-
PDF
- Split View
-
Views
-
Cite
Cite
Yu-Jin Wang, Jian-Quan Liu, Georg Miehe, Phylogenetic Origins of the Himalayan Endemic Dolomiaea , Diplazoptilon and Xanthopappus (Asteraceae: Cardueae) Based on Three DNA Regions , Annals of Botany, Volume 99, Issue 2, February 2007, Pages 311–322, https://doi.org/10.1093/aob/mcl259
Close - Share Icon Share
Abstract
It is an enduring question as to the mechanisms leading to the high diversity and the processes producing endemics with unusual morphologies in the Himalayan alpine region. In the present study, the phylogenetic relationships and origins of three such endemic genera were analysed, Dolomiaea , Diplazoptilon and Xanthopappus , all in the tribe Cardueae of Asteraceae.
The nuclear rDNA internal transcribed spacer (ITS) and plastid trnL-F and psbA-trnH regions of these three genera were sequenced. The same regions for other related genera in Cardueae were also sequenced or downloaded from GenBank. Phylogenetic trees were constructed from individual and combined data sets of the three types of sequences using maximum parsimony, maximum likelihood and Bayesian analyses.
The phylogenetic tree obtained allowed earlier hypotheses concerning the relationships of these three endemic genera based on gross morphology to be rejected. Frolovia and Saussurea costus were deeply nested within Dolomiaea , and the strong statistical support for the Dolomiaea–Frolovia clade suggested that circumscription of Dolomiaea should be more broadly redefined. Diplazoptilon was resolved as sister to Himalaiella , and these two together are sister to Lipschitziella . The clade comprising these three genera is sister to Jurinea , and together these four genera are sister to the Dolomiaea–Frolovia clade. Xanthopappus , previously hypothesized to be closely related to Carduus , was found to be nested within a well-supported but not fully resolved Onopordum group with Alfredia , Ancathia , Lamyropappus , Olgaea , Synurus and Syreitschikovia , rather than the Carduus group. The crude dating based on ITS sequence divergence revealed that the divergence time of Dolomiaea – Frolovia from its sister group probably occurred 13·6–12·2 million years ago (Ma), and the divergence times of the other two genera, Xanthopappus and Diplazoptilon , from their close relatives around 5·7–4·7 Ma and 2·0–1·6 Ma, respectively.
The findings provide an improved understanding of the intergeneric relationships in Cardueae. The crude calibration of lineages indicates that the uplifts of the Qiinghai–Tibetan Plateau since the Miocene might have served as a continuous stimulus for the production of these morphologically aberrant endemic elements of the Himalayan flora.
INTRODUCTION
The Qinghai–Tibetan Plateau (QTP) is the highest and largest plateau in the world, having a mean elevation of >4·0 km and an area of about 2·5×10 6 km 2 ( Zheng, 1996 ). Plants occurring there comprise major components of the Himalayan alpine flora ( Wu, 1987 ; Wu et al. , 1995 ). This region, with southeast China, forms the Himalayan biodiversity hotspot, and has been designated as one of the 34 most important centres of biodiversity because of its high species richness and abundance of endemic species ( Wilson, 1992 ; Myers et al. , 2000 ). The mechanisms leading to this high diversity and the processes producing endemics, often with unusual morphologies, have been the subject of much interest ( Wulff, 1943 ; Wu, 1987 ; Wu et al. , 1995 ; Liu et al. , 2002 , 2006 ).
