-
PDF
- Split View
-
Views
-
Cite
Cite
Regina Z. Cer, Duncan E. Donohue, Uma S. Mudunuri, Nuri A. Temiz, Michael A. Loss, Nathan J. Starner, Goran N. Halusa, Natalia Volfovsky, Ming Yi, Brian T. Luke, Albino Bacolla, Jack R. Collins, Robert M. Stephens, Non-B DB v2.0: a database of predicted non-B DNA-forming motifs and its associated tools, Nucleic Acids Research, Volume 41, Issue D1, 1 January 2013, Pages D94–D100, https://doi.org/10.1093/nar/gks955
Close - Share Icon Share
Abstract
The non-B DB, available at http://nonb.abcc.ncifcrf.gov, catalogs predicted non-B DNA-forming sequence motifs, including Z-DNA, G-quadruplex, A-phased repeats, inverted repeats, mirror repeats, direct repeats and their corresponding subsets: cruciforms, triplexes and slipped structures, in several genomes. Version 2.0 of the database revises and re-implements the motif discovery algorithms to better align with accepted definitions and thresholds for motifs, expands the non-B DNA-forming motifs coverage by including short tandem repeats and adds key visualization tools to compare motif locations relative to other genomic annotations. Non-B DB v2.0 extends the ability for comparative genomics by including re-annotation of the five organisms reported in non-B DB v1.0, human, chimpanzee, dog, macaque and mouse, and adds seven additional organisms: orangutan, rat, cow, pig, horse, platypus and Arabidopsis thaliana. Additionally, the non-B DB v2.0 provides an overall improved graphical user interface and faster query performance.
INTRODUCTION
In addition to the canonical right-handed double helix, repetitive DNA sequences have the ability to form alternative conformations (non-B) [reviewed in (1)]. Specifically, inverted repeats can adopt hairpin and cruciform structures; homopurine sequences with mirror symmetry may fold into intramolecular triplexes; alternating purine–pyrimidine bases can switch from right-handed B- to left-handed Z-DNA conformations; sets of three, four or five guanines, each separated by approximately one to seven bases, can form quadruplex structures and direct repeats can give rise to loops or hairpins through the misalignment of complementary strands [reviewed in (2)]. These underlying structural motifs have been found to play a role in the control of diverse biological functions, including replication, transcription (3,4), immune response (5), recombination (6), chromosomal fragility (1,7) and antigenic variation in human pathogens (1,8). There is also increasing evidence associating non-B DNA structures with human disease (9–13). Thus, a deeper functional annotation of these motifs is warranted to help uncover their contribution to specific disease processes.
We introduced the original non-B DB to provide a comprehensive resource for non-B DNA-forming sequence motifs (14). The original database provided the first complete annotations of non-B DNA-forming sequence motifs that covered genomes of mammalian organisms, including human, mouse, chimpanzee, dog and macaque. We recently added the non-B DNA Motif Search Tool (nBMST) (15), where users can submit their own sequences, up to 20 Mb, for motif searching. A streamlined interface, additional visualization options and algorithmic enhancements in v2.0 improve the utility of non-B DB as a tool aimed at understanding how alternative DNA structures may impact biological function, genomic instability and disease across a variety of species. The goal of the non-B database is to allow investigators to view their own genomic feature annotations in the context of these predicted motifs and to aid in the formation of hypotheses regarding the potential role of these structures in other cellular processes.
REFINEMENT OF SEARCH CRITERIA
The addition of the web server and the inclusion of additional species provided an opportunity to re-evaluate the available literature on the in vivo formation of non-B DNA-forming motifs and re-implement an updated algorithm in the C programming language conforming to the American National Standards Institute (ANSI C), rather than the Perl predecessors. The definitions of the non-B DNA motifs have been updated based on a re-evaluation of current experimental and theoretical studies on the formation of non-B DNA. The updated algorithm is roughly an order of magnitude faster than the original Perl version.
