | Endoplasmic reticulum–nucleus signalling pathway, MAPK cascade, negative regulation of defence response | |
| AT1G72360 | 3.76 | 11466.3b | Ethylene-responsive element- binding protein | Cellular response to ethylene stimulus, regulation of transcription | |
| AT3G28580 | 3.76 | 8339.31b | AAA-type ATPase family protein | Response to abscisic acid stimulus, response to ethylene stimulus | |
| AT5G46080 | 3.62 | 890.14b | Protein kinase family protein | Ethylene biosynthetic process, protein phosphorylation | |
| AT5G01550 | 3.08 | 1208.89b | LECRKA4.2 (LECTIN RECEPTOR KINASE A4.1) | Abscisic acid-mediated signalling pathway, protein phosphorylation, response to chitin | |
| AT3G13100 | 2.66 | 2294.74b | ATMRP7; ATPase | Response to other organisms, salicylic acid biosynthetic process | |
| Root development | |||||
| AT4G31500 | 3.44 | 17621.3b | CYP83B1 (CYTOCHROME P450 MONOOXYGENASE) | Adventitious root development, callose deposition in cell wall during defence response | |
| AT1G67980 | 3.42 | 1164.11b | CCoAMT; caffeoyl-CoA O-methyltransferase | Lignin biosynthetic process | |
| AT3G45960 | 2.83 | 1535.55b | ATEXLA3 (Arabidopsis thaliana expansin-like a3) | Plant-type cell wall loosening, plant-type cell wall organization | |
| AT5G13080 | 2.58 | 3789.3b | WRKY75; transcription factor | Cellular response to phosphate starvation, lateral root development, response to ethylene stimulus | |
| Signalling/defence | |||||
| AT2G17060 | 3.79 | 561.85b | Disease resistance protein (TIR- NBS-LRR class) | Defence response, signal transduction | |
| AT4G28350 | 3.66 | 1223.16b | Lectin protein kinase family protein | Defence response to fungus, protein phosphorylation, response to chitin | |
| AT1G64400 | 3.09 | 2202.11b | Long-chain-fatty-acid–CoA ligase | Defence response to insect, fatty acid biosynthetic process | |
| AT3G21650 | 2.74 | 900.6b | Serine/threonine protein phosphatase 2A (PP2A) | Signal transduction | |
a Lateral root.
b Root.
FLS2 is highly expressed in emerging lateral roots
In soil, roots are exposed to a variety of microorganisms, both pathogenic and beneficial. Interestingly, when plants were grown under non-sterile conditions, an up-regulation of FLS2 promoter expression was observed in the endodermis and cortical cells but not in epidermal cells, showing that the pFLS2::GUS expression in roots is not restricted to the vascular cylinder but can expand at least to the cortical cell layer (Supplementary Fig. S5A, B at JXB online).
This expansion of the promoter activity to different tissues also became apparent during the developmental process of LR growth. The pFLS2::GUS lines exhibited significant staining in the LR primordia and outgrowing LRs (Fig. 3A–D). When they reached a certain developmental stage, the promoter activity was restricted again to the vascular cylinder of the developed LR and no staining was found in the tip of the LR, similarly to what was observed for the primary root tip (data not shown). Outgrowing LRs provide prominent entry points of bacterial pathogens as the outgrowth from the pericycle to the outer epidermis is accompanied by epidermal cracks, where bacteria can easily attach and gain access to root tissues (Supplementary Fig. S4 at JXB online; Dong et al., 2003; Tyler and Triplett, 2008). Thus, similarly to leaves, promoter activity can be found in cells vulnerable to bacterial infection.
Flg22 affects growth of FLS2-expressing lateral roots and auxin distribution. (A) pFLS2::GUS seedlings (10 d after germinationg) show prominent GUS staining in outgrowing lateral roots (LRs) (arrows); bar=50 μm. (B) Cross-section of LR outgrowth (arrows); bar=10 μm. (C) Promoter activity is present in a developed LR; bar=50 μm. (D) Cross-section of a developed LR; bar=10 μm. (E) Col-0 and fls2 seedlings 12 d after germination with and without flg22 (1 μM) treatment; red arrows indicate LRs. (F) Graph showing quantification of LR per cm root length in Col-0 and fls2 seedlings with and without flg22 treatment (1 μM); bars represent the average of three independent experiments; error bars represents the SD; statistical significance is represented by Student’s t-test (P-value >0.001). (G) Confocal micrographs show roots of DR5:GFP transgenic seedling roots (10 d after germination) incubated for 72h with or without flg22 (1 μM); arrowheads indicate GFP signals in epidermal cells of flg22-treated seedlings; middle and bottom panels depict different developmental stages of LR formation along the axis of 10-day-old roots; arrows indicate DR5:GFP signals marking LR primordia; bar=50 μm.
