-
PDF
- Split View
-
Views
-
Cite
Cite
Quhao Wei, Qingfeng Hu, Shanshan Li, Huoyang Lu, Guoqiang Chen, Beiqiong Shen, Ping Zhang, Yonglie Zhou, A novel functional class 2 integron in clinical Proteus mirabilis isolates, Journal of Antimicrobial Chemotherapy, Volume 69, Issue 4, April 2014, Pages 973–976, https://doi.org/10.1093/jac/dkt456
Close - Share Icon Share
Abstract
To describe a novel functional class 2 integron that was found in clinical Proteus mirabilis isolates.
Class 1 and 2 integrons were screened by PCR in 153 clinical Proteus isolates. The variable regions of class 1 and 2 integrons were determined by restriction analysis and sequencing. The mutations of internal stop codons in class 2 integrons and their common promoters were also determined by sequencing. Enterobacterial repetitive intergenic consensus (ERIC)–PCR was used to analyse the phylogenetic relations of class 2 integron-positive P. mirabilis isolates.
Class 1 integrons were detected in 96 (63%) of 153 Proteus isolates: eight different gene cassette arrays were detected, including dfrA32-ereA1-aadA2, which was detected for the first time in P. mirabilis. Class 2 integrons were detected in 101 (66%) of 153 Proteus isolates: four different gene cassette arrays were detected, including dfrA1-catB2-sat2-aadA1, which was detected for the first time in a class 2 integron. A novel functional class 2 integron was detected in 38 P. mirabilis isolates with a common promoter (−35 TTTAAT|16 bp|−10 TAAAGT). The variable region of this functional class 2 integron contained dfrA14 and three novel open reading frames with unknown functions. Very similar ERIC–PCR fingerprinting patterns were detected in these 38 P. mirabilis isolates and were different from other class 2 integron-positive isolates.
A novel functional class 2 integron was found for the first time in P. mirabilis. These functional class 2 integron-harbouring P. mirabilis isolates were likely to be clonally spread in our hospital.
Introduction
Integrons, as a natural cloning and expression system, can capture exogenous gene cassettes by site-specific recombination and ensure the expression of the genes within them, and therefore play important roles in the acquisition and lateral transfer of antibiotic resistance genes.1–5 Integrons are usually classified according to the primary structure of their integrases.5 Class 1 integrons are the dominant type detected in clinical isolates and are most correlated with antibiotic resistance; therefore they have been comprehensively studied.6 Class 2 integrons are the second major type of integrons obtained from clinical isolates. Usually, class 2 integrons are embedded in the non-replicative transposon Tn7 and its relatives. Most class 2 integron integrase (IntI2) genes present an internal stop codon, TAA, that results in the production of a truncated integrase, IntI2*, which cannot catalyse gene cassette integration and excision.7,8 In contrast to class 1 integrons, class 2 integrons have gene cassette arrays that are usually conserved because of inactive integrase.
Hansson et al.7 changed the internal stop codon TAA to GAG to encode glutamic acid experimentally and restore activity to IntI2*. The active IntI2*179E can efficiently excise cassettes between two attCs (gene cassette recombination sites) and integrate the cassettes by attI2 (class 2 integron recombination site) and attC recombination. Until now, the natural functional class 2 integron from clinical isolates has been reported only by Márquez et al.9 They found a class 2 integron with functional IntI2, in which the internal stop codon TAA mutated to glutamine codon CAA, from a pathogenic Escherichia coli strain. In this study, we describe a novel functional class 2 integron that was found in clinical Proteus mirabilis isolates.
Materials and methods
Bacterial strains
One hundred and fifty-three Proteus strains were isolated from clinical specimens of patients in the Zhejiang Provincial People's Hospital, a tertiary hospital with 2200 beds, during 2011 and 2012. These clinical isolates included 140 P. mirabilis isolates, 12 Proteus vulgaris isolates and 1 Proteus penneri isolate.
Bacterial DNA preparation and integron analysis
Total genomic DNA was isolated from stationary-phase broth cultures that were grown overnight in Luria–Bertani broth (Oxoid, UK) with the EZ-10 Spin Column Bacterial Genomic DNA MiniPreps Kit (Bio Basic Inc., Canada) according to the manufacturer's instructions. Primers used in this study are all listed in Table S1 (available as Supplementary data at JAC Online). rTaq DNA polymerase (TaKaRa, Japan) was used in PCRs to screen class 1 and class 2 integrons. LA Taq DNA polymerase (TaKaRa, Japan) was used in PCRs to amplify the variable regions of class 1 and class 2 integrons.
