-
PDF
- Split View
-
Views
-
Cite
Cite
Duncan M. Baird, Bethan Britt-Compton, Jan Rowson, Nazar N. Amso, Linda Gregory, David Kipling, Telomere instability in the male germline, Human Molecular Genetics, Volume 15, Issue 1, 1 January 2006, Pages 45–51, https://doi.org/10.1093/hmg/ddi424
Close - Share Icon Share
Abstract
Telomeres play a key role in upholding the integrity of the genome, and telomerase expression in spermatogonial stem cells is responsible for the maintenance of telomere length in the human male germline. We have previously described extensive allelic variation in somatic cell telomere length that is set in the zygote, the ultimate source of which may be the germline. This implies that despite telomerase activity, substantial telomere length variation can be generated and tolerated in the germline; in order to investigate this further, we have examined the nature of telomere length variation in the human male germline. Here, we describe an analysis of both genome-wide telomere length and single molecule analysis of specific chromosome ends in human sperm. We observed individual specific differences in genome-wide telomere length. This variation may result from genetic differences within the components that determine the telomere length setting of each individual. Superimposed on the genome wide telomere length setting was a stochastic component of variation that generates germ-cells containing severely truncated telomeres. If not re-lengthened during early embryogenesis, such telomeres may limit the replicative capacity of cells derived from the zygote and have the potential to create fusagenic chromosomes, unbalanced translocations and terminal micro-deletions. These data may have implications for the genetic determination of ageing, genetic disease and fertility.
INTRODUCTION
Replication of linear chromosomes results in the loss of terminal sequences with each cell division (1). These end-replication losses are compensated for by the enzyme telomerase, which synthesises telomere repeats de novo (2). Expressed in stem cell compartments, telomerase is stringently repressed in the majority of human somatic tissues (3). With ongoing cell division, telomere erosion will ultimately result in the loss of telomeric function and the triggering of the double-stranded break repair apparatus, leading to a p53-dependent growth arrest termed replicative senescence (4). This occurs after a specific number of cell divisions (5) that is dependent upon the telomere length of the starting cell population (6). By controlling the replicative capacity of human cells, this phenomenon has the potential to act as a potent tumour suppressive mechanism. In the absence of a functional DNA damage response, telomere erosion and the subsequent uncapping of the telomere can result in fusogenic chromosome ends. The resulting cycles of anaphase bridge-breakage and fusion generate chromosomal rearrangements that can drive early-stage neoplasia (7) or underlie genetic disease (8), these would include unbalanced translocations and terminal deletions.
We have previously described extensive allelic variation in the length of the XpYp telomere in somatic cells, hypothesizing that this is established in the zygote (9). This implies that the ultimate source of this variation is likely to be found within the germline, with differing maternal and paternal telomeric contributions to the zygote. This situation appears inconsistent with the telomerase activity that has been detected in oocytes (10) and at high levels in the testis (3,11–13). It is presumed that this activity is sufficient to maintain telomeres in the germline at their maximum length for subsequent generations. To address this issue, we used high-resolution telomere length analysis to investigate the nature of telomere dynamics in the human male germline. We show that genome-wide telomere length setting is variable, with mean telomere lengths ranging from <9 to over 17 kb. This variation appears to be sufficient to account for the allelic telomere length differences that have been observed in somatic cells (9). Telomere analysis at the single molecule level revealed that in addition to a genome-wide telomere length setting, telomeres are subjected to addition mutational mechanisms that generate sperm cells containing severely truncated telomeres. Such telomeres were often shorter than the telomeres observed in senescent cells and thus have the potential to limit the replicative capacity of cells derived from the zygote. Furthermore, such truncated telomeres may contribute to genomic instability and genetic disease via telomere uncapping resulting in chromosomal fusion or by aberrant meiotic synapsis.