A full and detailed description of the motif definitions is beyond the scope of this update and will be published elsewhere. Briefly, we have extended direct repeat definition to contain repeats up to 300 nucleotides from the original 50-nucleotide repeat length. The inverted repeats have a minimum length of 6 nucleotides as opposed to the original definition of 10 nucleotides (14). In addition, the inverted repeats have no maximum repeat length and can have up to 100 nucleotide spacers separating the arms of the repeat. The subsets of cruciforms are defined as those inverted repeats with up to four nucleotides spacers. Minimum repeat length for mirror repeats is 10 nucleotides; the subset of mirror repeats with >90% pyrimidines on one strand and less than eight nucleotide spacers are flagged as triplex-forming motifs. The G-quadruplex algorithm identifies four or more individual G-runs of at least three nucleotides in length. The algorithm requires at least one nucleotide between each run and considers up to seven nucleotides as spacer, including guanines. The Z-DNA algorithm searches for alternative purine–pyrimidine tracts of at least 10 nucleotides with the exclusion of AT/TA dinucleotides. The A-phased motifs are defined as three or more tracts of four to nine adenines or adenines followed by thymines, with centers separated by 11–12 nucleotides. In addition to the motifs described in the original database release, we have added short tandem repeats (STRs) as a separate sequence motif. The STR algorithm searches for repeats of one to nine nucleotides repeated at least three times with no spacers between repeat units and could be as simple as a mononucleotide repeat or a complex nine-nucleotide repeat such as GATCAACGG GATCAACGG GATCAACGG. The algorithm only counts the perfect repeats and does not allow for interruptions in the repeat units. In refining the motif definitions, particular care has been taken to collapse overlapping motifs of the same type where possible. For example, where a long dinucleotide repeat is broken up partially along its length, the smaller contiguous repeat sequence may form several valid direct repeats, mirror repeats or inverted repeats with the remainder of the repeat. Analogous overlapping repeats are possible, but more rare, with more complicated sequences. In all cases, only one result is returned for a given motif, and the number of valid permutations and overall motif length are reported in the output field.
RE-ANNOTATION OF V1.0 ORGANISMS
We have re-annotated the five organisms, human, mouse, chimpanzee, macaque and dog, reported in the original non-B DB (14). As the new definitions of A-phased repeats are more restrictive (they do not allow thymine-adenine-TpA-steps), the total number of this motif decreased from ∼1 million to ∼300k. Overall, the numbers of inverted, direct and mirror repeats all increased in the new version, as the algorithm now searches for longer mirror and direct repeats, and inverted repeats start at a minimum of 6 nucleotides as opposed to 10 in version 1.0. The updated annotations also include the newly added STRs. Version 1.0 is still available under ‘nonbpDAS’, whereas version 2.0 is listed as ‘nonbDAS’.