With respect to variable regions of class 1 and class 2 integrons, same-sized amplicons were compared by restriction analysis with HinfI (TaKaRa, Japan) and Cfr13I (Fermentas, Canada) respectively. Primer walking was used to sequence at least one representative of each type of amplicon, as described previously.10 Sequence analysis was conducted using Vector NTI Advance 10 (Invitrogen, USA).
Determination of the internal stop codon mutation in the intI2 gene
In intI2 gene-positive strains, primer intI2F combined with dfrA1R or dfrA14R (Table S1, available as Supplementary data at JAC Online) was used to amplify the intI2 gene together with attI2 and the common promoter Pc (Figure S1, available as Supplementary data at JAC Online), which was located in the attI2 site and can drive the transcription of the downstream gene cassette in the variable region of class 2 integrons. PCR products were sequenced directly, and the mutations of the internal stop codon in the intI2 genes as well as the types of common promoters of class 2 integrons were determined manually according to their sequences.
Typing of intI2-positive P. mirabilis by enterobacterial repetitive intergenic consensus (ERIC)–PCR
The phylogenetic relations of intI2 gene-positive P. mirabilis were analysed using ERIC–PCR with primer ERIC2 (Table S1, available as Supplementary data at JAC Online) as described previously.11 The following conditions of amplification were used: denaturation for 4 min at 94°C; 40 cycles of 40 s at 94°C, 1 min at 40°C and 5 min at 72°C; and a final extension step of 10 min at 72°C. Products were separated by electrophoresis in 1% agarose containing ethidium bromide (0.5 μg/mL) in 1× Tris/acetate/EDTA buffer for 1 h at 80 V. The generated fingerprints were compared visually.
GenBank accession numbers
The partial sequences of class 2 integrons with the dfrA1-catB2-sat2-aadA1 and dfrA1-sat2 gene cassette arrays, the functional class 2 integron and the class 1 integron with the dfrA32-ereA1-aadA2 gene cassette array from this study were deposited in GenBank with accession numbers JX867126–JX867129.
Results and discussion
Class 1 integrons and their gene cassettes among Proteus isolates
Class 1 integrons were detected in 96 (63%) of 153 Proteus isolates. In these 96 class 1 integron-positive strains, variable regions were successfully amplified in 70 isolates. Eight different gene cassette arrays were detected (Table 1). The most prevalent gene cassette arrays were aadB-aadA2 and dfrA17-aadA5, which were detected in 37 and 17 isolates, respectively. dfrA32-ereA1-aadA2, which was only previously reported in Laribacter hongkongensis (GenBank accession number GU726907) and Salmonella enterica (GenBank accession number GU067642), was also detected in four P. mirabilis isolates in this study.12,13 To the best of our knowledge, this is the first report of the dfrA32 gene cassette in clinical P. mirabilis.
Class 1 and class 2 integrons in 153 clinical Proteus isolates