RESULTS
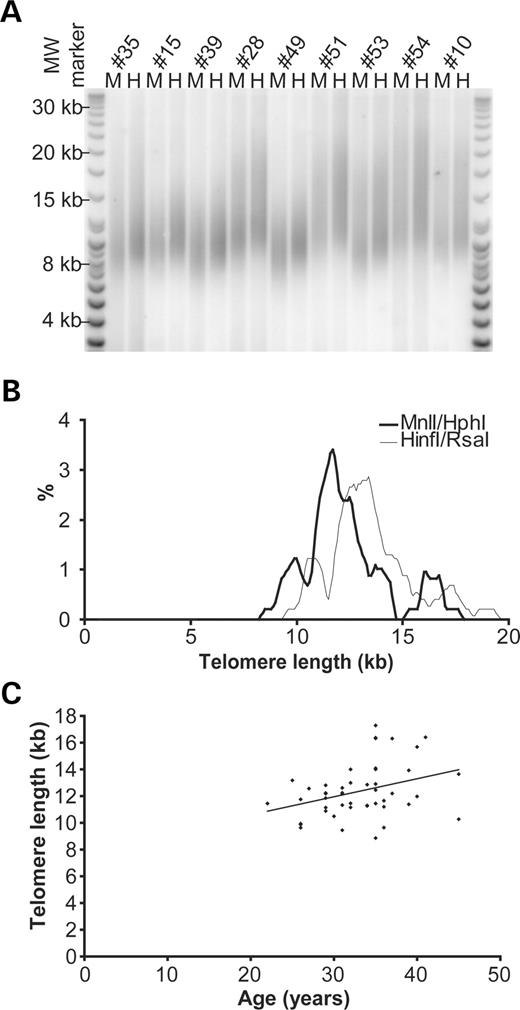
Genome-wide terminal restriction fragment (TRF) length was estimated by field inversion gel electrophoresis (FIGE) of DNA obtained from a panel of 54 human semen samples (Fig. 1A). Each sample was analysed following two separate restriction digests with HinfI/RsaI (typically used for telomere length analysis) and HphI/MnlI (Fig. 1A). Unlike HinfI/RsaI, HphI and MnlI cut DNA at the telomere repeat variants TGAGGG and TCAGGG, respectively, that are present in the proximal regions of human telomeres, but do not cut within the canonical telomere repeat unit TTAGGG. Thus, we were able to obtain an accurate estimation of the length of TRF containing predominantly TTAGGG repeats without the confounding influence of subtelomeric sequence variation within HinfI/RsaI generated TRFs (14). Telomere length within the population was variable and the distribution appeared to be trimodal, with a group of individuals exhibiting short homogeneous telomere lengths (<9 kb) with others displaying long heterogeneous telomeres (>16 kb) (Fig. 1B). The mean telomere length of the population was 12.48±2.00 kb (SD) with MnlI/RsaI and 13.57±1.99 kb (SD) with HinfI/RsaI. There was a weak correlation (r=0.34, P<0.02) between age and telomere length such that telomere length may increase with age at a rate of 135 bp/year (Fig. 1C). This data is consistent with previous observations (15) and may be sufficient to account for the correlation between paternal age and telomere length of offspring (16). However, the weak correlation with age appeared insufficient to fully account for the observed telomere length variation; as for example, the nine individuals aged 35 displayed the full range of telomere sizes (8.86–17.29 kb, Fig. 1C). The differences in length between the two digests of each individual revealed that the subtelomeric sequences and telomere repeat variants of HinfI/RsaI generated TRFs are also variable in length and comprise around 1.09±0.48 kb (mean±SD). These data indicate that the individual specific differences in TRF length are a consequence of two independently variable components; subtelomeric sequences and TTAGGG repeat content.
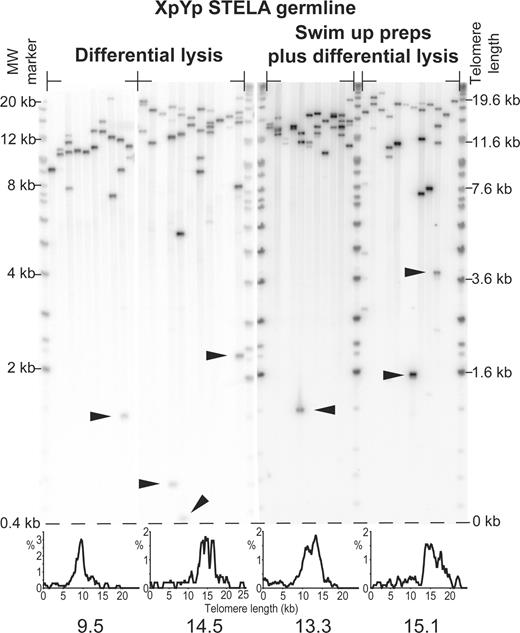
Because TRF analysis is limited in resolution and biased towards the detection of long telomeres, we undertook single telomere length analysis (STELA) at the XpYp telomere on a subset of the DNAs used for TRF analysis (n=10). STELA allows telomere lengths to be determined from single DNA molecules and can detect short telomeres that cannot be detected by Southern blot and other hybridization-based approaches. All the samples were analysed using the XpYpE2 primer (9) and a subset was analysed with two additional primers XpYpM and XpYpC (1666 and 887 bp from the telomere, respectively); all primers revealed identical XpYp telomere length distributions (shifted by the expected amounts), indicating that these distributions were representative of a single telomere. Telomere length estimates from the TRF analysis correlated strongly with the STELA data (r=0.91, P<0.001), suggesting that XpYp telomere length reflects genome-wide telomere length. Again, as with TRF analysis, STELA revealed a high level of inter-individual telomere length variation. XpYp lengths ranged from 9.5 kb in individuals exhibiting short TRF lengths, to 19.3 kb in individuals with long heterogeneous TRFs (Table 1). Most surprising, however, was that despite telomerase activity maintaining genome-wide telomere length, a significant proportion of severely truncated telomeric molecules were detected (Fig. 2). These molecules occurred with mean frequency of 3.5% greater than 2.33 SDs below the mean of the distributions when only 1% would be expected by chance (one-tailed level of significance of 0.01, critical z=2.3263); the overall frequency of these molecules displayed a significant departure from the numbers of such molecules expected based upon a normal distribution (χ2 test P<0.0001, Table 1). It was striking that the truncated XpYp telomeres were as short as those observed in senescent fibroblast populations (9) and thus may not be capable of forming a functional telomeric structure. If not re-lengthened during early embryogenesis, such telomeres have the potential to severely limit the replicative potential of cells derived from a zygote that received such shortened telomeres. Furthermore, critically eroded telomeres are fusagenic and thus have the potential to initiate genomic instability in the zygote.
