-
PDF
- Split View
-
Views
-
Cite
Cite
Ralph van Berloo, GGT 2.0: Versatile Software for Visualization and Analysis of Genetic Data, Journal of Heredity, Volume 99, Issue 2, March/April 2008, Pages 232–236, https://doi.org/10.1093/jhered/esm109
Close - Share Icon Share
Abstract
Ever since its first release in 1999, the free software package for visualization of molecular marker data, graphical genotype (GGT), has been constantly adapted and improved. The GGT package was developed in a plant-breeding context and thus focuses on plant genetic data but was not intended to be limited to plants only. The current version has many options for genetic analysis of populations including diversity analyses and simple association studies. A second release of the GGT package, GGT 2.0 (available through http://www.plantbreeding.wur.nl), is therefore presented in this paper. An overview of existing and new features that are available within GGT 2.0, and a case study in which GGT 2.0 is applied to analyze an existing set of plant genetic data, are presented and discussed.
Recently, Excoffier and Heckel (2006) presented an interesting overview and review of available software in the field of population genetics. The free software package graphical genotypes (GGTs) was unfortunately not discussed in this paper, probably because historically GGT is more associated with the visualization of data, rather than population genetic analyses. But recent improvements and extensions of the package have now brought many of the analyses available in other software to the set of tools present in GGT, retaining the user-friendly (point and click) approach of the program. GGT was developed in a plant-breeding research environment and is therefore biased toward plant-breeding types of problems and data. However, its scope also includes other types of genetic data. Ever since the first publication on the GGT software package (van Berloo 1999), the amount of users has grown considerably, to at least several hundreds of users worldwide. A growing amount of papers have been published in which GGT was used for analysis, visualization, or obtaining marker statistics (e.g., Von Korff et al. 2004; Iban et al. 2005; Yun et al. 2006; De Vos et al. 2007; Huang et al. 2007; Poormohammad Kiani et al. 2007). These papers mainly deal with application in plant genetics but also applications in fungal and animal genetics have been described. Over the past years, the functionality of the GGT software was extended considerably. This has led to the release of an official new version of this versatile package for visualization and analysis of genetic data. The new features present in the new release: GGT 2.0 are discussed in this paper. A more detailed discussion of all features can be found in the GGT 2.0 user manual (van Berloo 2007) that is distributed together with the GGT 2.0 executable.
Input Data
Molecular Marker Data
GGT 2.0 deals with visualization and analyses that involve molecular marker scores. Common input data consist of a matrix of marker scores with markers arranged in rows and genotypes arranged in columns. In most cases, GGT 2.0 will be used to visualize data of markers with known map positions on a genetic map, allowing GGT 2.0 to display estimated lengths of genomic compositions as colored chromosome bar segments (an example is shown in Figure 1). However, not all analyses available within GGT 2.0 strictly require map positions of markers to be known, and an option to impute dummy positions when these are not known is therefore present. GGT 2.0 accepts single and composite (multiple chromosome) .ggt datafiles, which consist of a simple text format. GGT 2.0 has a special module to prepare .ggt datafiles from commonly used locus marker files and map files, which conform to the Joinmap (van Ooijen 2006) style of encoding genetic data. It also accepts data prepared in a spreadsheet format. Special attention has been given to a stepwise compilation of data into GGT 2.0, thereby allowing users to join data from several sheets of a spreadsheet, which can be quite useful when the data are too large to fit in a single sheet. There are no hard coded limits to the amount of data that can be handled by GGT 2.0. Commonly used population sizes in plant breeding (e.g., 150 individuals each scored for a few hundred markers) can be used comfortably on modern computer hardware. Larger data sets can still be used but will take longer for analyses and drawing to complete. For large data sets, it is recommended to disable the more advanced drawing options. At most 80 different alleles per locus may be indicated.