Flg22 regulates lateral root growth and auxin distribution
Long-term treatment with flg22 leads to inhibition of root growth in wild-type seedlings (Gomez-Gomez et al., 1999). This study was extended and it was observed that the flg22-dependent inhibition of root growth (Supplementary Fig. S5C at JXB online) was accompanied by a reduced number of LRs (Fig. 3E, F). As LR initiation is strongly dependent on auxin accumulation in the cells primed for LR outgrowth (Dubrovsky et al., 2008), experiments were carried out to determine whether flg22 treatment might interfere with auxin distribution and maxima during root and LR growth. DR5::GFP (auxin-responsive GFP) lines were treated with flg22 and it was found that the auxin maxima in the LR primordia are reduced after 72h of flg22 treatment compared with the control line, which was mock treated during this period (Fig. 3G). In addition, in the flg22-treated DR5::GFP seedlings, GFP signals were observed in the root epidermal cells, which were not present in control lines (Fig. 3G). Thus, these data showed that flg22 influences auxin distribution in a cell type-specific manner. The ectopic up-regulation of auxin in the epidermal cells as well as the down-regulation of auxin in the LR primordia might contribute to the flg22-dependent inhibition of root and LR growth. This correlates with the identification of AT1G59500 and AT1G68765 from the transcriptome data set, which are known auxin-responsive genes [The Arabidopsis Information Resource (TAIR)], and is in agreement with previous studies showing that auxin and auxin-responsive genes are also regulated by flg22 (Zipfel et al., 2004; Navarro et al., 2006). The findings are also consistent with reduced DR5–GUS expression in roots and inhibition of auxin-mediated adventitious root growth when triggered with oligogalacturonides, components of the plant cell wall known to trigger plant defences similar to PAMPs (Savatin et al., 2011).
Hormones and stress signals regulate FLS2 expression in different root tissues
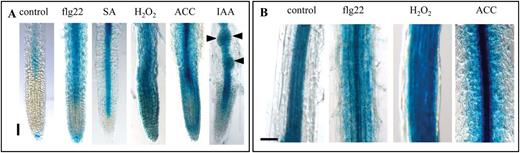
PAMP-triggered immunity (PTI) is highly regulated by the action of phytohormones such as salicylic acid (SA), ethylene, and jasmonate (JA) (Bari and Jones, 2009). In this context, the different hormones and abiotic stresses which are known to play important roles in PTI responses were studied for their effect on FLS2 promoter activity. In mock-treated roots, pFLS2::GUS expression was visible in the root late elongation zone, as described above (Figs 2A, 5). Additionally in ~20% of the control roots, a distinct GUS staining in root cap cells directly underneath the root meristem was observed (Fig. 4A). Incubation with flg22 led to an increased FLS2 promoter activity in the root tip starting at the transition zone and extending to cortical cells in the differentiation zone (Fig. 4A, B). When treated with SA, pFLS2::GUS roots showed a strong blue staining in the vasculature, which started close to the meristematic zone (Fig. 4A), but did not extend to the cortex or to the differentiation zone (data not shown). Treatment with H2O2 or the ethylene precursor ACC provoked an almost uniform promoter activity in the root cap, root meristem, and root epidermal cells (Fig. 4A, B). However, ACC induced pFLS2::GUS activity in the vasculature to a much higher extent compared with H2O2 or mock treatment (Fig. 4B).
Induced FLS2 expression in roots is regulated in a tissue-dependent manner. (A) Promoter activity in the root tip of pFLS2::GUS seedlings (8 d after germination) after treatment with flg22 (10 μM), SA (50 μM), H2O2 (1mM), ACC (10 μM), and IAA (10 μM). (B) Promoter activity in the root differentiation zone after flg22 (10 μM), H2O2 (1mM), and ACC (10 μM) treatment; (A, B) bar=100 μm.