ADDITION OF NEW ORGANISMS
With the ever increasing number of sequenced whole genomes, we have updated the non-B DB with the annotations of thale cress, cow, horse, orangutan, pig, rat and platypus. The addition of platypus and thale cress (Arabidopsis thaliana), the first plant genome in the database, extends the evolutionary reach of the database (Table 1).
Non-B DB v2.0 motif counts by organism
| Species . | A-phased repeat . | G-quadruplex . | Z-DNA . | Direct repeat . | Inverted repeat . | Mirror repeat . | STR . |
|---|---|---|---|---|---|---|---|
| Horse 2.2 | 351 711 | 332 283 | 340 218 | 512 314 | 4 823 814 | 894 099 | 1 599 735 |
| Human 37.1 | 404 289 | 361 419 | 412 600 | 1 501 567 | 6 365 102 | 1 895 545 | 3 025 648 |
| Mouse 37.1 | 324 077 | 482 833 | 877 676 | 2 393 352 | 5 244 776 | 2 801 083 | 3 649 837 |
| Rat 4.2 | 318 619 | 417 278 | 953 455 | 2 265 975 | 4 895 917 | 2 570 060 | 3 118 948 |
| Dog 2.1 | 310 071 | 487 193 | 375 567 | 1 519 410 | 5 362 953 | 1 977 051 | 3 602 105 |
| Chimp 2.1 | 390 907 | 326 097 | 389 608 | 1 355 391 | 6 150 067 | 1 761 969 | 2 849 252 |
| Macaque 1.1 | 373 161 | 293 036 | 387 648 | 1 358 285 | 5 746 000 | 1 760 255 | 2 827 111 |
| Orangutan 1.2 | 386 971 | 314 395 | 378 166 | 1 297 200 | 6 038 798 | 1 700 296 | 2 800 053 |
| Cow 4.1 | 284 375 | 3 9 834 | 355 796 | 751 541 | 5 131 526 | 979 843 | 1 960 468 |
| Pig 4.1 | 308 331 | 414 700 | 342 753 | 1 077 302 | 4 856 703 | 1 518 336 | 2 737 708 |
| Platypus 1.1 | 47 493 | 74 309 | 35 765 | 118 410 | 718 314 | 149 526 | 451 324 |
| A. thaliana | 24 909 | 1219 | 6299 | 33 311 | 283 522 | 62 634 | 125 949 |
| Species . | A-phased repeat . | G-quadruplex . | Z-DNA . | Direct repeat . | Inverted repeat . | Mirror repeat . | STR . |
|---|---|---|---|---|---|---|---|
| Horse 2.2 | 351 711 | 332 283 | 340 218 | 512 314 | 4 823 814 | 894 099 | 1 599 735 |
| Human 37.1 | 404 289 | 361 419 | 412 600 | 1 501 567 | 6 365 102 | 1 895 545 | 3 025 648 |
| Mouse 37.1 | 324 077 | 482 833 | 877 676 | 2 393 352 | 5 244 776 | 2 801 083 | 3 649 837 |
| Rat 4.2 | 318 619 | 417 278 | 953 455 | 2 265 975 | 4 895 917 | 2 570 060 | 3 118 948 |
| Dog 2.1 | 310 071 | 487 193 | 375 567 | 1 519 410 | 5 362 953 | 1 977 051 | 3 602 105 |
| Chimp 2.1 | 390 907 | 326 097 | 389 608 | 1 355 391 | 6 150 067 | 1 761 969 | 2 849 252 |
| Macaque 1.1 | 373 161 | 293 036 | 387 648 | 1 358 285 | 5 746 000 | 1 760 255 | 2 827 111 |
| Orangutan 1.2 | 386 971 | 314 395 | 378 166 | 1 297 200 | 6 038 798 | 1 700 296 | 2 800 053 |
| Cow 4.1 | 284 375 | 3 9 834 | 355 796 | 751 541 | 5 131 526 | 979 843 | 1 960 468 |
| Pig 4.1 | 308 331 | 414 700 | 342 753 | 1 077 302 | 4 856 703 | 1 518 336 | 2 737 708 |
| Platypus 1.1 | 47 493 | 74 309 | 35 765 | 118 410 | 718 314 | 149 526 | 451 324 |
| A. thaliana | 24 909 | 1219 | 6299 | 33 311 | 283 522 | 62 634 | 125 949 |
Non-B DB v2.0 motif counts by organism