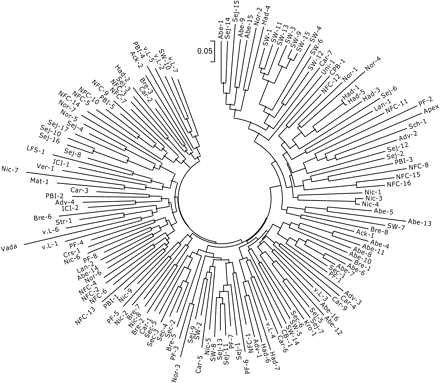
Neighbor-Joining dendrogram derived from Jaccard similarity estimates using the amplified fragment length polymorphism (AFLP) fingerprint data scored in a set of 148 barley cultivars.
Trait Data
GGT 2.0 now has several options to perform analyses using numerical trait data in combination with molecular marker data. A properly formatted matrix of trait data, containing column and row headers can be copied and pasted from a spreadsheet program into GGT 2.0. Alternatively, the .qua textual file description of trait observations (a format used in quantitative trait locus [QTL] mapping studies) is also accepted.
Methods Implemented in GGT 2.0: General Overview
Visualization, Graphical Images of Estimated Genomic Composition
GGT was originally created for visualization of molecular marker data. This is still the core functionality and the display of data have been considerably improved. It now features an overview of the data one chromosome at a time, one genotype at a time, or all chromosomes for all genotypes in a single image. On the fly, allele statistics are calculated and an estimate of the most commonly present allele for all marker loci is determined and shown graphically as a “consensus genotype”, which can be useful for comparison purposes.
Sorting and Filtering of Data
GGT 2.0 can be used to sort the genotypes based on a number of criteria, for instance allelic composition. In this way, those genotypes that have the most desirable composition can be more easily identified. More specific selection of genotypes is possible through the filter and selection option. This allows users to specify exact criteria for individual markers and to perform a simultaneous filtering on several loci of interest. Genotypes that meet all criteria will be marked and shown. This feature can be useful to select genotypes bearing a desired combination of genes or QTLs that can be selected through flanking markers.
Generation of Reports
GGT 2.0 provides reports on allelic composition arranged in a per-genotype or per-marker format. Direct exports to a spreadsheet format are available for these numerical reports.
Diversity Analysis
GGT 2.0 has options not only to determine similarity of genotypes, based on marker patterns, but also similarity of markers can be assessed. Three commonly used similarity parameters (Simple Matching, Jaccard, and Euclidean similarity/distance) are available, and a full diallel matrix of the complementary genetic distances can be computed. This matrix can then be stored for use in other software, like MEGA (Kumar et al. 2004) or Splitstree (Huson 1998), or it can be used as source data to calculate a Neighbor-Joining or unweighted pair group method with arithmetic mean dendrogram.
Analysis of Linkage Disequilibrium
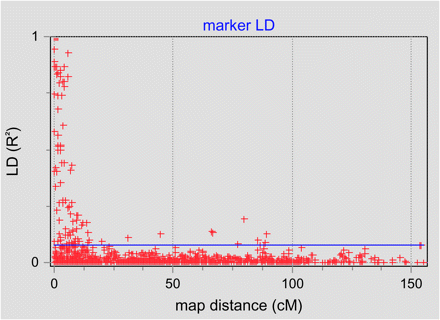
When analyzing marker data that are scored in unrelated germplasm or germplasm where relationships are presumed but not well defined, one of the statistics of interest is the amount of linkage disequilibrium (LD; for more background see e.g., Gaut and Long 2003). The presence of intermarker LD and the amount of LD are indicative for the prospects of usage of the material in, for instance, association mapping studies. GGT 2.0 uses numerical allele values to calculate a number of popular LD statistics including R2, Lewontins D′ and an adjusted χ2 value for multiallelic markers (see Zhao et al. 2005 for definitions of these parameters). The results of LD analyses are reported in 3 ways: a matrix of pairwise LD observations, a graph plotting observed LD against pairwise marker distances, and a heat map, which plots the observed LD matrix in a graphical way with color intensities depending on LD values.