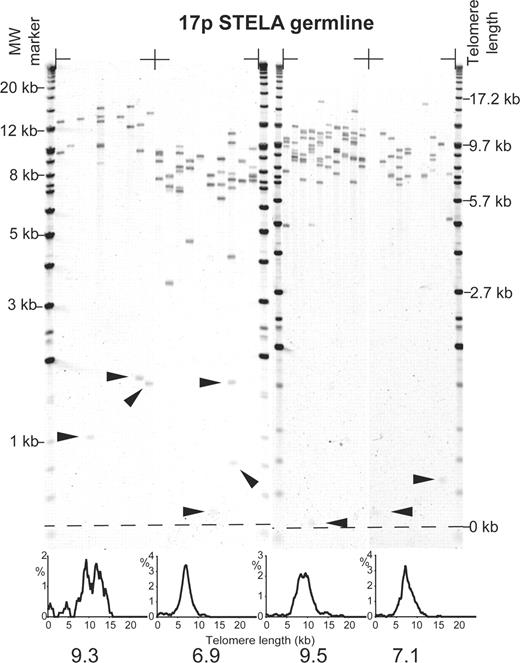
Although telomere lengths at the XpYp telomere correlate with genome-wide telomere lengths, the possibility remained that the unexpected dynamics described here may be specific to the XpYp telomere. We, therefore, investigated the telomere dynamics of an autosomal telomere. We developed an assay based on the published sequence of the end of 17p (17), a chromosome implicated in genomic rearrangements during early-stage neoplasia (18). Two different primers were employed for 17p STELA; 17p6 and 17pseq1rev1 (3078 and 311 bp from the telomere, respectively) both revealed identical telomere length distributions shifted by the expected amounts. The germline 17p telomere length distributions were determined from four individuals, which revealed that the 17p telomere was also variable in length (Fig. 3, Table 1). As with the XpYp telomere, we observed that the mean telomere length at 17p was superimposed by stochastically shortened telomeres, with 4.0% greater than 2.33 SDs below the mean of the distributions (Fig. 3, Table 1). Consistent with previous work in cultured lymphocytes (19) and our own observations in fibroblasts strains (unpublished data), in all the individuals analysed, the 17p telomere was shorter than the XpYp by a mean of 4.3 kb (Table 1). Furthermore, the telomere-length distributions within each individual was significantly less heterogeneous at 17p compared with that of XpYp (P<0.001, Table 1). These data indicate that cis-acting determinants may be operating at this telomere to create a telomere length differential and to modulate stability.
We performed a series of experiments to address whether the outlying short telomeres were the result of somatic contamination of germline DNA or shearing during DNA preparation. Somatic cells contain telomeres of the same size range as the short outlying telomeres detected in the male germline. The genomic DNA preparations analysed by STELA employed differential lysis steps to remove somatic contamination (20). However, despite these steps, it was possible that some degree of somatic cell-derived DNA could contaminate the germline DNA preparations. Thus, we separated highly motile spermatozoa by sperm swim-up preparations (21) and extracted DNA using the same differential lysis procedure. These highly pure male germline DNA preparations were subjected to STELA and no differences in the telomere length spectrums were observed compared with DNA prepared by differential lysis alone (Fig. 2). Although the shearing of DNA during preparation could create shortened telomeric molecules, such molecules would not be predicted to be ligatable without further modification and thus would not be detected by STELA. To confirm this, we sheared semen-derived high-molecular weight DNA by passing it multiple times through a 0.5 mm diameter hypodermic needle; again, no differences were observed in the telomere length spectrums (data not shown). As an additional negative control for sheared DNA, we designed an oligonucleotide linker with six bases of homology to the sequence of the tandem repeats that constitute the minisatellite locus D1S8 (MS32) (20). This linker contained the same GC content and 5′ Teltail sequence as the telorette linkers used in STELA (9). Ligation of these linkers to germline DNA, followed by polymerase chain reaction (PCR) amplification with Teltail and MS32 adjacent primers, yielded no detectable products upon Southern hybridization with a probe containing the MS32 adjacent sequence (data not shown). As expected, amplification products were detected between primers situated in the MS32 adjacent DNA. Taken together, these data indicate the ligation mediated PCR used in STELA does not detect the presence of sheared DNA molecules. We, therefore, consider the telomere length distributions obtained by STELA to be representative of those in the human male germline.