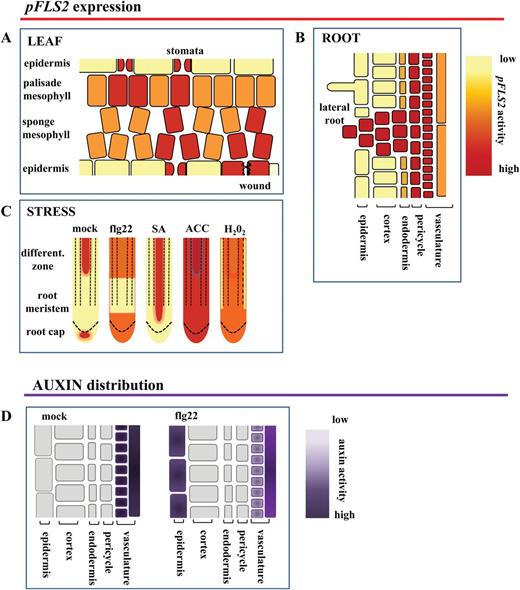
Model summarizing FLS2 cell-type and tissue-specific expression patterns. The cartoon depicts the promoter activity of FLS2 in leaves (A) and roots (B); (C) stress responsiveness of the promoter in roots; and (D) flg22-dependent ectopic up-regulation of auxin in root epidermal cells.
It was also tested whether the promoter of FLS2 is auxin responsive. The emergence of LR primordia becomes highly induced by incubation with the auxin analogue IAA, which exhibited clear FLS2 promoter activity (Fig. 4A). However, GUS accumulation was specific to LR primordia and the vasculature in IAA-treated roots, and no GUS staining was observed in cortical cells. These experiments revealed that flg22, SA, H2O2, and ethylene all influence the expression activity of the FLS2 promoter, but the responses are specific to different tissue layers in the root (Fig. 4). In summary, this identifies an unexpected level of tissue-dependent regulation of FLS2 expression in response to a variety of different stresses (Fig. 5).
Discussion
The prevailing view in plant immunity is that all plant cells are capable of pathogen perception and initial defence responses. This would require constitutive expression of at least the primary sensors of the immune system. Based on plant-scale expression analysis, FLS2 was found in all plant organs including flowers, leaves, stems, and roots (Gomez-Gomez and Boller, 2000; this study). However, cell type-specific responses might play an important role in the context of how plants initiate defence responses against potentially invasive pathogens, but do not fend off beneficial microbes that are often needed for plant growth especially in low nutrient conditions (Bulgarelli et al., 2013). It was therefore proposed that the cellular and tissue location of immune components is essential to mount the appropriate defence responses, and that they should be best located at putative entry sites of pathogens to inhibit their invasion efficiently (Faulkner and Robatzek, 2012). In this study, the FLS2 promoter activity was followed and it was found that while FLS2 is generally expressed in all tissues, there are remarkable differences in the level of the expression regulated in a cell type-specific and developmental manner. In addition, the FLS2 promoter activity is responsive to several hormones playing roles in plant immunity such as SA and ethylene, which themselves are induced upon flg22 elicitation (Felix et al., 1999; Tsuda et al., 2008). Consistently, these and the present observations show that FLS2 is subject to positive regulation between receptor expression and the immune response.
Prominent entry sites of potential pathogens are guarded by high FLS2 expression
Hydathodes are pores at the leaf margin that are continuous with the xylem. Hydathodes are targeted by pathogenic bacteria such as Xanthomonas campestris pv. campestris, as points of access into plant tissue (Hugouvieux et al., 1998). The stomatal pores (Zeng and He, 2010) represent another prominent entry route of bacterial pathogens. Stomata close upon PAMP perception to restrict pathogen entry, and successful pathogens secrete effectors such as HopM1, syringoline, and coronatine that inhibit the closure and/or actively induce re-opening (Melotto et al., 2006; Schellenberg et al., 2010; Zeng and He, 2010; Lozano-Duran et al., 2014). Both cell types are characterized by a high promoter activity of FLS2 compared with the surrounding mesophyll cells (Fig. 1C, E), suggesting that cells at tissue entry points are particularly well equipped to detect invading pathogens. Further, the mesophyll cells forming the substomatal cavity also exhibit a higher FLS2 promoter activity (Figs 1F, 5). Previous data show that FLS2 mediates immunity at the level of stomatal entry (Zipfel et al., 2004; Zeng and He, 2010). In agreement with this, stomatal expression of FLS2 is enhanced upon bacterial infection. GUS staining is more intense in guard cells relative to the surrounding cells, indicating a guard cell-specific regulation of FLS2 promoter activity (Fig. 1H). Although the possibility that the prominent GUS staining at hydathodes might be unspecific cannot be ruled out, FLS2 expression at this location is consistent with the fact that hydathodes mark the end-points of the vasculature, another tissue exhibiting high FLS2 expression and, importantly, responsive to flg22. The overall patterns of pFLS2::GUS expression observed (Fig. 5A–C) are in agreement with publicly available expression data (eFPBrowser; Faulkner and Robatzek, 2012).