Association Mapping
Simple procedures for establishing associations of genomic regions with traits of interest have grown in popularity in recent years. GGT 2.0 can facilitate preliminary association analyses in a user-friendly way. These analyses require, in addition to marker data, trait observations on the same genotypes. When dominant or codominant biallelic markers are available, a simple correlation analysis of markers and traits can be performed by GGT 2.0. In the case of multiallelic markers, a more complex analysis of variance is available. Depending on the association method, correlation significances are reported and a correction for the number of performed tests, through a false discovery rate approach, is calculated. Resulting associations can be displayed in a graphic fashion, plotting observed associations along the chromosome bars.
Population Subset Selection
The most recent addition to GGT 2.0 is a module to select subsets from larger populations in such a way that a maximum amount of genetic diversity is retained in the selected set of lines. This is done using a nested iterative procedure (for details see Bataillon et al. 1996) or through a simulated annealing approach. These procedures try to search the complex solution space in an efficient way and will not in all cases reach the global optimal solution. However, an exhaustive search for a global solution would require far too much time. The implemented search methods are expected to come up with a solution that is a close approximation of the global optimum, in an acceptable amount of time.
Data Input and Output
Most user requests for improvement of the GGT package were in the field of usability and simplicity of use, especially with regard to an easy input and output/export of relevant data. The original, text-based input of single linkage groups has been supplemented with options to load data on several groups from a single file. Input of data from a spread sheet was added as an option and also recoding of multiple character allele codes to the internally used single character style of coding. Numerical values that are associated with alleles (e.g., band intensity values rather than discrete band scores) can be imported as well. Export options to external software now include: spreadsheet matrices, MEGA (Kumar et al. 2004) or Splitstree (Huson 1998) distance matrices, and Mapchart (Voorrips 2002) linkage map descriptions. Plots, charts, and images created by GGT 2.0 can be saved in a metafile or jpeg format.
A Case Study of the Use of GGT 2.0 That Focuses on Recent GGT Features: Association Mapping in Unrelated Barley Genotypes
Using GGT 2.0, we reanalyze parts of the barley dataset discussed in Kraakman et al. (2004, 2006). Kraakman et al. focused on a set of approximately 150 cultivated barley varieties and genotyped these varieties with Amplified Fragment Length Polymorphism (AFLP) markers. Phenotype data were obtained from variety trials and national variety evaluations, as well as field evaluations. Here GGT 2.0 was used to transform the (binary) AFLP marker data and the linkage map to a GGT datafile. A spreadsheet file with phenotypic observations was also prepared. In order to perform association mapping, we wanted to investigate:
a) Possible indications of population structure in the dataset.
b) The amount of Linkage Disequilibrium in the dataset.
c) Associations between markers and some of the resistance scores observed in the set of cultivars.
These analyses were all possible within GGT 2.0 using built in functionality.
Diversity Analysis
The “genetic distances” module of GGT 2.0 was used to calculate a matrix of pairwise genotype distances, where these distances were derived using the Jaccard similarity coefficient. Next, this matrix was used to derive a Neighbor-Joining dendrogram. The dendrogram viewer of the MEGA 3 package (Kumar et al. 2004) was used to visualize the tree, which is shown in Figure 1. From this figure, we do not observe indications for the presence of clear substructures among the set of cultivars (in the original paper by Kraakman et al., this finding is confirmed using Bayesian clustering).
LD Analysis
Next, we evaluated the presence of LD and the decay of LD with map distance. For binary AFLP data, it is appropriate to assign a numerical value “1” for the presence of a marker band and ‘0’ when a band is absent. We entered these values in the “settings” module as new allele properties within GGT 2.0. LD analyses were performed and R2 values for pairs of markers reported. The LD decay plot created by GGT 2.0, in which observed LD is plotted against map distance, was exported to a file and is shown in Figure 2. As a rough estimate of the rate of LD decay, we observe that most LD disappears for genetic distances over 10 cM. A marker map with a density of one marker every 10 cM or less would therefore be advisable for association mapping.
LD decay plot showing results of LD analysis performed on genetic data in a set of 148 barley cultivars.
Association Analysis
Although the available marker map does not have the desired density, we can still perform association analysis. Not all genomic associations will be revealed by this analysis, but for our purpose of a first genomic scan, the results are still of interest.