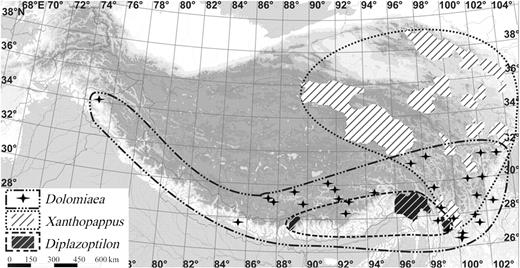
Cardueae sensu lato ( s.l. ) is a large tribe of the family Asteraceae, with about 2500 species in >80 genera mainly distributed in the Northern Hemisphere of the Old World. It is traditionally classified into four sub-tribes (Echinopsidinae, Carlininae, Carduinae and Centaureinae; Bremer, 1994 ). There is considerable controversy regarding the tribal delimitation of Cardueae, especially relating to the segregation of sub-tribes Echinopsidinae and Carlininae as separate tribes (e.g. Wagenitz, 1976 ; Dittrich, 1977 ; Bremer, 1994 ). However, molecular evidence supports the maintenance of Cardueae in its broad sense, and has also revealed paraphyly of sub-tribe Carduinae ( Susanna et al. , 1995 , 2006 ; Häffner and Hellwig, 1999 ; García-Jacas et al. , 2002 ). Carduinae is the largest sub-tribe of Cardueae, comprising nearly 40 genera and about 1600 species ( Bremer, 1994 ). Within this sub-tribe, some genera show extensive diversification and are species rich, for example Cousinia (approx. 600 species) and Saussurea (approx. 400 species). However, there are far fewer species in genera Dolomiaea , Diplazoptilon and Xanthopappu s, and all these three genera are endemic to the Himalayan region (Fig. 1 ; Ling, 1965 ; Lipschitz, 1979 ; Chen, 1987 ; Shi, 1987 , 1994 ; Ying and Zhang, 1994 ; Liu, 1996 ).
Distributions of Dolomiaea , Diplazoptilon and Xanthopappus in the Himalayan–Tibetan region. The distributions are outlined according to specimen collections in the field and in herbaria.
Dolomiaea , a small genus with approx. 14 species, is restricted to alpine habitats between 2900 and 4800 m of the QTP ( Ling, 1965 ; Shi, 1987 , 1994 ). It has been suggested that it could be related to Carduus , Jurinea or Saussurea s.l. , and it is distinguished from these related genera by the presence of scabrid pappus bristles and alveolate receptacles without scales ( Ling, 1965 ; Shi, 1986 ). Two sections were suggested within the genus based on their style differences (long and acute vs. short and round) ( Shi, 1986 ), and these two sections were previously treated as two distinct genera by Ling (1965) , Dolomiaea and Vladimiria. Diplazoptilon contains only two species, D. picridifolium and D. cooperi , and is distributed in NW Yunnan and SE Tibet on the QTP at an elevation of 3600–4100 m. This genus has double rows of plump pappus bristles as a diagnostic feature, and was suggested to be related to Dolomiaea or Saussurea ( Ling, 1965 ; Shi and Jin, 1983 ; Shi, 1986 , 1987 ). Xanthopappus is a monotypic genus which is commonly found on the dry slopes and degraded alpine meadows of the QTP between 2400 and 4000 m. This genus was always considered to have a close relationship to Carduus ( Dittrich, 1977 ; Shi, 1986 , 1987 ; Bremer, 1994 ; Wu et al. , 2003 ). Xanthopappus differs from Carduus in having yellow florets and smooth filaments, but Smith (1917) considered that these differences were insufficient to establish a separate genus, and he suggested reducing this monotypic genus into Carduus . However, this proposal was not adopted by later researchers (e.g. Dittrich, 1977 ; Shi, 1987 ; Bremer, 1994 ).