DISCUSSION
Here, we have demonstrated that in the human germline, telomeres are highly dynamic structures. The variation in XpYp telomere length observed between fibroblast strains from unrelated donors (up to 6 kb) (9) is within the range of variation of the genome-wide telomere length setting revealed by the TRF analysis described here. This variation appears to mirror that observed between strains and cultivars of different species including mice (22,23) and Caenorhabditis elegans (24). A number of proteins have been identified that influence steady-state telomere length setting in human cells (25) and there is a strong genetic component to human telomere length determination (26,27) with putative determinants of telomere length mapped to a locus on human ch. 12 (28), the X-chromosome (29) and murine ch. 2 (30). Furthermore, mutations within the RNA component of the telomerase complex result in the autosomal dominant form of dyskeratosis congenita (31). The resulting haploinsufficiency for telomerase causes a progressive shortening of telomeres between generations and disease anticipation, whereby the severity of the condition increases and the age of onset decreases (32). These data are consistent with the concept that variants within the genes that determine genome-wide telomere length may underlie the variation in germline telomere length described here. Furthermore, as telomerase activity is considered to maintain telomere length in germ-cell lineages derived from the zygote (11,33), these data may reflect zygotic telomere length of the individuals analysed, which may in turn influence the heritability of longevity.
The extreme variation observed by STELA in the germline, in the form of severely truncated telomeres, is greater than that observed in somatic cells where the allelic variation is up to 6 kb (9). Two interpretations of these observations are possible. First, the shortened telomeres might be re-lengthened during embryogenesis (but not sufficiently to homogenize allelic telomere length). There is a precedent for this because nuclear transfer appears to reset telomere length in cows (34), although this was not observed in sheep or goats (35,36). Alternatively, zygotes that receive such truncated telomeres might not proceed to full-term, as a consequence of the cellular responses to critically shortened telomeres or genomic instability.
The nature of the mutational mechanisms that underlie the telomeric instability we describe here are as yet unclear. The observed heterogeneity is not specific to the germline, but is observed in all telomerase-positive cells analysed to date (DMB unpublished observations). The influence, if any, of inter-allelic meiotic recombination is, therefore, likely to be small. This is consistent with our previous data that revealed high levels of linkage disequilibrium at human telomeres, indicating that telomeres are recombinationally suppressed (37,38). An analogous process termed telomere-rapid deletion (TRD) has been described in the meiotic yeast cells (39). TRD is dependent upon the MRX complex (40), and it has been postulated to result from the processing of Holliday junctions formed at the base of T-loops (41,42). The mechanistic basis of the large-scale telomere truncation events described here may be similar to that proposed for TRD, with the caveat that the mechanism in yeast resets previously lengthened telomeres to the genome average (40); this is not the case with the events described here. Other intra-allelic mutational mechanisms that could result in the mutation profile we observed include, telomeric sister chromatid exchange, which occurs with an estimated frequency 20 times that of the genome average (43), and oxidative damage (44).
We have observed telomere truncation events at two human telomeres with a mean frequency of 3.6%. Assuming that these events occur with a similar frequency at all telomeres, 96.4% of telomeres at any one chromosome end would be within the normal range, thus we predict that only 19% of germ-cells (0.96446×100) will contain a full complement of chromosomes with telomeres of the genome-wide length. Telomere truncation can limit replicative potential, and the subsequent loss of end capping can result in genomic instability (7,45). This raises the possibility that zygotic telomere dysfunction maybe a significant contributor to the estimated 70% of conceptions that are lost prior to live birth (46) and the 50% of spontaneous abortions that have detectable chromosomal abnormalities (47). Consistent with this, cryptic terminal rearrangements including those involving Yp have been observed in couples that suffer multiple miscarriages (48). Furthermore, chromosomes with shortened telomeres suffer from aberrant meiotic synapsis (49), and telomere truncation in the male germline may, therefore, contribute towards the observed levels of aneuploidy (up to 1.55%) in human sperm (50). Together, these phenomena may explain the relatively low fecundity of humans (51). Whatever the causal mechanisms that underlie these events, it seems likely that they have the potential to impact upon fertility, genomic instability and genetic disease.
MATERIALS AND METHODS
Sample preparation and DNA extraction
Semen samples were obtained from men undertaking routine semen analysis according to WHO guidelines (52) and the study was approved by the Local Research Ethics Committee. DNA was prepared from samples displaying a normal semen analysis using a differential lysis procedure as described previously (20). Prior to DNA preparation by differential lysis, a subset of semen samples were subjected to the sperm swim-up procedure as described previously (21). Semen was mixed with culture medium and centrifuged for 10 min at 250g. The supernatant was discarded and the pellet resuspended in a further 5 ml of culture medium, the centrifugation process repeated and the supernatant discarded. One millilitre of culture medium was layered onto the sperm pellet and the tube incubated at 37°C for 30 min to allow the active sperm to swim-up from the pellet into the medium. After incubation, the top layer of medium was removed with a sterile pipette and transferred to a sterile tube. This isolates the motile, viable sperm for telomere analysis, after which DNA was extracted using the differential lysis procedure (20).
