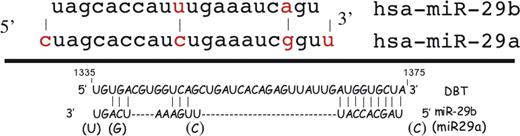-
PDF
- Split View
-
Views
-
Cite
Cite
Benjamin D. Mersey, Peng Jin, Dean J. Danner, Human microRNA (miR29b) expression controls the amount of branched chain α-ketoacid dehydrogenase complex in a cell, Human Molecular Genetics, Volume 14, Issue 22, 15 November 2005, Pages 3371–3377, https://doi.org/10.1093/hmg/ddi368
Close - Share Icon Share
Abstract
Branched chain amino acids (BCAAs) play critical roles in cell and tissue functions in addition to being important components of protein structure. The multifunctional roles speak to the need for maintaining tight control of their concentration within cells. As the BCAA cannot be made de novo in mammals, their cellular concentration is a function of dietary intake, endogenous protein turnover and catabolism of the three amino acids. The branched chain α-ketoacid dehydrogenase (BCKD) complex commits the BCAA to degradation and thus is vital in controlling their concentration within a cell. In mammals, BCKD activity state depends on the presence of a covalently bound phosphate on one protein component of the complex. Phosphate is added to the protein through the action of the complex-specific kinase and results in BCKD inactivation. Here, we demonstrate that another reaction plays a role in determining the total amount of BCKD present in a cell. The microRNA (miR29b) molecule tested is targeted to the mRNA for the dihydrolipoamide branched chain acyltransferase component of BCKD and prevents translation when bound. This is the first demonstration of the use of a microRNA to exert control on a metabolic pathway of amino acid catabolism in mammals and offers an explanation for the observed differences in the amount of the BCKD complex present in different tissues and under varying nutritional states.
INTRODUCTION
Expression of small non-coding RNA (ncRNA) molecules is now recognized to have important roles in development and maintenance of cells and tissues in organisms from plants to humans (1). One member group of these ncRNAs is the microRNAs (miR) whose roles were first recognized in Caenorhabditis elegans and Drosophila. In mammals, miRs control gene expression either by interfering with production, stability or translation of the mRNA target (2–5). MicroRNAs can be a product of their own gene or originate from within introns of protein encoding transcripts (6,7). The primary miR transcript is cut by the RNaseIII enzyme known as DROSHA in the nucleus to yield a 70–100 nucleotide product that forms a hairpin structure that is exported to the cytosol by EXPORTIN 5 (8). In the cytosol, these hairpin structures are cut by the endonuclease DICER to a double-stranded product of 20–25 bp. This double-stranded RNA is incorporated into the RNA-induced silencing complex (RISC) where the dominant miR is retained and used to direct RISC to the target mRNA (3,9–11). Identical pairing of the entire miR with its target results in target degradation, whereas imperfect pairing beyond the first 7–9 bases at the 5′ end of the miR blocks translation of the targeted mRNA but does not destroy the mRNA (12).
Surveys to determine tissue distribution and expression level for miRs in mammals report varying results even with the same tissues and miRs (13–16). This variation remains to be explained. It has been suggested that miRs can have multiple targets and the preference is related to the binding strength to the different targets (17–19). Binding of the miR with its target is strongly influenced by the first 7–9 nucleotides at the 5′ end of the miR. In mammals, the binding site in the mRNA target is frequently found in the 3′ untranslated region (UTR) (17). Further, it has been suggested that multiple miRs may bind to the same mRNA and act in concert with one another (20,21). The number of miRs that are present and can recognize a specific target within a cell type remains a question. Whether these short interfering RNAs are used for control of constitutively expressed genes or to fine tune expression of inducible genes is not known (1).