| Species . | A-phased repeat . | G-quadruplex . | Z-DNA . | Direct repeat . | Inverted repeat . | Mirror repeat . | STR . |
|---|---|---|---|---|---|---|---|
| Horse 2.2 | 351 711 | 332 283 | 340 218 | 512 314 | 4 823 814 | 894 099 | 1 599 735 |
| Human 37.1 | 404 289 | 361 419 | 412 600 | 1 501 567 | 6 365 102 | 1 895 545 | 3 025 648 |
| Mouse 37.1 | 324 077 | 482 833 | 877 676 | 2 393 352 | 5 244 776 | 2 801 083 | 3 649 837 |
| Rat 4.2 | 318 619 | 417 278 | 953 455 | 2 265 975 | 4 895 917 | 2 570 060 | 3 118 948 |
| Dog 2.1 | 310 071 | 487 193 | 375 567 | 1 519 410 | 5 362 953 | 1 977 051 | 3 602 105 |
| Chimp 2.1 | 390 907 | 326 097 | 389 608 | 1 355 391 | 6 150 067 | 1 761 969 | 2 849 252 |
| Macaque 1.1 | 373 161 | 293 036 | 387 648 | 1 358 285 | 5 746 000 | 1 760 255 | 2 827 111 |
| Orangutan 1.2 | 386 971 | 314 395 | 378 166 | 1 297 200 | 6 038 798 | 1 700 296 | 2 800 053 |
| Cow 4.1 | 284 375 | 3 9 834 | 355 796 | 751 541 | 5 131 526 | 979 843 | 1 960 468 |
| Pig 4.1 | 308 331 | 414 700 | 342 753 | 1 077 302 | 4 856 703 | 1 518 336 | 2 737 708 |
| Platypus 1.1 | 47 493 | 74 309 | 35 765 | 118 410 | 718 314 | 149 526 | 451 324 |
| A. thaliana | 24 909 | 1219 | 6299 | 33 311 | 283 522 | 62 634 | 125 949 |
| Species . | A-phased repeat . | G-quadruplex . | Z-DNA . | Direct repeat . | Inverted repeat . | Mirror repeat . | STR . |
|---|---|---|---|---|---|---|---|
| Horse 2.2 | 351 711 | 332 283 | 340 218 | 512 314 | 4 823 814 | 894 099 | 1 599 735 |
| Human 37.1 | 404 289 | 361 419 | 412 600 | 1 501 567 | 6 365 102 | 1 895 545 | 3 025 648 |
| Mouse 37.1 | 324 077 | 482 833 | 877 676 | 2 393 352 | 5 244 776 | 2 801 083 | 3 649 837 |
| Rat 4.2 | 318 619 | 417 278 | 953 455 | 2 265 975 | 4 895 917 | 2 570 060 | 3 118 948 |
| Dog 2.1 | 310 071 | 487 193 | 375 567 | 1 519 410 | 5 362 953 | 1 977 051 | 3 602 105 |
| Chimp 2.1 | 390 907 | 326 097 | 389 608 | 1 355 391 | 6 150 067 | 1 761 969 | 2 849 252 |
| Macaque 1.1 | 373 161 | 293 036 | 387 648 | 1 358 285 | 5 746 000 | 1 760 255 | 2 827 111 |
| Orangutan 1.2 | 386 971 | 314 395 | 378 166 | 1 297 200 | 6 038 798 | 1 700 296 | 2 800 053 |
| Cow 4.1 | 284 375 | 3 9 834 | 355 796 | 751 541 | 5 131 526 | 979 843 | 1 960 468 |
| Pig 4.1 | 308 331 | 414 700 | 342 753 | 1 077 302 | 4 856 703 | 1 518 336 | 2 737 708 |
| Platypus 1.1 | 47 493 | 74 309 | 35 765 | 118 410 | 718 314 | 149 526 | 451 324 |
| A. thaliana | 24 909 | 1219 | 6299 | 33 311 | 283 522 | 62 634 | 125 949 |
ADDITION OF KEY VISUALIZATION TOOLS
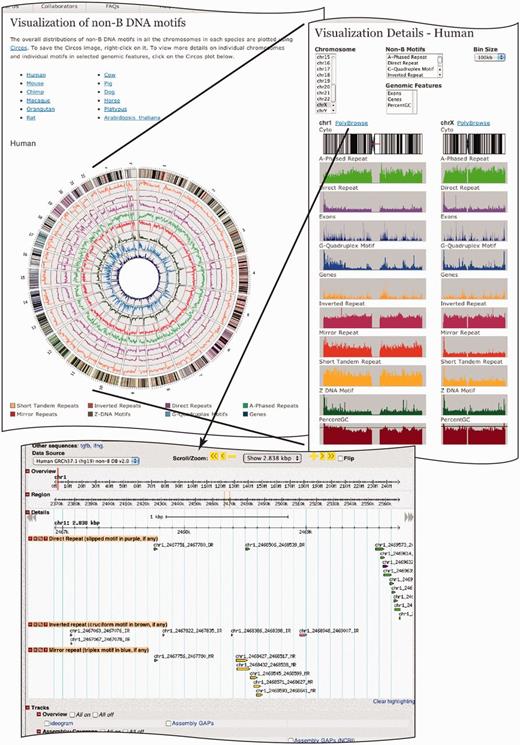
The most exciting enhancement in non-B DB v2.0 is the addition of graphical tools (http://nonb.abcc.ncifcrf.gov/apps/site/visualization/) to better visualize and interpret the genome-wide distributions of non-B DNA motifs. Non-B DB v2.0 allows for customized visual browsing of the non-B DNA motifs with three levels of visualization: (i) whole genome-level visualization using Circos plots (16); (ii) chromosomal-level visualization with custom histogram plots; and (iii) nucleotide-level visualization using the existing PolyBrowse interface.