TRF analysis
DNA was digested with HinfI/RsaI or MnlI/HphI according to the manufacturer's instructions (New England Biolabs, MA, USA) and 1 µg size fractionated by FIGE. Telomeric fragments were detected by Southern hybridization using a telomere repeat containing probe, and mean TRF length was determined as described previously (1). As a control for incomplete digests, 1 µg of the DNA was resolved by 0.7% agarose electrophoresis; discrete products were detected by Southern hybridization with a probe (29C1) that detects the minisatellite locus DXYS14 (53), indicating complete digestion.
Polymerase chain reaction
STELA was undertaken as described previously (9), with the following modifications for the analysis of the long germline telomeres: thermal cycling was as follows: 94°C for 15 s; primer annealing at 65°C for the XpYp specific primers, 59°C for the 17p6 and 17pseq1rev1 primer and 66.5°C for MS32; extension at 68°C for 12 min; for 22 cycles. Furthermore, the Taq/Pwo ratio was adjusted from 25:1 to 10:1 and typically a maximum of five amplifiable telomeric molecules were analysed in each separate PCR; thus to obtain a statistically significant sample of molecules, each DNA sample was subjected to at least 40 separate STELA reactions. Amplified products were resolved by 0.5% agarose Tris-acetate–EDTA gel electrophoresis and detected by two separate Southern hybridizations with 33P-labelled telomere-adjacent probe and a TTAGGG repeat containing probe. The XpYp telomere-adjacent probe was as described in (9), the 17p telomere-adjacent probes were generated by PCR between primers 17p6 and 17p2 or 17prev1 and 17pseq1.
As a negative control against DNA shearing, an oligonucleotide linker was synthesized to mimic the sequence content of telorette linker used for STELA. This linker, based on the repeat sequence of the minisatellite locus D1S8 (MS32), contained the same length of homology, GC content and 5′ Teltail sequence as the telorette linker. This linker was ligated to the test DNA under the same conditions as employed in STELA. PCR was undertaken (with the thermal cycling conditions described earlier) with MS32 adjacent primers MS32B and Teltail, and as a positive PCR control, the MS32 adjacent DNA was amplified with MS32B and MS32OR. Products were resolved by agarose gel electrophoreses and detected by Southern hybridization using a probe generated by PCR between MS32B and MS32OR.
PCR primers and linkers
MS32B 5′-TAAGCTCTCCATTTCCAGTTTCTGG-3′; MS32OR 5′-ACCACCCTTCCCACCAAACTACTC-3′; MS32linker 5′-TGCTCCGTGCATCTGGCATCGAGTCAC-3′; XpYpM 5′-ACCAGGTTTTCCAGTGTGTT-3′; XpYpC 5′-CAGGGACCGGGACAAATAGAC-3′; 17p2 5′-GAGTCAATGATTCCATTCCTAGC-3′; 17p6 5′-GGCTGAACTATAGCCTCTGC-3′; 17pseqrev1 5′-GAATCCACGGATTGCTTTGTGTAC-3′; 17pseq1 5′-CCTCAGCCTCTCAACCTGCTTG-3′.
ACKNOWLEDGEMENTS
This work was supported by a personal fellowship award to D.M.B. from Research into Ageing and Cancer Research, UK. D.M.B. is a Cancer Research UK Senior Cancer Research Fellow.
Conflict of Interest statement. The authors have no conflict of interest.
Figure 1. TRF analysis of human male germline. (A) Representative TRF Southern blot; each individual is analysed in duplicate, H, HinfI/RsaI; M, MnlI/HphI and the DNA resolved by FIGE. (B) Telomere length distribution, thin line represents HinfI/RsaI data mean 13.57±1.99 kb (SD), the thick line represents MnlI/HphI 12.48±2.00 kb (SD). (C) MnlI/HphI TRF estimates plotted as a function of age, with regression line showing a slope of +135 bp/year, correlation coefficient r=0.34, P<0.02, n=47.
Figure 2. XpYp STELA on male germline DNA obtained from four unrelated individuals, products detected by Southern hybridization with a (TTAGGG)n containing probe. Preparations using the differential lysis DNA isolation procedure and sperm swim-up preparations for the isolation of motile spermatozoa are indicated. Arrows show examples of telomeres that are shorter than 2.33 SDs from the mean of their respective telomere length distributions. Note that for bands that are detected by Southern hybridization with a telomere repeat containing probe, the low-molecular weight bands which contain less telomere repeats will, therefore, display a weaker hybridization signal. Histograms displaying the telomere length distributions and the modal telomere length are shown below. Molecular weight markers are shown on the left and corrected for telomere length on the right.