The branched chain amino acids (BCAAs), leucine, isoleucine and valine, account for up to 20% of the component amino acids in most proteins. In addition, BCAAs play important roles in synapse function, insulin secretion and protein turnover (22–24). BCAAs are essential components of the diet of metazoans, because they cannot be synthesized de novo in these organisms. Cellular concentration of the BCAA is determined by dietary protein intake, endogenous protein turnover and the catabolism of the individual BCAA, which occurs in all cells with mitochondria as the entire catabolic pathway is located in these organelles. Branched chain α-ketoacid dehydrogenase (BCKD) catalyzes the irreversible step in BCAA catabolism and thus commits to a decrease in the concentration of these amino acids within that cell. It was found that activity of the existing BCKD complex within a cell is regulated by a complex-specific kinase (BCkinase) that adds a phosphate group to the E1α subunit. That portion of BCKD with a phosphorylated subunit is catalytically inactive (25). The activity state of BCKD shows a direct relationship to the amount of BCkinase present in mitochondria within the cell. A recent report implicated that Drosophila use miR to control the expression of the amount of BCKD complex present in a cell and the authors speculate that miR expression responds to dietary intake of protein for the flies (26). A gene for a BCkinase cannot be found in the Drosophila genomic database. The absence of a BCkinase supports the idea that the fly BCKD is regulated by miR277 expression. Here we asked if any of the human miRs have as targets one or more mRNAs that encode human BCKD subunits. Several miRs were identified with this potential (http://www.sanger.ac.uk/Software/Rfam/mirna/) (27). Our results identify that endogenously expressed human miR29b recognizes the mRNA for the dihydrolipoyl branched chain acyltransferase (DBT) component of the BCKD complex in HEK293 cells. These findings suggest that as in Drosophila the amount of BCKD present within human cells is determined by the expression of miR29b and potentially other miRs.
RESULTS
BCKD subunit expression and activity
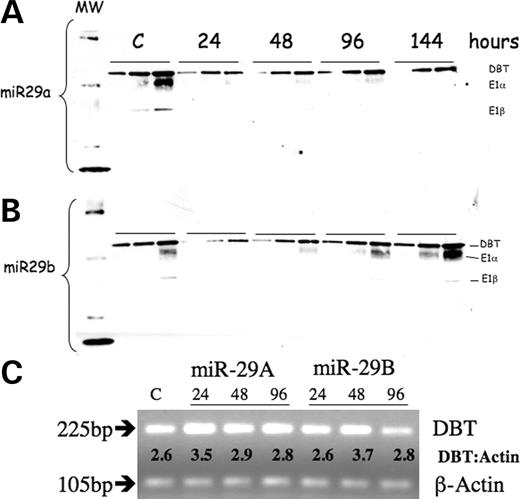
HEK293 cells express BCKD activity and the activity state within these cells reflects the expression level of the BCkinase in human kidney tissue (28). Under normal culture conditions, about 50% of the BCKD complex in HEK293 cells is catalytically inactive due to BCkinase expression [22.6±11.7 pmol CO2/(mg protein/min) out of 43.1±15.7 pmol CO2/(mg protein/min) total activity] (28). This cell line therefore is appropriate for assessing whether the miR control for the amount of BCKD expression reported in flies has been retained in humans along with the addition of the BCkinase control of activity state of existing BCKD. Members of the human miR29 family have, among their predicted mRNA targets, the one that produces the DBT protein that forms the core of the BCKD complex. The predicted pairing of miR29a and miR29b occurs in the 3′ end of the mRNA but lies within the coding sequence of the terminal exon 11 and binds in the region that is 70 nucleotides upstream of the stop codon (Fig. 1). The pairing for miR29b conforms to the predicted canonical pattern for a functional miR, as the first nine bases directly match the nucleotide sequence within the mRNA for DBT (17). miR29b and miR29a were synthesized by us and used to transfect HEK293 cells. A fluorescent labeled, randomly generated double-stranded RNA of 22 bp was co-transfected along with the specific miR and the percentage of cells with fluorescent expression was estimated by microscopic visual inspection. Control cells for each protocol received the tagged randomly generated RNA and served as mock transfection for the test miR. Data reported were obtained from cell populations where transfection efficiency exceeded 85% (Fig. 2).