Within a paraphyletic Carduinae, molecular data have revealed four distinct groups represented by Saussurea , Carduus, Onopordum and Xeranthemum , together with some genera without known affinities such as Berardia or Staehelina ( Susanna et al. , 1995 , 2006 ; Häffner and Hellwig, 1999 ; García-Jacas et al. , 2002 ). Although the Himalayan endemic Dolomiaea , Diplazoptilon and Xanthopappus were all ascribed to the traditional sub-tribe Carduinae, their hypothesized close relatives were evidently placed in the different clades identified by these molecular analyses. In addition, within the Saussurea group, intergeneric relationships remain unresolved. A recent investigation of the species-rich Saussurea s.l. , which has abundant endemics in the QTP, revealed that this genus is heterogeneous, and three genera were separated: Himalaiella , Frolovia and Lipschitziella ( Raab-Straube, 2003 ). Both Himalaiella and Lipschitziella are endemic to the Himalayan region, and Frolovia occurs in the adjacent highlands. Their phylogenetic and taxonomic relationships to other endemic genera from the Himalayan region, especially Diplazoptilon and Dolomiaea , remain unclear, although Susanna et al. (2006) proposed that Himalaiella , Lipschitziella and Frolovia should be reduced to synonymy with Jurinea and Dolomiaea , respectively. Apart from these taxa, section Jacea of sub-genus Saussurea and section Aucklandia of sub-genus Frolovia should also be excluded from Saussurea (Y.-J. Wang et al ., unpubl. res.). The remaining species from other sections and sub-genera of the Saussurea s.l. , designated as Saussurea sensu stricto ( s.s. ), comprise a well-supported clade, despite poor resolution within it ( Raab-Straube, 2003 ; Wang and Liu, 2004 a , b ; Wang et al. , 2005 b ; Y.-J. Wang et al ., unpubl. res.), and is possibly sister to Hemistepta or Polytaxis ( Kita et al. , 2004 ; Susanna et al. , 2006 ).
DNA data, particularly DNA sequences, have greatly contributed to the understanding of the phylogenetics, evolution and taxonomy of Asteraceae (e.g. Jansen and Kim, 1996 ; Bayer and Starr, 1998 ; Goertzen et al. , 2003 ). This is particularly true for elucidating generic delimitation and phylogenetic relationships of the problematic genera in Cardueae, for which morphological data are lacking or ambiguous ( Susanna et al. , 1995 , 1999 , 2006 ; Häffner and Hellwig, 1999 ; García-Jacas et al. , 2001 , 2002 ; Raab-Straube, 2003 ; Hellwig, 2004 ; Kita et al. , 2004 ; Wang and Liu, 2004 a , b ; Martins and Hellwig, 2005 a , b ; Wang et al. , 2005 b ; Hidalgo et al. , 2006 ). Molecular reconstructions of evolutionary relationships between living organisms are increasingly used to infer the putative causes of diversification processes, and origin of endemics within historic and geographic contexts (e.g. Kelch and Baldwin, 2003 ; Pennington et al. , 2004 a , b ). Based on the new plastid and nuclear DNA sequence data for representative species of the Himalayan endemic genera Dolomiaea , Diplazoptilon and Xanthopappus and related genera of Cardueae, the main objectives of this study were to: ( a ) evaluate the systematic positions of these three genera; ( b ) estimate possible time scales for the origin of these endemic genera within the same phylogenetic frame; and ( c ) infer possible corresponding geological correlations with uplifts of the QTP, which are also useful for seeking general rules that can account for origins of the alpine flora with aberrant morphology occurring there.
MATERIALS AND METHODS
Sampling strategy, plant materials and data sets
According to Susanna et al. (2006) , Diplazoptilon , Himalaiella and Lipschitziella should be reduced into Jurinea and Frolovia into Dolomiaea . In addition, they further suggested that the monotypic Hemistepta should be incorporated into Saussurea . However, they were tentatively treated as distinct genera following the available taxonomic treatments for two reasons. First, the sampled species are limited and an extensive investigation of more species is needed. Secondly, their taxonomic relationships are complex and the final taxonomic treatments have to depend on more morphological evidence in addition to molecular support. Up to now, morphological investigations are lacking, and how broadly to circumscribe these genera still remains elusive.