Figure 3. STELA at the telomere of 17p in four unrelated individuals using the 17pseq1rev1 primer, products detected by Southern hybridization with a (TTAGGG)n containing probe. Filled arrows indicate telomeres shorter than 2.33 SDs from the mean of the distribution. Histograms displaying the telomere length distributions, with the modal telomere length are shown below. Molecular weight markers are shown on the left and corrected for telomere length on the right.
Summarizing telomere length data of individuals analyzed with both TRF and STELA.
| Sample number . | TRF . | XpYp STELA . | 17p STELA . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | HinfI/RsaI . | MnlI/HphI . | Mean (±SD) . | Mode . | n . | Outliers (%) . | Mean (±SD) . | Mode . | n . | Outliers (%) . |
| 101 | 13.35 | 12.81 | 11.10 (4.47) | 13.1 | 284 | 7 (2.5) | — | — | — | — |
| 102 | 12.34 | 11.14 | 9.83 (3.28) | 11.0 | 162 | 8 (4.9) | — | — | — | — |
| 103 | 12.60 | 11.75 | 14.17 (4.54) | 14.5 | 52 | 2 (3.9) | — | — | — | — |
| 104 | 10.77 | 9.45 | 10.92 (3.96) | 12.8 | 302 | 10 (3.3) | — | — | — | — |
| 105 | 10.89 | 9.64 | 12.17 (4.25) | 13.7 | 177 | 7 (4.0) | — | — | — | — |
| 106 | 9.97 | 8.86 | 9.64 (3.19) | 9.5 | 70 | 2 (2.9) | 7.17 (1.97) | 6.9 | 315 | 15 (4.8) |
| 107 | 10.53 | 9.93 | 9.51 (4.45) | 9.7 | 72 | 2 (2.8) | — | — | — | — |
| 108 | 18.09 | 17.28 | 14.46 (6.28) | 19.3 | 25 | 0 (0.0) | 10.47 (3.41) | 9.3 | 77 | 4 (5.2) |
| 109 | 13.09 | 11.79 | 11.35 (4.20) | 13.3 | 308 | 20 (6.5) | 7.94 (2.01) | 7.1 | 174 | 5 (2.9) |
| 110 | 14.70 | 14.09 | 14.72 (4.52) | 15.1 | 99 | 4 (4.0) | 9.64 (2.64) | 9.5 | 365 | 12 (3.3) |
| Mean | 13.57a | 12.48a | 11.79 (4.2) | 13.24 | 155 | 6.2 (3.5) | 8.80 (2.51) | 8.2 | 233 | 9.0 (4.0) |
| Sample number . | TRF . | XpYp STELA . | 17p STELA . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | HinfI/RsaI . | MnlI/HphI . | Mean (±SD) . | Mode . | n . | Outliers (%) . | Mean (±SD) . | Mode . | n . | Outliers (%) . |
| 101 | 13.35 | 12.81 | 11.10 (4.47) | 13.1 | 284 | 7 (2.5) | — | — | — | — |
| 102 | 12.34 | 11.14 | 9.83 (3.28) | 11.0 | 162 | 8 (4.9) | — | — | — | — |
| 103 | 12.60 | 11.75 | 14.17 (4.54) | 14.5 | 52 | 2 (3.9) | — | — | — | — |
| 104 | 10.77 | 9.45 | 10.92 (3.96) | 12.8 | 302 | 10 (3.3) | — | — | — | — |
| 105 | 10.89 | 9.64 | 12.17 (4.25) | 13.7 | 177 | 7 (4.0) | — | — | — | — |
| 106 | 9.97 | 8.86 | 9.64 (3.19) | 9.5 | 70 | 2 (2.9) | 7.17 (1.97) | 6.9 | 315 | 15 (4.8) |
| 107 | 10.53 | 9.93 | 9.51 (4.45) | 9.7 | 72 | 2 (2.8) | — | — | — | — |
| 108 | 18.09 | 17.28 | 14.46 (6.28) | 19.3 | 25 | 0 (0.0) | 10.47 (3.41) | 9.3 | 77 | 4 (5.2) |
| 109 | 13.09 | 11.79 | 11.35 (4.20) | 13.3 | 308 | 20 (6.5) | 7.94 (2.01) | 7.1 | 174 | 5 (2.9) |
| 110 | 14.70 | 14.09 | 14.72 (4.52) | 15.1 | 99 | 4 (4.0) | 9.64 (2.64) | 9.5 | 365 | 12 (3.3) |
| Mean | 13.57a | 12.48a | 11.79 (4.2) | 13.24 | 155 | 6.2 (3.5) | 8.80 (2.51) | 8.2 | 233 | 9.0 (4.0) |
aMean derived from full sample; n number of molecules analysed; Outliers, number of molecules 2.33 SDs below the mean. All lengths in kb; —, not determined.