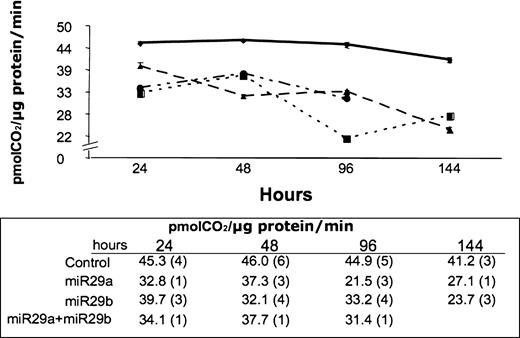
If transfection of cells with miR29a and/or miR29b alters the amount of BCKD within a cell, total BCKD activity will reflect the change. Cells are treated with the BCkinase inhibitor α-chloroisocaproate to determine the total BCKD activity within the cells (28,29). Total BCKD activity decreased over a 6-day period following a single transfection with either miR29a or miR29b. Control cells contain only the random 22mer RNA and BCKD activity did not change during this time course. The decrease in total activity was evident by 24-h post-transfection and continued to decrease over the 144 h tested (Fig. 3). The decrease in total activity amounted to about 40% by 144 h, suggesting that miR29b has retained function in humans as in the Drosophila. Growth restrictions of the cells in the 24-well plates prohibited further extension of the time course. Transfection with both miRs did not produce an additive effect and therefore this part of the protocol was not continued. The loss of BCKD total activity implies a decrease in the amount of total BCKD that is present in the mitochondria and presumably due to a decrease in DBT protein. The amount of DBT depends on production and degradation of this protein. A previous report has predicted that the half-life of the BCKD proteins is 22–26 h (30). Another factor that might play a role in DBT production is the amount of miR present in a cell and the half-life of these molecules. No information is available regarding the stability of exogenously added double-stranded microRNA, but endogenously produced mature miRs are reported to exist for more than 3 days within a cell (31).
When the miR exhibits imperfect pairing with its target, mRNA beyond the first nine bases translation of the message is blocked, but not destroyed. The antigenic presence of DBT was determined by western blots using increasing amounts of mitochondrial protein to estimate the relative concentration (Fig. 4). A reduced amount of DBT in the mitochondria occurred with the addition of either miR29a or miR29b by 24-h post-transfection. The amount of DBT returned to its original level by 144 h. Loss of DBT reduces the other BCKD subunits, as their presence depends on binding to DBT. Therefore, even though DBT is found to be back to normal amount 144 h after transfection with miR29b, the total activity of BCKD is still lower than normal because the complex has not been reformed. Control cells were harvested at different time points in individual experiments following this protocol. In the example shown here, control cells were harvested at the 96-h time point; however, the amount of DBT protein in the control cells did not change over the entire time course. Recall that control cells always contain the random sequence for a 22mer RNA. During this same time course, the amount of mRNA for DBT did not change (Fig. 4C), indicating that mRNA was not destroyed and suggests that translation of the DBT mRNA was blocked when miR29b is bound to the mRNA.
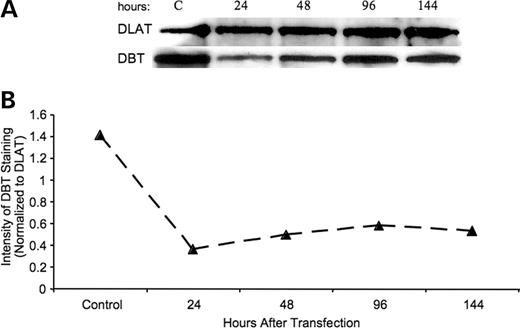
To demonstrate that the observed loss of activity and protein was not through a direct interaction of the miR with the protein, a western blot for the dihydrolipoyl acyltransferase (DLAT) protein was done with the same membrane used for western blots to assess DBT in these mitochondrial preparations. DLAT is the core protein of the pyruvate dehydrogenase complex that also functions as a multienzyme complex in mitochondria. As with the BCKD complex, the proteins forming the pyruvate dehydrogenase complex are encoded by nuclear genes. DBT and DLAT are structurally similar in amino acid sequence, but exhibit a high degree of substrate specificity in catalytic function (32). Comparison of the two nucleic acid sequences for the mRNA for DBT and DLAT revealed no sequence identity. There is no sequence pairing between the miR29a and miR29b for the mRNA for DLAT as shown for the mRNA encoding DBT (Fig. 1). The observed decrease in DBT but not in DLAT (Fig. 5) supports the function of the miRs used here specifically affect translation of mRNA for DBT. As variation between membranes prepared from individual experiments will not allow for averaging of the data, results shown in Figure 5 are from a single experiment representative of three repeats of this protocol.