Dolomiaea in its current circumscription contains about 15 species, of which 14 species were previously recorded ( Ling, 1965 ; Shi, 1987 , 1994 ) and one was newly found during the present field studies (unpublished new species). Because two infrageneric sections ( Dolomiaea and Vladimiria ) had been treated as two separate genera ( Ling, 1965 ), the sampling strategy was also designed to test whether they should still be segregated. Section Dolomiaea comprises three species: D. calophylla , D. wardii and one new unpublished species ‘ D. tibetica ’, and section Vladimiria contains the remaining 11 species. Dolomiaea calophylla and ‘ D. tibetica ’ were chosen to represent section Dolomiaea , and three species ( D. scabrida , D. souliei and D. edulis ) to represent section Vladimiria. Diplazoptilon comprises two closely related species, and of these only D. cooperi was sampled. The monotypic genus Xanthopappus was collected from two localities, and its hypothesized related genera were mainly chosen according to their distribution range and possible systematic positions within the whole of Cardueae, based on morphology and recent molecular phylogenetic analyses ( Shi, 1987 ; Bremer, 1994 ; Susanna et al. , 1995 , 2006 ; Häffner and Hellwig, 1999 ; Häffner, 2000 ; García-Jacas et al. , 2002 ; Raab-Straube, 2003 ; Kita et al. , 2004 ; Wang and Liu, 2004 a , b ; Wang et al. , 2005 b ). The analysis of these genera covered the major clades identified in the recent molecular analyses of this tribe, and all genera of Cardueae occurring in the QTP were sampled. For those genera with abundant species (e.g. Cousinia , Jurinea and Onopordum ), only one species was chosen as a representative of each genus. Saussurea has been the subject of extensive sampling for molecular analyses ( Raab-Straube, 2003 ; Kita et al. , 2004 ; Wang and Liu, 2004 a , b ; Wang et al. , 2005 b ), and the type species S. alpina was therefore well suited to represent the monophyletic assemblage of Saussurea s.s. However, S. costus , a species in section Aucklandia of sub-genus Frolovia and S. forestii in section Jacea of sub-genus Saussurea were also sampled because unpublished data revealed that these species should be excluded from Saussurea . The other genera involved in the present analyses might not be monophyletic, but this should not affect the assessment of relationships and origins of three endemic genera because they are mainly distributed in Central Asia or Europe, far away from the QTP. Most genera of Cardopatiinae, Carlininae, Centaureinae and Echinopsidinae were found to comprise corresponding monophyletic lineages, except for a few aberrant genera with unresolved positions ( Häffner and Hellwig, 1999 ; Häffner, 2000 ; García-Jacas et al. , 2002 ; Susanna et al. , 2006 ), and therefore only one genus was chosen to represent each lineage.
Among 22 newly sampled taxa, from 16 genera, leaves were collected from herbaria specimens for nine taxa, and leaves of the remaining species were directly collected in the field and dried with silica gel. Their origins are listed in Table 1 , and the voucher specimens are deposited in the Northwest Plateau Institute of Biology, Chinese Academy of Sciences. Data sets are based on the sequences of the nuclear ribosomal internal transcribed spacer (ITS), plastid trnL intron and trnL-F intergenic spacer ( trnL-F ), and plastid psbA-trnH intergenic spacer ( psbA-trnH ) regions. Newly available sequences, accession numbers and the origin of materials are also given in Table 1 . The other sequences for analyses were downloaded from GenBank, and their origins are referred to in Susanna et al. (1995 , 2006 ), Häffner and Hellwig (1999) , O'Hanlon and Peakall (2000) , García-Jacas et al. (2002) , Kelch and Baldwin (2003) and Kita et al. (2004) . Because of the poor DNA extracted from the specimens and unavailable DNA for those species for which the sequences were downloaded from GenBank, all three fragments were not obtained for all species.