Summarizing telomere length data of individuals analyzed with both TRF and STELA.
| Sample number . | TRF . | XpYp STELA . | 17p STELA . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | HinfI/RsaI . | MnlI/HphI . | Mean (±SD) . | Mode . | n . | Outliers (%) . | Mean (±SD) . | Mode . | n . | Outliers (%) . |
| 101 | 13.35 | 12.81 | 11.10 (4.47) | 13.1 | 284 | 7 (2.5) | — | — | — | — |
| 102 | 12.34 | 11.14 | 9.83 (3.28) | 11.0 | 162 | 8 (4.9) | — | — | — | — |
| 103 | 12.60 | 11.75 | 14.17 (4.54) | 14.5 | 52 | 2 (3.9) | — | — | — | — |
| 104 | 10.77 | 9.45 | 10.92 (3.96) | 12.8 | 302 | 10 (3.3) | — | — | — | — |
| 105 | 10.89 | 9.64 | 12.17 (4.25) | 13.7 | 177 | 7 (4.0) | — | — | — | — |
| 106 | 9.97 | 8.86 | 9.64 (3.19) | 9.5 | 70 | 2 (2.9) | 7.17 (1.97) | 6.9 | 315 | 15 (4.8) |
| 107 | 10.53 | 9.93 | 9.51 (4.45) | 9.7 | 72 | 2 (2.8) | — | — | — | — |
| 108 | 18.09 | 17.28 | 14.46 (6.28) | 19.3 | 25 | 0 (0.0) | 10.47 (3.41) | 9.3 | 77 | 4 (5.2) |
| 109 | 13.09 | 11.79 | 11.35 (4.20) | 13.3 | 308 | 20 (6.5) | 7.94 (2.01) | 7.1 | 174 | 5 (2.9) |
| 110 | 14.70 | 14.09 | 14.72 (4.52) | 15.1 | 99 | 4 (4.0) | 9.64 (2.64) | 9.5 | 365 | 12 (3.3) |
| Mean | 13.57a | 12.48a | 11.79 (4.2) | 13.24 | 155 | 6.2 (3.5) | 8.80 (2.51) | 8.2 | 233 | 9.0 (4.0) |
| Sample number . | TRF . | XpYp STELA . | 17p STELA . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | HinfI/RsaI . | MnlI/HphI . | Mean (±SD) . | Mode . | n . | Outliers (%) . | Mean (±SD) . | Mode . | n . | Outliers (%) . |
| 101 | 13.35 | 12.81 | 11.10 (4.47) | 13.1 | 284 | 7 (2.5) | — | — | — | — |
| 102 | 12.34 | 11.14 | 9.83 (3.28) | 11.0 | 162 | 8 (4.9) | — | — | — | — |
| 103 | 12.60 | 11.75 | 14.17 (4.54) | 14.5 | 52 | 2 (3.9) | — | — | — | — |
| 104 | 10.77 | 9.45 | 10.92 (3.96) | 12.8 | 302 | 10 (3.3) | — | — | — | — |
| 105 | 10.89 | 9.64 | 12.17 (4.25) | 13.7 | 177 | 7 (4.0) | — | — | — | — |
| 106 | 9.97 | 8.86 | 9.64 (3.19) | 9.5 | 70 | 2 (2.9) | 7.17 (1.97) | 6.9 | 315 | 15 (4.8) |
| 107 | 10.53 | 9.93 | 9.51 (4.45) | 9.7 | 72 | 2 (2.8) | — | — | — | — |
| 108 | 18.09 | 17.28 | 14.46 (6.28) | 19.3 | 25 | 0 (0.0) | 10.47 (3.41) | 9.3 | 77 | 4 (5.2) |
| 109 | 13.09 | 11.79 | 11.35 (4.20) | 13.3 | 308 | 20 (6.5) | 7.94 (2.01) | 7.1 | 174 | 5 (2.9) |
| 110 | 14.70 | 14.09 | 14.72 (4.52) | 15.1 | 99 | 4 (4.0) | 9.64 (2.64) | 9.5 | 365 | 12 (3.3) |
| Mean | 13.57a | 12.48a | 11.79 (4.2) | 13.24 | 155 | 6.2 (3.5) | 8.80 (2.51) | 8.2 | 233 | 9.0 (4.0) |
aMean derived from full sample; n number of molecules analysed; Outliers, number of molecules 2.33 SDs below the mean. All lengths in kb; —, not determined.