| Species . | Number of isolates . | intI1 . | Variable region of class 1 integron . | intI2 . | Variable region of class 2 integron . | ERIC type (number) . |
|---|---|---|---|---|---|---|
| P. mirabilis (n = 140) | 35 | − | ND | − | ND | ND |
| 2 | + | —a | − | ND | ND | |
| 3 | + | dfrA12-orfF-aadA2 | − | ND | ND | |
| 1 | + | dfrA32-ereA1-aadA2 | − | ND | ND | |
| 2 | + | dfrA1-orfC | − | ND | ND | |
| 1 | + | aadA2 | − | ND | ND | |
| 11 | − | ND | + | dfrA1-sat2-aadA1 | C (8), D (2), E (1) | |
| 1 | + | —a | + | dfrA1-catB2-sat2-aadA1 | C (1) | |
| 37 | + | aadB-aadA2 | + | dfrA14-(orfP-orfQ-orfR)b | A (37) | |
| 1 | + | aadA2 | + | dfrA14-(orfP-orfQ-orfR)b | A (1) | |
| 23 | + | —a | + | dfrA1-sat2-aadA1 | B (21), C (2) | |
| 16 | + | dfrA17-aadA5 | + | dfrA1-sat2-aadA1 | C (16) | |
| 3 | + | dfrA32-ereA1-aadA2 | + | dfrA1-sat2-aadA1 | B (1), C (2) | |
| 2 | + | dfrA12-orfF-aadA2 | + | dfrA1-sat2-aadA1 | B (1), C (1) | |
| 1 | + | aacA4-cmlA1 | + | dfrA1-sat2-aadA1 | C (1) | |
| 1 | + | aadB | + | dfrA1-sat2-aadA1 | C (1) | |
| P. vulgaris (n = 12) | 6 | − | ND | − | ND | ND |
| 1 | + | dfrA12-orfF-aadA2 | − | ND | ND | |
| 2 | − | ND | + | dfrA1-sat2-aadA1 | ND | |
| 1 | − | ND | + | dfrA1-sat2 | ND | |
| 1 | − | ND | + | dfrA1-catB2-sat2-aadA1 | ND | |
| 1 | + | dfrA17-aadA5 | + | dfrA1-sat2-aadA1 | ND | |
| P. penneri (n = 1) | 1 | − | ND | − | ND | ND |
| Species . | Number of isolates . | intI1 . | Variable region of class 1 integron . | intI2 . | Variable region of class 2 integron . | ERIC type (number) . |
|---|---|---|---|---|---|---|
| P. mirabilis (n = 140) | 35 | − | ND | − | ND | ND |
| 2 | + | —a | − | ND | ND | |
| 3 | + | dfrA12-orfF-aadA2 | − | ND | ND | |
| 1 | + | dfrA32-ereA1-aadA2 | − | ND | ND | |
| 2 | + | dfrA1-orfC | − | ND | ND | |
| 1 | + | aadA2 | − | ND | ND | |
| 11 | − | ND | + | dfrA1-sat2-aadA1 | C (8), D (2), E (1) | |
| 1 | + | —a | + | dfrA1-catB2-sat2-aadA1 | C (1) | |
| 37 | + | aadB-aadA2 | + | dfrA14-(orfP-orfQ-orfR)b | A (37) | |
| 1 | + | aadA2 | + | dfrA14-(orfP-orfQ-orfR)b | A (1) | |
| 23 | + | —a | + | dfrA1-sat2-aadA1 | B (21), C (2) | |
| 16 | + | dfrA17-aadA5 | + | dfrA1-sat2-aadA1 | C (16) | |
| 3 | + | dfrA32-ereA1-aadA2 | + | dfrA1-sat2-aadA1 | B (1), C (2) | |
| 2 | + | dfrA12-orfF-aadA2 | + | dfrA1-sat2-aadA1 | B (1), C (1) | |
| 1 | + | aacA4-cmlA1 | + | dfrA1-sat2-aadA1 | C (1) | |
| 1 | + | aadB | + | dfrA1-sat2-aadA1 | C (1) | |
| P. vulgaris (n = 12) | 6 | − | ND | − | ND | ND |
| 1 | + | dfrA12-orfF-aadA2 | − | ND | ND | |
| 2 | − | ND | + | dfrA1-sat2-aadA1 | ND | |
| 1 | − | ND | + | dfrA1-sat2 | ND | |
| 1 | − | ND | + | dfrA1-catB2-sat2-aadA1 | ND | |
| 1 | + | dfrA17-aadA5 | + | dfrA1-sat2-aadA1 | ND | |
| P. penneri (n = 1) | 1 | − | ND | − | ND | ND |
ND, not detected.
aClass 1 integrons for which PCR failed to amplify the gene cassette array.
borfP, orfQ and orfR were three ORFs; whether they were gene cassettes and their functions remain to be determined.