To perform association analysis, a spreadsheet file with quantitative resistance score observations on all available cultivars was prepared. As row headers in the first column, we used the same genotypic labels that were used for the marker data, whereas the column headers were used to specify trait names. The phenotypic data were imported into GGT 2.0 by copying and pasting the relevant block of data from the spreadsheet. Next, association analysis was performed. Because we were dealing with binary AFLP markers, regular Pearson correlation analysis was used. The results of this analysis reported the observed correlations and squared correlations, as well as the associated probabilities. Also adjusted threshold levels, using a false discovery rate approach to account for multiple testing issues, were calculated and reported. As this case study is merely meant to illustrate the options in GGT 2.0, we only present here a small part of the results. Table 1 shows an excerpt of the association results. On chromosome 1, we can clearly observe significant associations of marker E38M54-472 with barley yellow dwarf virus (BYDV) and marker E39M61-255 with infection type (IT) and area under disease progress curve (AUDPC); a disease severity parameter. These results are fully in line with results reported earlier by Kraakman et al. (2006), and full details on these and other results can be obtained from this paper. Note that using GGT 2.0, we were able to obtain these results without the need to apply statistical software and with the use of very simply formatted data files as input.
Association analyses results for barley disease related traits and plant height that were scored among the set of 148 barley cultivars
| IT | LP | AUDPC | BYDV | HEIGHT | |
| Marker | |||||
| E38M54-472 | 1 | 1.9 | 1.6 | 8.6 | 0 |
| E37M33-311 | 0.1 | 0.6 | 0.2 | 0.4 | 0 |
| E38M55-205 | 0.9 | 0.2 | 0.9 | 1.8 | 1.3 |
| E33M54-214 | 0.2 | 0.5 | 0.4 | 0.3 | 0.6 |
| E38M50-119 | 0.4 | 1.9 | 0.5 | 1.4 | 3.3 |
| E39M61-255 | 26.0 | 0.5 | 20.4 | 0.2 | 0.2 |
| E39M61-222 | 0.4 | 0.3 | 0.8 | 0.3 | 0.5 |
| E38M50-284 | 0.1 | 0.7 | 0.2 | 0.1 | 0.4 |
| E35M54-183 | 0.3 | 0.1 | 0.1 | 3.1 | 0.8 |
| E35M54-180 | 0.2 | 0.1 | 0.1 | 3.1 | 0.8 |
| IT | LP | AUDPC | BYDV | HEIGHT | |
| Marker | |||||
| E38M54-472 | 1 | 1.9 | 1.6 | 8.6 | 0 |
| E37M33-311 | 0.1 | 0.6 | 0.2 | 0.4 | 0 |
| E38M55-205 | 0.9 | 0.2 | 0.9 | 1.8 | 1.3 |
| E33M54-214 | 0.2 | 0.5 | 0.4 | 0.3 | 0.6 |
| E38M50-119 | 0.4 | 1.9 | 0.5 | 1.4 | 3.3 |
| E39M61-255 | 26.0 | 0.5 | 20.4 | 0.2 | 0.2 |
| E39M61-222 | 0.4 | 0.3 | 0.8 | 0.3 | 0.5 |
| E38M50-284 | 0.1 | 0.7 | 0.2 | 0.1 | 0.4 |
| E35M54-183 | 0.3 | 0.1 | 0.1 | 3.1 | 0.8 |
| E35M54-180 | 0.2 | 0.1 | 0.1 | 3.1 | 0.8 |
Displayed are the −10Log(P) values (−10Log transformations of the association probabilities) for markers on chromosome 1. Markers are arranged (in map order) in rows, traits are arranged in columns. Marker E38M54-472 shows a strong association with BYDV. Marker E39M61-225 shows a very strong association with the traits IT and AUDPC. False discovery rate correction of critical values (bold numbers) indicates that associations with a −10Log (P) value higher than 3.1 should be considered significant in this analysis.