Circos plots
Circos plots have been created for all the genomes annotated in non-B DB along with gene density. Where cytogenetic band information is available, it is included as the outermost circle in the Circos plot and the gene features and non-B DNA motifs are color coded (Figure 1).
A screen shot of the visualization page in non-B DB. The top left panel displays clickable anchor text links to all the available genomes in non-B DB, whereas the bottom panel displays the Circos plot for the human genome. The motifs are color coded as shown in the panel. Clicking on the Circos plots takes the user to the chromosome-wide non-B DNA motif histograms on the top right panel. Users are able to choose the chromosome and non-B DNA motif of interest and compare with available genomic features such as exons, genes and percent GC content. The histograms are available in 100-, 500- and 1000-kb bin sizes. Chromosome 1 and chromosome X are compared side by side as an example. The bottom panel displays the PolyBrowse tracks showing subset motifs for a region of chromosome 1. In ‘direct repeats’ tracks, the main motifs are in green, whereas the subset slipped motifs are in purple. In ‘inverted repeats’ tracks, the main motifs are in pink, whereas the subset cruciform motifs are in brown (not shown). Similarly, in ‘mirror repeats’ tracks, the main motifs are in yellow, whereas the subset triplexes are in blue.
Histogram plots
Clicking on a particular Circos plot takes the user to chromosomal-level histogram plots for that species (Figure 1, top right panel). Histogram plots have been generated for all non-B DNA motifs and the genomic annotation features genes, exons and percent GC content, for all the chromosomes of all non-B DB annotated genomes, and are available in three pre-computed bin sizes: 100, 500 and 1000 kb. The chromosome details page allows the user to select one or multiple chromosomes, non-B DNA motifs and genomic features, where the web layouts are dynamically generated with a chromosome cytogenetic band plot followed by each chromosome’s feature plots. The widths of the chromosomal plots are scaled accordingly to the chromosome length (Figure 1, top right panel). Any one of the three bin sizes, 100, 500 and 1000 kb, can also be selected from a drop-down box. The dynamic plot layout capability allows for customized visualization and comparison of genomic distributions across motif types, features and chromosomes. Users may save a high-resolution image by clicking on an individual histogram that opens up in a new browser tab.
PolyBrowse
Each chromosome in the histogram display page is also linked to the PolyBrowse web interface (http://pbrowse3.abcc.ncifcrf.gov/cgi-bin/gb2/gbrowse/), where the chromosomal region to be visualized can be controlled by the user, and all non-B DNA motifs, their subsets and many genomic annotations are displayed as color-coded tracks (Figure 1, bottom panel). This functionality was described in v1.0 in detail. Recent studies on G-quadruplexes (17) and inverted repeats (7) have shown an association between these non-B motifs and gene features (Figure 1), such as transcription start sites. By facilitating the comparison between non-B DNA motif and gene feature distributions, the new genome- and chromosome-wide visualization features in v2.0 of non-B DB provide researchers an opportunity to generate new hypotheses regarding the role and association of non-B DNA motifs with cellular processes.
UPDATED nBMST
As we still lack complete reference genomes for most organisms, we recognize that users may wish to annotate and visualize DNA segments specific to their research; thus, we provide an accessory tool, nBMST, where users can submit sequences of up to 20 Mb for annotation of potential non-B DNA-forming regions (15). Specifically, nBMST is intended to enhance the applicability of the motif-finding algorithms to viral and bacterial genomes.