Endogenous expression of miR29a and miR29b
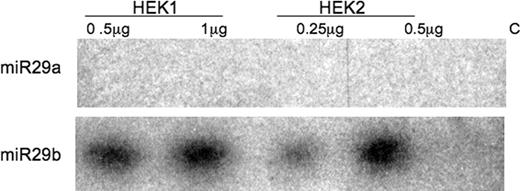
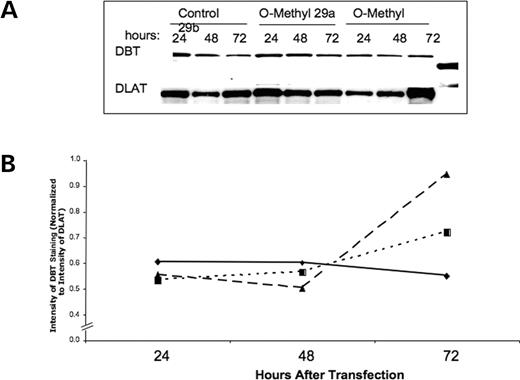
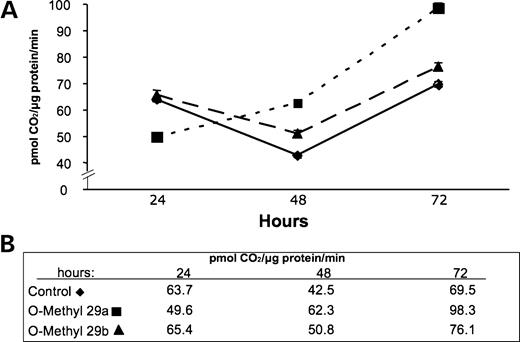
Although demonstration that exogenous addition of these miRs decreases DBT synthesis and results in a reduced level of BCKD function, the results do not address whether these miRs exist and function endogenously within the HEK293 cells. To address this question, RNA was isolated under conditions that preserve the small RNA from two independent cultured cell preparations collected on separate days. The isolated RNA was hybridized in separate reactions with a radiolabeled antisense RNA against miR29a or miR29b. As illustrated by the autoradiograph of the RNase-protected products, only miR29b was detected in each of the two separate preparations of the HEK293 cells (Fig. 6). To begin assessing a functional role for this miR on BCKD, antisense RNA with 2′-O-methyl oligoribonucleotides was used for transfection followed by BCKD activity measurements and western blots as earlier. The O-methyl modification has the ability to inactivate endogenously produced miR function in cultured human cells (33). With either antisense RNA, a small change in DBT protein concentration occurred (Fig. 7) that resulted in a small increase in total BCKD activity (Fig. 8). This increase in total BCKD activity was 20% above control cells that received only the random sequence 22mer RNA. Although endogenous miR29a is not detected, the similarity in nucleotide sequence for the two antisense RNAs may result in hybridization to block endogenous miR29b.
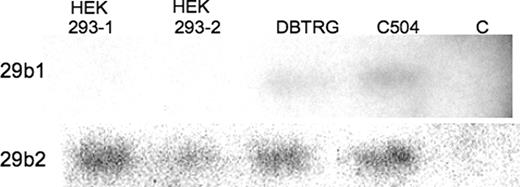
The miRNA Registry Web site assigns the human chromosome location for the mi29b-1 gene to 7q32.2–32.3 and that for miR29b-2 to 1q32.2. The hairpin loop pre-miR structure is similar in nucleotide sequence for each gene product with diversity only in the sequence forming the hairpin. Antisense RNAs to the loop region were synthesized and used for RNase protection assays to determine which gene site was producing the miR29b found in these cells. As seen in Figure 9, only the miR29b-2 showed the predicted hybridization product in the HEK293 and two other human cell types implicating expression from the region on chromosome 1.
DISCUSSION
The speculation that Drosophila use miR expression in response to dietary protein to control BCAA metabolism by regulating expression of the BCKD complex raised the question of whether this mechanism for controlling the amount of BCKD in a cell was retained in human cells (26). Numerous human miRs exist that have as predicted targets mRNAs for components of the BCKD complex along with a large number of mRNAs for other proteins (17). The strongest predicted pairing occurs between members of the miR29 family and the mRNA for DBT. DBT forms the core of the BCKD complex and provides the binding site for all other proteins in the complex including the BCkinase (34). Reducing DBT will therefore reduce the amount of total BCKD, as all the other subunits must bind to this core component. There are no reports that describe procedures for formation of the BCKD complex through subunit interactions.