Association analyses results for barley disease related traits and plant height that were scored among the set of 148 barley cultivars
| IT | LP | AUDPC | BYDV | HEIGHT | |
| Marker | |||||
| E38M54-472 | 1 | 1.9 | 1.6 | 8.6 | 0 |
| E37M33-311 | 0.1 | 0.6 | 0.2 | 0.4 | 0 |
| E38M55-205 | 0.9 | 0.2 | 0.9 | 1.8 | 1.3 |
| E33M54-214 | 0.2 | 0.5 | 0.4 | 0.3 | 0.6 |
| E38M50-119 | 0.4 | 1.9 | 0.5 | 1.4 | 3.3 |
| E39M61-255 | 26.0 | 0.5 | 20.4 | 0.2 | 0.2 |
| E39M61-222 | 0.4 | 0.3 | 0.8 | 0.3 | 0.5 |
| E38M50-284 | 0.1 | 0.7 | 0.2 | 0.1 | 0.4 |
| E35M54-183 | 0.3 | 0.1 | 0.1 | 3.1 | 0.8 |
| E35M54-180 | 0.2 | 0.1 | 0.1 | 3.1 | 0.8 |
| IT | LP | AUDPC | BYDV | HEIGHT | |
| Marker | |||||
| E38M54-472 | 1 | 1.9 | 1.6 | 8.6 | 0 |
| E37M33-311 | 0.1 | 0.6 | 0.2 | 0.4 | 0 |
| E38M55-205 | 0.9 | 0.2 | 0.9 | 1.8 | 1.3 |
| E33M54-214 | 0.2 | 0.5 | 0.4 | 0.3 | 0.6 |
| E38M50-119 | 0.4 | 1.9 | 0.5 | 1.4 | 3.3 |
| E39M61-255 | 26.0 | 0.5 | 20.4 | 0.2 | 0.2 |
| E39M61-222 | 0.4 | 0.3 | 0.8 | 0.3 | 0.5 |
| E38M50-284 | 0.1 | 0.7 | 0.2 | 0.1 | 0.4 |
| E35M54-183 | 0.3 | 0.1 | 0.1 | 3.1 | 0.8 |
| E35M54-180 | 0.2 | 0.1 | 0.1 | 3.1 | 0.8 |
Displayed are the −10Log(P) values (−10Log transformations of the association probabilities) for markers on chromosome 1. Markers are arranged (in map order) in rows, traits are arranged in columns. Marker E38M54-472 shows a strong association with BYDV. Marker E39M61-225 shows a very strong association with the traits IT and AUDPC. False discovery rate correction of critical values (bold numbers) indicates that associations with a −10Log (P) value higher than 3.1 should be considered significant in this analysis.
This case study shows the use of GGT 2.0 in a population for which we may expect, based on knowledge of the natural way of propagation of barley and knowledge on germplasm sources and history, presence of sufficient LD, and absence of population structure. In other types of research germplasm like for instance interbred populations, the analyses described in this paper may not be appropriate. It remains up to the researcher using GGT 2.0 and his prior knowledge of sources, origins, and other information of the genetic materials at hand to make this judgment for himself. A rough first analysis of data is presented in the presented case study. A more extensive analysis on the presence of population substructure, for instance using the Structure package (Pritchard 2000) is advisable (and is described in Kraakman et al. 2004, 2006). More detailed follow-up analyses using higher marker densities could be a next step. Alternatively, parents could be selected from the germplasm at hand and new populations, expected to segregate for the traits of interest, could be developed and analyzed using conventional QTL mapping.
Conclusions
The GGT 2.0 package provides users interested in plant genetics worldwide with a free and versatile package that is able to assist in a number of steps in plant-breeding programs or genetic analyses, make selection based on markers easier, and perform a range of analyses on molecular data gathered in experimental or other populations.
Availability
The GGT package runs under the Microsoft Windows operating system and is freely available from the Web site of the Laboratory of Plant Breeding of Wageningen University and Research Centre in the Netherlands. The installer package includes several example data files and a user manual. URL: http://www.plantbreeding.wur.nl and http://www.pbr.wur.nl.
Funding
Centre for Biosystems Genomics (to R.v.B).
Dr. Arnold Kraakman and Dr Rients Niks are acknowledged for providing the data used in the presented case study. Keygene NV supported publication of this paper in the final stages.
References
Author notes
Corresponding Editor: Leif Andersson