Wounds and cracks in the epidermal layers represent sites of vulnerability with respect to pathogen infection. The bacterial colonization beyond these primary infection sites is dependent on secreted effectors such as syringoline promoting distant tissue colonization (Misas-Villamil et al., 2013). The FLS2 promoter is responsive to wounding in leaves (Fig. 1I; Supplementary Fig. S3C at JXB online), suggesting that cells at these sites might depend on higher FLS2 levels to fend off pathogen invasion of neighbouring tissues. This is consistent with a previous study which revealed that higher protein levels of FLS2 contributes to more flg22 binding and are positively associated with reduced Pto DC3000 proliferation (Vetter et al., 2012).
Plants also have ‘natural’ wounds, which occur during the emergence of LRs. These manifest as ruptures in the epidermal cell layer around the LR meristem. Detailed observations of bacterial colonization of roots led to the assumption that bacteria use these LR emergence sites as entry routes to the roots (Dong et al., 2003; Tyler and Triplett, 2008) (Supplementary Fig. S4B at JXB online). Although in developed roots FLS2 expression was not present in the meristem, the FLS2 promoter exhibited a strong activity in the LR primordia and outgrowing LRs (Fig. 3A–D). These observations indicate that the FLS2 expression is highly dynamic and regulated in a cell type- and development-dependent manner (Fig. 5B, C). Considering that LRs do not possess a root cap, which can also function as a PAMP-reactive physical barrier to the root meristem (Plancot et al., 2013), it might be essential for a plant to guard the LR meristem.
The vasculature is a tissue with high FLS2 expression
Evidently, the vasculature provides excellent means for pathogens to spread throughout the plant. Together with the vasculature being rich in nutrients and water, this makes the vasculature a very attractive target tissue for pathogens. In plant interactions with a fungal pathogen, strong lignification of vascular bundles is associated with a compromised infection (Tanaka et al., 2014). One significant observation of the present study is the defined and high activity of the FLS2 promoter in the root stele, which is correlated with a high abundance of the FLS2–GFP fusion protein (Fig. 2B–E). Interestingly, high promoter activity in vascular tissue was also found for PEPR1 and PEPR2, receptors associated with damage-elicited responses and immunity (Bartels et al., 2013). In addition, it was observed that the vasculature contributes to the flg22-induced increase in [Ca2+]i (Fig. 2F). It has been described that flg22 induces the production of lignin (Schenke et al., 2011), but whether lignification is part of the FLS2-mediated immunity to prevent colonization and spread through the vasculature remains to be addressed. In the leaf, Pseudomonas bacteria colonize distant tissues along the vasculature (Misas-Villamil et al., 2011), whereas, in the root, the bacterial pathogen Ralstonia solanacearum directly utilizes plant xylem vessels to move through the plant (Digonnet et al., 2012). It is tempting to speculate that the absence of elicitor-active flagellin promotes the infection success of R. solanacearum bypassing FLS2-mediated defences in the vasculature (Pfund et al., 2004).
While FLS2 expression is restricted to the stele under normal conditions, expression can be expanded to the cortex under certain stresses (Figs 4, 5C) and it is shown that roots are sensitive to flg22 initiating typical defence responses (Millet et al., 2010; Jacobs et al., 2011; this study). It is possible that low expression of FLS2 in the root cortex allows the colonization of this tissue by beneficial bacteria without triggering defence. High constitutive expression of FLS2 in the stele might provide an additional barrier to bacterial invasion of the vascular tissue beyond the cortex, and stress-induced expansion of this zone of expression might reflect increased vulnerability of the tissue. Flg22-dependent gene induction was quite specifically activated in the elongation zone, whereas flg22-induced callose deposition was observed over the entire root length (Millet et al., 2010). However, whether these immune response are initiated in epidermal cells, cortical cells, or inner cylinder cells needs to be addressed in the future.