OVERALL GRAPHICAL USER INTERFACE IMPROVMENT
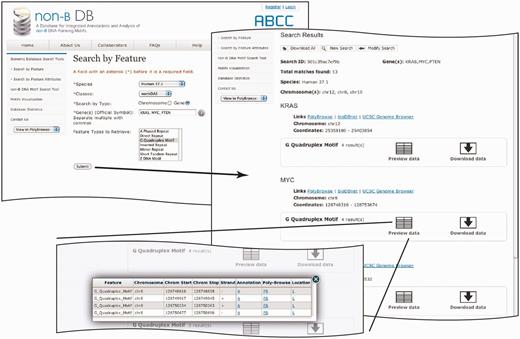
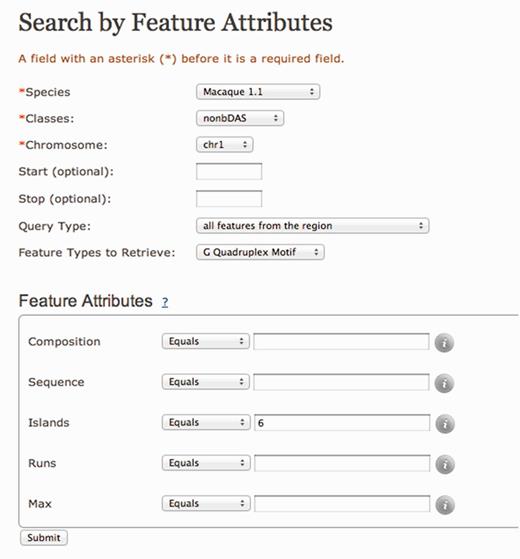
In addition to updates to the algorithmic methods, content and visualization tools, non-B DB v2.0 incorporates an enhanced graphical user interface to allow streamlined access to the data. Version 2.0 has eliminated ‘Search by Location’ and consolidated it with ‘Search by Feature’ (Figure 2). In addition, ‘Search by Feature Attributes’ (Figure 3) has been improved significantly by including more details on the type of query a user can make (http://nonb.abcc.ncifcrf.gov/apps/faq/publicFaqPage#146-20). This allows users to make specific questions, such as how many G-quadruplexes with six islands are in chromosome 1 of macaques. This can be performed by specifying the number of islands as shown in Figure 3.
Search by Feature Attributes page flow. The improved graphical interface of the query result page in the top right panel shows the overall summary at the top followed by details on three different genes, KRAS, MYC and PTEN. Each gene has a separate section. Preview data for each motif give the top results for each query in the bottom left panel.
Search by Feature Attributes page with G-quadruplex motif as an example. Search by features allows for multiple filters for each feature. In the case of G-quadruplexes, users can filter the results based on base composition, sequence, number of G islands and number of G runs, and the largest G-quadruplex can be formed. Each filter can have one or more values, such as ‘equal’, ‘not equal’, ‘less than’ and ‘greater than’ allowing flexibility in the filtering process.
The updated query results page includes a ‘download all link’, ‘search criteria’, ‘species’, ‘chromosome number’ and ‘search identification number’ (Figure 2). In addition, each motif result is provided with a preview and the download links (Figure 2, top left and bottom panels). The preview link will only show a small number of results; to retrieve all the results, users must use download data links. The preview data contain individual motifs with links to the location, PolyBrowse and annotation. The new annotation page contains the base composition of each motif, the sequence, total sequence length, spacer length, repeat unit and number of permutations where applicable.
HOW TO ACCESS SUBSETS
Cruciform structures, triplex motifs and slipped structures are subsets of inverted, mirror and direct repeats, respectively. These subsets can be accessed through the motif tracks in PolyBrowse, where the subset motifs are colored differently than their parent motif (Figure 1, bottom panel).
FUTURE DIRECTIONS
We plan to update the database as reference genomes are revised and new genomes become available. In addition, we are working on introducing imperfect repeats to our algorithm, adding additional annotations to PolyBrowse, such as chromosomal break points, locations of cancer translocations, DNA copy number variations and structural variants and fragile sites, all of which may correlate with the existence of various subclasses of non-B DNA-forming repeats. We are also working to improve interspecies navigation and comparison capabilities.
CONTACTING US
Given the ability to produce large genome assemblies, the 20-Mb limit of the user input sequence annotation site may be insufficient in some cases. If this is the case, or if specific parameter subsets are desired, please contact us. Because we anticipate the addition of new genomes at periodic intervals, we have added a mailing list to the website. Users may supply their email address to receive notification of these updates. The mailing access can be accessed by following the link on the left of the main web page.
FUNDING
Funding for open access charge: This work was supported in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E and funding from CBIIT/caBIG ISRCE yellow task #09-260 to NCI-Frederick.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
REFERENCES
Author notes
Present address: Regina Z. Cer, Biological Defense Research Directorate, Naval Medical Research Center, Henry Jackson Foundation for the Advancement of Military Medicine, Frederick, MD 21702, USA.




Comments