References
Harley, C.B., Futcher, A.B. and Greider, C.W. (
Greider, C.W. and Blackburn, E.H. (
Kolquist, K.A., Ellisen, L.W., Counter, C.M., Meyerson, M., Tan, L.K., Weinberg, R.A., Haber, D.A. and Gerald, W.L. (
d'Adda di Fagagna, F., Reaper, P.M., Clay-Farrace, L., Fiegler, H., Carr, P., Von Zglinicki, T., Saretzki, G., Carter, N.P. and Jackson, S.P. (
Hayflick, L. (
Allsopp, R.C. and Harley, C.B. (
Artandi, S.E., Chang, S., Lee, S.L., Alson, S., Gottlieb, G.J., Chin, L. and DePinho, R.A. (
Flint, J. and Knight, S. (
Baird, D.M., Rowson, J., Wynford-Thomas, D. and Kipling, D. (
Wright, D.L., Jones, E.L., Mayer, J.F., Oehninger, S., Gibbons, W.E. and Lanzendorf, S.E. (
Wright, W.E., Piatyszek, M.A., Rainey, W.E., Byrd, W. and Shay, J.W. (
Kim, N.W., Piatyszek, M.A., Prowse, K.R., Harley, C.B., West, M.D., Ho, P.L., Coviello, G.M., Wright, W.E., Weinrich, S.L. and Shay, J.W. (
Fujisawa, M., Tanaka, H., Tatsumi, N., Okada, H., Arakawa, S. and Kamidono, S. (
Allshire, R.C., Dempster, M. and Hastie, N.D. (
Allsopp, R.C., Vaziri, H., Patterson, C., Goldstein, S., Younglai, E.V., Futcher, A.B., Greider, C.W. and Harley, C.B. (
Unryn, B.M., Cook, L.S. and Riabowol, K.T. (
Riethman, H., Ambrosini, A., Castaneda, C., Finklestein, J., Hu, X.L., Mudunuri, U., Paul, S. and Wei, J. (
Reid, B.J., Prevo, L.J., Galipeau, P.C., Sanchez, C.A., Longton, G., Levine, D.S., Blount, P.L. and Rabinovitch, P.S. (
Martens, U.M., Zijlmans, J.M., Poon, S.S., Dragowska, W., Yui, J., Chavez, E.A., Ward, R.K. and Lansdorp, P.M. (
Jeffreys, A.J., Tamaki, K., MacLeod, A., Monckton, D.G., Neil, D.L. and Armour, J.A. (
Kuzan, F.B., Hillier, S.L. and Zarutskie, P.W. (
Kipling, D. and Cooke, H.J. (
Hemann, M.T. and Greider, C.W. (
Cheung, I., Schertzer, M., Baross, A., Rose, A.M., Lansdorp, P.M. and Baird, D.M. (
Smogorzewska, A. and de Lange, T. (
Slagboom, P.E., Droog, S. and Boomsma, D.I. (
Graakjaer, J., Pascoe, L., Der-Sarkissian, H., Thomas, G., Kolvraa, S., Christensen, K. and Londono-Vallejo, J.A. (
Vasa-Nicotera, M., Brouilette, S., Mangino, M., Thompson, J.R., Braund, P., Clemitson, J.R., Mason, A., Bodycote, C.L., Raleigh, S.M., Louis, E. et al. (
Nawrot, T.S., Staessen, J.A., Gardner, J.P. and Aviv, A. (
Zhu, L., Hathcock, K.S., Hande, P., Lansdorp, P.M., Seldin, M.F. and Hodes, R.J. (
Vulliamy, T., Marrone, A., Goldman, F., Dearlove, A., Bessler, M., Mason, P.J. and Dokal, I. (
Vulliamy, T., Marrone, A., Szydlo, R., Walne, A., Mason, P.J. and Dokal, I. (
Yashima, K., Maitra, A., Rogers, B.B., Timmons, C.F., Rathi, A., Pinar, H., Wright, W.E., Shay, J.W. and Gazdar, A.F. (
Lanza, R.P., Cibelli, J.B., Blackwell, C., Cristofalo, V.J., Francis, M.K., Baerlocher, G.M., Mak, J., Schertzer, M., Chavez, E.A., Sawyer, N. et al. (
Shiels, P.G., Kind, A.J., Campbell, K.H., Waddington, D., Wilmut, I., Colman, A. and Schnieke, A.E. (
Betts, D.H., Perrault, S.D., Petrik, J., Lin, L., Favetta, L.A., Keefer, C.L. and King, W.A. (
Baird, D.M., Coleman, J., Rosser, Z.H. and Royle, N.J. (
Baird, D.M., Jeffreys, A.J. and Royle, N.J. (
Li, B. and Lustig, A.J. (
Bucholc, M., Park, Y. and Lustig, A.J. (
Lustig, A.J. (
Wang, R.C., Smogorzewska, A. and de Lange, T. (
Bailey, S.M., Brenneman, M.A. and Goodwin, E.H. (
von Zglinicki, T., Saretzki, G., Docke, W. and Lotze, C. (
Hemann, M.T., Strong, M.A., Hao, L.Y. and Greider, C.W. (
Chard, T. (
Eiben, B., Borgmann, S., Schubbe, I. and Hansmann, I. (
Cockwell, A.E., Jacobs, P.A., Beal, S.J. and Crolla, J.A. (
Liu, L., Franco, S., Spyropoulos, B., Moens, P.B., Blasco, M.A. and Keefe, D.L. (
Macas, E., Imthurn, B. and Keller, P.J. (
Macklon, N.S., Geraedts, J.P. and Fauser, B.C. (
WHO (