List of taxa and sources of plant material analysed for the first time in the present study and the sequence accession numbers in GenBank
| Species . | Origins . | ITS . | trnL-F . | psbA-trnH . |
|---|---|---|---|---|
| Ancathia igniaria (Spreng.) DC. | Xinjiang, Altai, China; Zhu6945 | AY914811 | — | — |
| Arctium lappa L. | Jiuzhi, Qinghai, China; Liu1834 | AY914812 | AY914854 | AY914834 |
| Carduus crispus L. | Tongren, Qinghai, China; Liu079 | AY914813 | AY914855 | AY914835 |
| Cirsium lidjiangense Petrak ex Hand.-Mazz. | Muli, Sichuan, China; Liu2137 | AY914828 | AY914856 | AY914836 |
| Diplazoptilon cooperi (Anthi.) Shih | Yadong, Xizang, China; 74–2338 | AY914814 | AY914857 | AY914837 |
| Dolomiaea calophylla Ling | Lasa, Xizang, China; Liu2565 | AY914816 | AY914859 | AY914839 |
| Dolomiaea edulis (Franch.) Shih | Hongyuan, Sichuan, China; Liu2191 | AY914817 | AY914860 | AY914840 |
| Dolomiaea scabrida (Shih et S. Y. Jin) Shih | Xizang, China; X102 | AY914818 | — | — |
| Dolomiaea souliei (Franch.) Shih | Lixian, Sichuan, China; Liu1942 | AY914815 | AY914858 | AY914838 |
| Dolomiaea tibetica S. W. Liu et J. Q. Liu | Sangri, Xizang, China; Liu1137 | AY914819 | — | — |
| Echinops przewalskii Iljin | Kunming, Yunnan, China; Liu2161 | AY914820 | AY914861 | AY914841 |
| Frolovia frolovii (Ledeb.) Raab-Straube | Altai, Xinjiang, China; A1194 | AY914822 | AY914862 | AY914842 |
| Hemistepta lyrata (Bunge) Bunge | Beijing, China; Wang040601 | AY914824 | AY914864 | AY914844 |
| Himalaiella deltoidea (DC.) Raab-Straube | Gonggashan, Sichuan, China; Liu2072 | AY914825 | AY914865 | AY914845 |
| Jurinea multiflora (L.) B. Fedtch. | Tacheng, Xinjiang, China; Guan4505 | AY914826 | — | AY914846 |
| Onopordum acanthium L. | Urumchi, Xinjiang, China; 79–149 | AY914827 | AY914866 | AY914847 |
| Saussurea alpina (L.) DC. | Upsaliensis, Suesica; 060802 | AY914829 | AY914867 | AY914848 |
| Saussurea costu s (Falc.) Raab-Straube | Weixi, Yunnan, China; Wang040602 | AY914821 | DQ874335 | DQ874336 |
| Saussurea forestii Diels | Cuona, Xizang, China; Qinghai-Tibet expedition, 3200 | DQ874337 | DQ874338 | DQ874339 |
| Serratula strangulata Iljin | Datong, Qinghai, China; Liu1608 | AY914830 | AY914868 | AY914850 |
| Silybum marianum (L.) Gaertn. | Wuding, Yunnan, China; Liu2143 | AY914831 | AY914869 | AY914849 |
| Xanthopappus subacaulis C. Winkl. | Henan, Qinghai, China; Liu050 (1) | AY914832 | AY914870 | AY914851 |
| Qilian, Qinghai, China; Liu1521 (2) | AY914833 | AY914871 | AY914852 |
| Species . | Origins . | ITS . | trnL-F . | psbA-trnH . |
|---|---|---|---|---|
| Ancathia igniaria (Spreng.) DC. | Xinjiang, Altai, China; Zhu6945 | AY914811 | — | — |
| Arctium lappa L. | Jiuzhi, Qinghai, China; Liu1834 | AY914812 | AY914854 | AY914834 |
| Carduus crispus L. | Tongren, Qinghai, China; Liu079 | AY914813 | AY914855 | AY914835 |
| Cirsium lidjiangense Petrak ex Hand.-Mazz. | Muli, Sichuan, China; Liu2137 | AY914828 | AY914856 | AY914836 |