Our results support predicted function of reduced translation for an miR with pairing of only the first 7–9 nucleotides in the 5′ end with the targeted mRNA (17). The remaining nucleotides potentially pair further into the mRNA with formation of loops in the mRNA. As HEK293 cells express BCkinase to a level that maintains the activity state of BCKD around 50%, these cells provide a model where we can ask if expression of DBT is reduced and the total activity of the BCKD complex is decreased. Addition of in vitro synthesized miR29a or miR29b to HEK293 cells separately or in combination resulted in a linear decrease in total BCKD activity. If miR expression blocks mRNA translation, the amount of protein found in the cells reflects a protein's half-life. Little is known about the half-life of components of the BCKD complex, but one report gave a 24-h half-life for all components (30). As the miR was added exogenously, the half-life of the added miR could contribute to the length of time for the observed change in proteins studied. No data is available for these in vitro synthesized double-stranded short RNAs (31).
Although there is no report for a direct interaction of an miR with a protein rather than an mRNA, we addressed that by investigating the effect of the miR29a and miR29b transfection on the expression of DLAT (35). Our findings support the proposal that mRNA for DBT is the target of miR29b and the resulting duplex blocks translation of this message.
Results with the exogenously added miR29a and miR29b do not address whether HEK293 cells show in vivo expression of these ncRNAs and when present if the miRs target the DBT mRNA. RNase protection analysis demonstrated the endogenous presence of only miR29b in HEK293 cells. The observed decrease in DBT does result in a decrease in total BCKD activity because the amount of complex will decrease without the core protein. It appears that other proteins of the BCKD complex also are lost when DBT is decreased. This is likely due to the fact that they bind to DBT and without DBT for binding, other proteins are degraded. It now becomes important to begin investigations to address factors that control expression of the miR29b gene. When the cells were transfected with antisense RNA against miR29a or miR29b, a modest increase in total BCKD activity was observed. The amount of increase is consistent with what we reported when cells were transfected with a plasmid containing the cDNA for DBT that resulted in overexpression of DBT (36). The amount of DBT protein produced in our previous paper was about 25% more than that in control cells and total BCKD activity was 15% higher than that in control cells (36). Increased expression of DBT did not result in an increased expression of the other subunits and therefore increase in total BCKD activity was minimal (36).
There are two human genes reported to encode miR29b (http://www.sanger.ac.uk/Software/Rfam/mirna/). The gene we expressed is that for miR29b-2 located on chromosome 1q32.2 and the human gene for DBT is also located on this chromosome but on the opposite arm at 1p31 (27,37). RNase protection analysis supports the finding that the gene for miR29b-2 on chromosome 1 is being expressed in the human kidney, glial and fibroblast cells (Fig. 9). The means by which expression of the miR29b gene in these cells is controlled needs to be investigated to better understand its observed function for blocking translation of the DBT mRNA.
Our finding represents the first report demonstrating regulation of the amount of BCKD that is present within a cell through the action of microRNAs. It is interesting to speculate that as the BCkinase must associate with the DBT protein to function efficiently, it may be necessary to regulate the amount of DBT produced (34). The demonstration that expression of microRNA can influence the amount of BCKD presents another site for investigation of inherited defects affecting BCKD function. Maple syrup urine disease (MSUD) is a condition in which BCKD activity is impaired by inherited mutations in the genes for the catalytic subunits of BCKD. Some individuals with MSUD present with a less severe clinical course and were missed in newborn screening programs but present with elevations in plasma BCAAs later in life (38). A molecular genetic basis for this intermittent condition remains to be defined. Changes in expression of the microRNAs that alter the amount of the BCKD present in a cell offer a possible explanation for this condition. We need to examine the tissue expression level of these and other microRNAs in cell lines from the individuals presenting with the intermittent MSUD.