Class 1 and class 2 integrons in 153 clinical Proteus isolates
| Species . | Number of isolates . | intI1 . | Variable region of class 1 integron . | intI2 . | Variable region of class 2 integron . | ERIC type (number) . |
|---|---|---|---|---|---|---|
| P. mirabilis (n = 140) | 35 | − | ND | − | ND | ND |
| 2 | + | —a | − | ND | ND | |
| 3 | + | dfrA12-orfF-aadA2 | − | ND | ND | |
| 1 | + | dfrA32-ereA1-aadA2 | − | ND | ND | |
| 2 | + | dfrA1-orfC | − | ND | ND | |
| 1 | + | aadA2 | − | ND | ND | |
| 11 | − | ND | + | dfrA1-sat2-aadA1 | C (8), D (2), E (1) | |
| 1 | + | —a | + | dfrA1-catB2-sat2-aadA1 | C (1) | |
| 37 | + | aadB-aadA2 | + | dfrA14-(orfP-orfQ-orfR)b | A (37) | |
| 1 | + | aadA2 | + | dfrA14-(orfP-orfQ-orfR)b | A (1) | |
| 23 | + | —a | + | dfrA1-sat2-aadA1 | B (21), C (2) | |
| 16 | + | dfrA17-aadA5 | + | dfrA1-sat2-aadA1 | C (16) | |
| 3 | + | dfrA32-ereA1-aadA2 | + | dfrA1-sat2-aadA1 | B (1), C (2) | |
| 2 | + | dfrA12-orfF-aadA2 | + | dfrA1-sat2-aadA1 | B (1), C (1) | |
| 1 | + | aacA4-cmlA1 | + | dfrA1-sat2-aadA1 | C (1) | |
| 1 | + | aadB | + | dfrA1-sat2-aadA1 | C (1) | |
| P. vulgaris (n = 12) | 6 | − | ND | − | ND | ND |
| 1 | + | dfrA12-orfF-aadA2 | − | ND | ND | |
| 2 | − | ND | + | dfrA1-sat2-aadA1 | ND | |
| 1 | − | ND | + | dfrA1-sat2 | ND | |
| 1 | − | ND | + | dfrA1-catB2-sat2-aadA1 | ND | |
| 1 | + | dfrA17-aadA5 | + | dfrA1-sat2-aadA1 | ND | |
| P. penneri (n = 1) | 1 | − | ND | − | ND | ND |
| Species . | Number of isolates . | intI1 . | Variable region of class 1 integron . | intI2 . | Variable region of class 2 integron . | ERIC type (number) . |
|---|---|---|---|---|---|---|
| P. mirabilis (n = 140) | 35 | − | ND | − | ND | ND |
| 2 | + | —a | − | ND | ND | |
| 3 | + | dfrA12-orfF-aadA2 | − | ND | ND | |
| 1 | + | dfrA32-ereA1-aadA2 | − | ND | ND | |
| 2 | + | dfrA1-orfC | − | ND | ND | |
| 1 | + | aadA2 | − | ND | ND | |
| 11 | − | ND | + | dfrA1-sat2-aadA1 | C (8), D (2), E (1) | |
| 1 | + | —a | + | dfrA1-catB2-sat2-aadA1 | C (1) | |
| 37 | + | aadB-aadA2 | + | dfrA14-(orfP-orfQ-orfR)b | A (37) | |
| 1 | + | aadA2 | + | dfrA14-(orfP-orfQ-orfR)b | A (1) | |
| 23 | + | —a | + | dfrA1-sat2-aadA1 | B (21), C (2) | |
| 16 | + | dfrA17-aadA5 | + | dfrA1-sat2-aadA1 | C (16) | |
| 3 | + | dfrA32-ereA1-aadA2 | + | dfrA1-sat2-aadA1 | B (1), C (2) | |
| 2 | + | dfrA12-orfF-aadA2 | + | dfrA1-sat2-aadA1 | B (1), C (1) | |
| 1 | + | aacA4-cmlA1 | + | dfrA1-sat2-aadA1 | C (1) | |
| 1 | + | aadB | + | dfrA1-sat2-aadA1 | C (1) | |
| P. vulgaris (n = 12) | 6 | − | ND | − | ND | ND |
| 1 | + | dfrA12-orfF-aadA2 | − | ND | ND | |
| 2 | − | ND | + | dfrA1-sat2-aadA1 | ND | |
| 1 | − | ND | + | dfrA1-sat2 | ND | |
| 1 | − | ND | + | dfrA1-catB2-sat2-aadA1 | ND | |
| 1 | + | dfrA17-aadA5 | + | dfrA1-sat2-aadA1 | ND | |
| P. penneri (n = 1) | 1 | − | ND | − | ND | ND |
ND, not detected.
aClass 1 integrons for which PCR failed to amplify the gene cassette array.
borfP, orfQ and orfR were three ORFs; whether they were gene cassettes and their functions remain to be determined.