Auxin-mediated root development is responsive to flg22
The long-term incubation with flg22 is known to inhibit root growth (Gomez-Gomez et al., 1999) and this inhibition of root growth is accompanied by a reduced development of LRs (Fig. 3E, F). Auxin, an important plant hormone involved in the regulation of root cell elongation and LR outgrowth, is found to be ectopically up-regulated in the epidermal cells of flg22-treated roots, whereas it is down-regulated in the LR primordia (Figs 3G, 5D). This is in agreement with studies describing an flg22-dependent antagonism for auxin activity, which leads to a rapid down-regulation of auxin-responsive genes and contributes to plant resistance against bacteria (Navarro et al., 2006). Ectopic up-regulation of auxin in root epidermal cells was also described to be involved in ethylene-dependent root growth arrest (Ruzicka et al., 2007). As ethylene production is triggered by flg22 (Felix et al., 1999), it might be possible that these hormones are together integrated in the flg22-induced inhibition of root growth, with a possible outcome being that flg22 reduces putative bacterial entry points at LRs.
This interplay between the flg22 responses and hormone signalling is also reflected at the level of the FLS2 promoter activity, as seen by the influence of IAA and ACC on the expression of FLS2. ACC treatment as well as the high induction around wound sites is consistent with a direct control of FLS2 transcription by ethylene signalling (Boutrot et al., 2010; Mersmann et al., 2010). Altogether, these findings show a positive regulation of FLS2 expression by hormones (ethylene and SA) and small signalling molecules such as reactive oxygen species, which are produced upon flg22 trigger (Bari and Jones, 2009). This positive transcriptional regulation might be important to deliver newly synthesized receptors to the plasma membrane since activated FLS2 is removed from the plasma membrane by endocytosis and degradation (Robatzek et al., 2006; Göhre et al., 2008; Smith et al., 2014).
Concluding remark
It is shown that the FLS2 promoter activity maps to vulnerable tissue targeted by bacteria for entry and colonization in plants. This will be useful to understand the tissue- and cell type-specific role of FLS2 in immune signalling, and will aid in strategies to enhance plant resistance by targeting of defence to relevant tissues.
Acknowledgements
We thank Kim Findlay and Elaine Barclay (the BioImaging Support, John Innes Centre, Norwich UK) for technical help with the microtome, and Christine Faulkner for critically reading the manuscript. MB was supported by a grant from the Deutsche Forschungsgemeinschaft. Research in the laboratory of SR is supported by the Gatsby Charitable Foundation and a grant from the European Research Council (ERC).
References



![Roots exhibit specific FLS2 expression patterns and tissue-specific responsiveness to flg22. In sterile-grown roots (8 d after germination) of pFLS2::GUS, the promoter activity is not present in root tips (A), but shows a high expression in the root stele (B) as revealed by root cross-section (C); bar=10 μm. (D) Confocal micrographs of pFLS2::FLS2–GFP show accumulation of GFP signal in the inner part of the stele (arrowheads point to inhibited uptake of propidium iodide at the endodermis; bar=10 μm. (E) Digital cross-section with plasma membrane localization of FLS2–GFP at cortical cells (arrowheads) and in the root cylinder (arrow). Autofluorescence of xylem is marked with asterisks. (F) Changes in [Ca2+]i values in mock-treated control (water, 35 s) or in response to flg22 (100nm, 35 s) in 35S::AEQ seedlings and the vasculature enhancer trap line KC274. Luminescence was measured over 1200 s. Data are presented as means ±SD, n=4 (mock), n=6 (flg22). (G) Immunoblot of detected FLS2 protein in roots and shoots. Samples were enriched for glycosylated proteins using ConA. (H) Immunoblot detection of phosphorylated MAPK present in Col-0 after 1 μM flg22 (10min) treatment but not in fls2. (I) Gene ontology of enriched genes specifically up-regulated in Ler roots after flg22 treatment (10 μM, 30min).](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/jxb/65/22/10.1093_jxb_eru366/4/m_exbotj_eru366_f0002.jpeg?Expires=1716377191&Signature=HHYQg3X1gcXkbWY5H8Cv9ZTaIn-Jm~QNjkgonVo-FU1rQ45yQHwdMu1PBIXtmzguWaC71H5e26iwX~qLtVYiNWxFihqmkDc~F0FMZYQGlkdVJ7mn0DKZnSFhXuQ4nBexv6TqIuhGF~VIkqOd66XfW3CNTNvkJJu9upKrn0bLIxFLfOBkWB8sQE3gjB3Rwr32UV8pbKp9IB-Cjod27f93Q31mpqymHmdXa-dqpJxU3aOFY4Z9U2xjraXVl5o3IVmYdpNoaqvzNm~VFp7gXxgQrJ44LVhpIw0Nkz3bdH-PtxyZEcTwbGBFacbNcGQT~i1NJwgtL9ZjXDR~qtq96buCxg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

Comments