MATERIALS AND METHODS
Cell culture
Human HEK293 cells were maintained in standard DMEM media supplemented with 10% FBS and antibiotics at 37°C and 5% CO2-humidified atmosphere. Prior to transfection with the miRs in Lipofectamine 2000, the cells were placed in standard media without antibiotics for 24 h. The individual miR was used at a concentration of 20 pmol/well for a 24-well plate or 100 pmol/well for a 6-well plate. When miR29a and miR29b were co-transfected, 10 pmol of each was used to produce the 20 pmol total per well. Transfection with the 2′-O-methyl oligoribonucleotides was done at a concentration of 10 pmol/well for the 24-well plates and 50 pmol/well for the 6-well plates. To estimate transfection efficiency, the Block-iT Fluorescent double-strand random 22mer RNA from Invitrogen (Carlsbad, CA) was co-transfected at one-half the concentration of the experimental miR. Control populations of cells contained only this tagged 22mer RNA. The FITC fluorescence was visualized by lex=494 nm and lem=519 nm to assess the number of cells that were transfected. Cells receiving only the tagged random sequence double-strand 22mer were used for non-specific reference at all data points. Cells were harvested and analyzed at indicted times post-transfection.
Synthesis of microRNA
MicroRNAs tested in these transfection experiments were synthesized using the Silencer siRNA Construction Kit from Ambion (Austin, TX) according to the manufacturer's instructions. The primers used for miR29a were 5′-CTAGCACCATCTGAAATCGGTTCCTGTCTC-3′ (forward) and 5′-AAAACCGATTTCAGATGGTGCTCCTGTCTC-3′ (reverse) and for the 29b duplex 5′-TAGCACCATTTGAAATCAGTCCTGTCTC-3′ (forward) and 5′-AAACTGATTTCAAATGGTGCCCTGTCTC-3′ (reverse). Concentration of the final product was estimated by absorption measurement at 260 nm after suspension in RNase-free water. Antisense oligoribonucleotides with all but three core nucleotides bearing the 2′-O-methyl modification were purchased from Integrated DNA Technologies (Coralville, IA).
Whole cell BCKD assay
Whole cell BCKD activity state, expressed as percent activity, was determined by measuring the 14CO2 released from [1−14C]-leucine per mg of total cellular protein under basal conditions relative to that produced after a 10-min pre- incubation with 1 mm α-chloroisocaproate, an inhibitor of BCKD-kinase, as previously described (39). Total activity values reported are an average±standard error (SE) of three to four experiments per cell line, each performed in triplicate.
Mitochondria isolation and western blot
Mitochondria were isolated from approximately 5×106 cells. Cells were released from the culture plates with trypsin/EDTA, centrifuged at 300×g, washed with phosphate- buffered saline (PBS) and resuspended in 1 ml mitochondrial isolation buffer (MIB; 220 mm mannitol, 70 mm sucrose, 2 mm HEPES, pH 7.4 with KOH). Cells were homogenized by douncing 25 times in a Wheaton Potter-Elvehjem tissue grinder and nuclei were pelleted by centrifugation at 2000×g for 10 min at 4°C. The mitochondria were pelleted from the supernatant by centrifugation for 10 min at 15 000×g at 4°C. Protein concentration in each sample was determined using the BCA Protein Assay (Pierce Biotechnology, Rockford, IL).
For western blot experiments, identical amounts of mitochondrial proteins for each condition were resolved through a 10% SDS–PAGE and then transferred to a nitrocellulose membrane by tank transfer in transfer buffer (24.7 mm Tris, 192 mm glycine and 20% methanol) at room temperature over 2 h. Equality of protein loading and transfer was determined by Ponceau-S staining of the membrane. Development of the immunoblot with antisera against the BCKD complex was done as previously described (36). Antisera against the dihydrolipoyl acetyltransferase (DLAT) was purchased from Santa Cruz Biotechnology, Inc. Visualization of the antibody binding was with West Pico Chemiluminescent Substrate detection protocol (Pierce). The chemiluminescent signal was exposed to Blue BioFilm (Denville Scientific, Inc., Metuchen, NJ) for up to 5 min.
Quantitative analysis of DBT mRNA
Total RNA from transfected cells was isolated using TRI Reagent (Sigma) according to the manufacturer's protocol. First-strand cDNA was synthesized using the SuperScript First Strand Synthesis System (Invitrogen) with Oligo-dT primers. PCR amplification of template cDNA was performed using Platinum Taq DNA Polymerase (Invitrogen). Samples were denatured at 94°C for 2 min followed by 28 cycles of denaturation at 94°C for 30 s, annealing at 51°C for 30 s and extension at 72°C for 30 s, with a 10-min final extension. Primers used for amplification of DBT and β-actin were purchased from Invitrogen and Integrated DNA Technologies, respectively. Sequences were as follows. DBT: 5′-CTCTCCGTGGACAGGTTGTT-3′ (forward), 5′-GTCTCCCCACATAGGCAATA-3′ (reverse); β-actin: 5′-GTGGCCGAGGACTTTGATTG-3′ (forward), 5′-AGTGGGGTGGCTTTTAGGATG-3′ (reverse).