Class 2 integrons and their gene cassettes among Proteus isolates
Class 2 integrons were detected in 101 (66%) of 153 Proteus iso-lates. Of them, 86 isolates were also positive for class 1 integrons. Variable regions of class 2 integrons were successfully amplified in all 101 isolates. Four different gene cassette arrays were detected (Table 1 and Figure S1, available as Supplementary data at JAC Online). The most prevalent of these gene cassette arrays was dfrA1-sat2-aadA1, which was detected in 60 isolates. dfrA1-catB2-sat2-aadA1 was detected in two isolates and dfrA1-sat2 was detected in one isolate. dfrA1-catB2-sat2-aadA1 is, to the best of our knowledge, reported in a class 2 integron for the first time in this study.
In the other 38 isolates, a novel variable region of class 2 integrons was obtained. The first gene cassette was dfrA14, which was followed by three novel open reading frames (ORFs) (Figure S1D, available as Supplementary data at JAC Online). These three novel ORFs were temporarily named orfP, orfQ and orfR in this study. orfP and orfQ translated in the same orientation as that of dfrA14, while orfR was translated in the reverse orientation and overlapped with orfQ (Figure S1D and Figure S2, available as Supplementary data at JAC Online). Sequence comparisons were carried out using the Basic Local Alignment Search Tool (BLAST) program (http://blast.ncbi.nlm.nih.gov/Blast.cgi); however, no matching sequence was retrieved with sequences of these three ORFs separately, indicating new exogenous sequences, which makes it difficult to identify the functions of these ORFs. Sequence analysis indicated that orfP and orfQ had a structure that was similar to the gene cassette, and the putative core sites are marked with dashed underlining in Figure S2, available as Supplementary data at JAC Online. However, IntI2- or IntI1-catalysed site-specific recombination should be conducted to identify whether orfP or orfQ was embedded as a gene cassette along with their attC sites.
Functional class 2 integrons in Proteus isolates
The mutations of the internal stop codon in the intI2 genes as well as the types of common promoters of class 2 integrons were also determined by sequences analysis. Results indicated that in the 38 class 2 integrons containing three novel ORFs, the internal stop codons in the intI2 genes were all mutated from TAA to the glutamine codon CAA (Figure S2, available as Supplementary data at JAC Online, marked with double underlining), implying that functional intI2 genes were present. The common promoters of these types of class 2 integrons were −35 TTTAAT|16 bp|−10 TAAAGT (Figure S2, available as Supplementary data at JAC Online). The first gene cassette of the functional class 2 integron that was detected in this study was also dfrA14, which was the same as that in the previously reported functional class 2 integron.9 Of the remaining 63 class 2 integrons, all possessed the internal stop codon, which implied that these were non-functional intI2 genes, and the common promoters of these class 2 integrons were −35 TTTAAT|16 bp|−10 TAAAAT, which was the same as that described previously.14
Clonal spread of functional class 2 integron-harbouring P. mirabilis
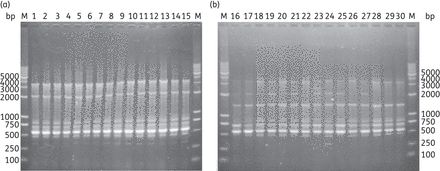
ERIC–PCR was used to type the phylogenetic relations of 96 class 2 integron-positive P. mirabilis isolates, and five ERIC types were obtained according to the electrophoresis patterns (Figure S3, available as Supplementary data at JAC Online). As shown in Table 1 and Figure 1, the 38 functional class 2 integron-positive P. mirabilis isolates all belonged to ERIC type A, and they were different from other class 2 integron-positive P. mirabilis isolates. All of these 38 functional class 2 integron-positive P. mirabilis isolates were also positive for class 1 integrons with the aadB-aadA2 gene cassette arrays, except one isolate, in which only aadA2 was detected in the variable region of the class 1 integron. These results indicated the clonal spread of functional class 2 integron-harbouring P. mirabilis in our hospital.
ERIC–PCR fingerprinting patterns of the functional class 2 integron-positive P. mirabilis isolates (a) and other class 2 integron-positive P. mirabilis isolates (b). Lane M, molecular size marker. Lanes 1–30, class 2 integron-positive isolates.
Conclusions
In conclusion, a novel functional class 2 integron was found, to the best of our knowledge, for the first time in P. mirabilis strains. This novel functional class 2 integron contained three novel ORFs with functions that remain to be determined. These functional class 2 integron-harbouring P. mirabilis were likely to be the result of clonal spread in our hospital.
Funding
This work was supported by Zhejiang Provincial Natural Science Foundation of China under Grant No. Y2110743 and by the National Natural Science Foundation of China under Grant No. 81101291.
Transparency declarations
None to declare.
References