RNase protection assay
Enriched small RNA from 107 non-transfected HEK293 cells was purified using the miRVana miRNA Isolation Kit (Ambion). Final concentration and purity of RNA was estimated using absorbencies at 260 and 280 nm. For microRNA detection, the miRvana miRNA Detection Kit (Ambion) was used according to the manufacturer's instructions. Antisense RNA probes for miR-29a 5′-AAAACCGAUUUCAGAUGGGUCUAGAAAACCAGAG-3′ and for miR29b 5′-AAACUGAUUUCAAAUGGUGCUACCAGAG-3′ and for hairpin loop probes 29b-1 5′-CCCAAGAACACUGAUUUCAAAUGGUGCUAGACCCAGAG-3′ and for 29b-2 5′-AAAAAGGUGCUAGAUACAAAGAUGGAAACCAGAG-3′ were obtained from Integrated DNA Technologies and end-labeled using γ-[32P]-ATP. After hybridization and RNase treatment, the double-strand products were resolved in a 15% polyacrylamide 8 m urea denaturing gel and visualized using phosphoimaging and autoradiography.
Conflict of Interest statement. None declared.
Figure 1. Nucleotide sequence for human miRNA29a and miRNA29b and the predicted base pairing with mRNA for DBT. Different bases are indicated by the red color of the base. The paired region in DBT mRNA covers nucleotides 1335–1375 that exist within the coding region of terminal exon 11. The dashes in the pairing indicate the distance between the regions of pairing of the miR with the mRNA to suggest where the loops are formed in the mRNA.
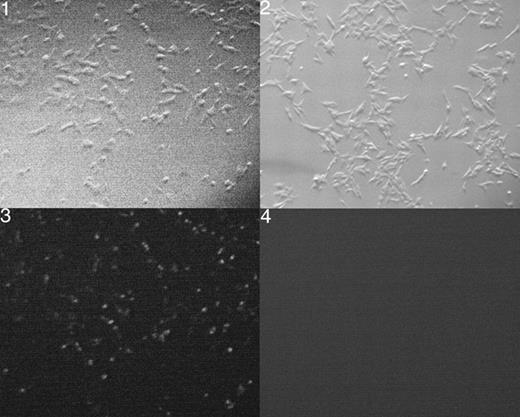
Figure 2. Photomicrograph of cells transfected with a random 22mer RNA labeled with FITC (1 and 3) or cells with lipofectamine alone (2 and 4). Panels 1 and 2 are dark field and 3 and 4 are viewed with a GFP filter. The figures show the greater than 80% transfection of the cells used in these studies. Magnification was 50×.
Figure 3. Total BCKD activity in HEK293 cells transfected with 20 pmol per 105 cells Block-IT Fluorescent RNA oligo (υ), miR29a (ν) or miR29b (σ), or 10 pmol per 105 cells both miR29a and miR29b (λ). Activity was measured at the indicated time points for cell growth after transfection and is reported as pmol CO2 released per µg of protein per minute. The numbers of independent experiments used to create the average values are reported in parentheses.
Figure 4. Western blot and PCR of DBT gene products in HEK293 cells transfected with miR29a or miR29b. Antisera for BCKD complex was used to probe mitochondrial proteins (4, 8 and 12 µg of mitochondrial protein at each time point) after transfection with miR29a (A) and miR29b (B). (C) Products from RT–PCR analysis (28 cycles) of transfected cells for DBT mRNA with β-actin as a loading control analyzed on a 2% agarose gel. The transfected miRNA and number of hours post-transfection when RNA harvest occurred are indicated. Ratio of DBT to β-actin density traces is indicated between the two products. The control value was done at different time points and does not change. The control value reported here was done at 96 h for (A) and (B) and 48 h for (C) and contain only the non-specific 22mer RNA.
Figure 5. DBT protein in HEK293 cells relative to DLAT protein after cell transfection with an miR29b. (A) Western blot for DBT and DLAT in mitochondria after transfection with miR29b at the indicated times after transfection. Control cells were taken after 96 h. (B) Calculations of the concentration of DBT to DLAT for the time course.
Figure 6. Endogenous expression of miR29a and miR29b in HEK293 cells. Radiolabeled antisense RNA against miR29a and miR29b were used to hybridize the isolated cellular RNA from two different cell preparations. RNase digestion was used to identify the double-stranded RNA products. Controls contain antisense RNA probe and tRNA, but no cellular RNA during the RNase treatment.
Figure 7. The use of O-methyl antisense RNA to block expression of endogenous miR29a and miR29b. (A) Western blot for DBT and DLAT for mitochondrial protein isolated at the indicated times after treatment with O-methyl antiRNA for 29a and 29b. (B) Relative concentration of DBT versus DLAT during this time course. The diamond is the control cells that only have the non-specific fluorescence labeled 22mer RNA. Squares are for antisense RNA 29a and triangles are for antisense RNA 29b.
Figure 8. Effect of O-methyl antisense RNA for miR29a and miR29b on BCKD total activity: (A) graphic representation of the activity; (B) absolute values for each time point.
Figure 9. Detection of specific endogenous gene products for miR29b in three different human cell lines. HEK293 cells (1 and 2) represent repeated analysis of this cell type and DBTRG is a human glial cell line and C504 a human dermal fibroblast cell line. Conditions are as described in Figure 6, with the antisense RNA used here is for the unique sequence of the hairpin loops for the gene encoding miR29b1 on chromosome 7 and the gene encoding miR29b2 on chromosome 1. Controls are as described in Figure 6.
References
Bartel, D.P. and Chen, C.Z. (
He, L. and Hannon, G.J. (
Hobert, O. (
Lee, R., Feinbaum, R. and Ambros, V. (
Lee, Y., Kim, M., Han, J., Yeom, K.H., Lee, S., Baek, S.H. and Kim, V.N. (
Ohler, U., Yekta, S., Lim, L.P., Bartel, D.P. and Burge, C.B. (
Zeng, Y. and Cullen, B.R. (
Doi, N., Zenno, S., Ueda, R., Ohki-Hamazaki, H., Ui-Tei, K. and Saigo, K. (
Xu, P., Guo, M. and Hay, B.A. (
Babak, T., Zhang, W., Morris, Q., Blencowe, B.J. and Hughes, T.R. (
Barad, O., Meiri, E., Avniel, A., Aharonov, R., Barzilai, A., Bentwich, I., Einav, U., Gilad, S., Hurban, P., Karov, Y. et al. (
Miska, E.A., Alvarez-Saavedra, E., Townsend, M., Yoshii, A., Sestan, N., Rakic, P., Constantine-Paton, M. and Horvitz, H.R. (
Nelson, P.T., Baldwin, D.A., Scearce, L.M., Oberholtzer, J.C., Tobias, J.W. and Mourelatos, Z. (
Lewis, B.P., Burge, C.B. and Bartel, D.P. (
Lewis, B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P. and Burge, C.B. (
Rehmsmeier, M., Steffen, P., Hochsmann, M. and Giegerich, R. (
Chang, S., Johnston, R.J., Jr, Frokjaer-Jensen, C., Lockery, S. and Hobert, O. (
Doench, J.G. and Sharp, P.A. (
Kimball, S.R. (
Malaisse, W.J., Hutton, J.C., Carpinelli, A.R., Herchuelz, A. and Sener, A. (
Ævarsson, A., Chuang, J.L., Wynn, R.M., Turley, S., Chuang, D.T. and Hol, W.G.J. (
Stark, A., Brennecke, J., Russell, R.B. and Cohen, S.M. (
Muller, E.A. and Danner, D.J. (
Paxton, R. and Harris, R.A. (
Honda, K., Ono, K., Mori, T. and Kochi, H. (
Kim, V.N. (
Wang, G.F., Kuriki, T., Roy, K.L. and Kaneda, T. (
Meister, G., Landthaler, M., Dorsett, Y. and Tuschl, T. (
Davie, J.R., Wynn, R.M., Meng, M., Huang, Y.S., Aalund, G., Chuang, D.T. and Lau, K.S. (
Patel, M.S., Naik, S., Johnson, M. and Dey, R. (
McConnell, B.B., McKean, M.C. and Danner, D.J. (
Herring, W.J., McKean, M., Dracopoli, N. and Danner, D.J. (
Peinemann, F. and Danner, D.J. (