| Diplazoptilon cooperi (Anthi.) Shih | Yadong, Xizang, China; 74–2338 | AY914814 | AY914857 | AY914837 |
| Dolomiaea calophylla Ling | Lasa, Xizang, China; Liu2565 | AY914816 | AY914859 | AY914839 |
| Dolomiaea edulis (Franch.) Shih | Hongyuan, Sichuan, China; Liu2191 | AY914817 | AY914860 | AY914840 |
| Dolomiaea scabrida (Shih et S. Y. Jin) Shih | Xizang, China; X102 | AY914818 | — | — |
| Dolomiaea souliei (Franch.) Shih | Lixian, Sichuan, China; Liu1942 | AY914815 | AY914858 | AY914838 |
| Dolomiaea tibetica S. W. Liu et J. Q. Liu | Sangri, Xizang, China; Liu1137 | AY914819 | — | — |
| Echinops przewalskii Iljin | Kunming, Yunnan, China; Liu2161 | AY914820 | AY914861 | AY914841 |
| Frolovia frolovii (Ledeb.) Raab-Straube | Altai, Xinjiang, China; A1194 | AY914822 | AY914862 | AY914842 |
| Hemistepta lyrata (Bunge) Bunge | Beijing, China; Wang040601 | AY914824 | AY914864 | AY914844 |
| Himalaiella deltoidea (DC.) Raab-Straube | Gonggashan, Sichuan, China; Liu2072 | AY914825 | AY914865 | AY914845 |
| Jurinea multiflora (L.) B. Fedtch. | Tacheng, Xinjiang, China; Guan4505 | AY914826 | — | AY914846 |
| Onopordum acanthium L. | Urumchi, Xinjiang, China; 79–149 | AY914827 | AY914866 | AY914847 |
| Saussurea alpina (L.) DC. | Upsaliensis, Suesica; 060802 | AY914829 | AY914867 | AY914848 |
| Saussurea costu s (Falc.) Raab-Straube | Weixi, Yunnan, China; Wang040602 | AY914821 | DQ874335 | DQ874336 |
| Saussurea forestii Diels | Cuona, Xizang, China; Qinghai-Tibet expedition, 3200 | DQ874337 | DQ874338 | DQ874339 |
| Serratula strangulata Iljin | Datong, Qinghai, China; Liu1608 | AY914830 | AY914868 | AY914850 |
| Silybum marianum (L.) Gaertn. | Wuding, Yunnan, China; Liu2143 | AY914831 | AY914869 | AY914849 |
| Xanthopappus subacaulis C. Winkl. | Henan, Qinghai, China; Liu050 (1) | AY914832 | AY914870 | AY914851 |
| Qilian, Qinghai, China; Liu1521 (2) | AY914833 | AY914871 | AY914852 |
List of taxa and sources of plant material analysed for the first time in the present study and the sequence accession numbers in GenBank
| Species . | Origins . | ITS . | trnL-F . | psbA-trnH . |
|---|---|---|---|---|
| Ancathia igniaria (Spreng.) DC. | Xinjiang, Altai, China; Zhu6945 | AY914811 | — | — |
| Arctium lappa L. | Jiuzhi, Qinghai, China; Liu1834 | AY914812 | AY914854 | AY914834 |
| Carduus crispus L. | Tongren, Qinghai, China; Liu079 | AY914813 | AY914855 | AY914835 |
| Cirsium lidjiangense Petrak ex Hand.-Mazz. | Muli, Sichuan, China; Liu2137 | AY914828 | AY914856 | AY914836 |
| Diplazoptilon cooperi (Anthi.) Shih | Yadong, Xizang, China; 74–2338 | AY914814 | AY914857 | AY914837 |
| Dolomiaea calophylla Ling | Lasa, Xizang, China; Liu2565 | AY914816 | AY914859 | AY914839 |
| Dolomiaea edulis (Franch.) Shih | Hongyuan, Sichuan, China; Liu2191 | AY914817 | AY914860 | AY914840 |
| Dolomiaea scabrida (Shih et S. Y. Jin) Shih | Xizang, China; X102 | AY914818 | — | — |
| Dolomiaea souliei (Franch.) Shih | Lixian, Sichuan, China; Liu1942 | AY914815 | AY914858 | AY914838 |
| Dolomiaea tibetica S. W. Liu et J. Q. Liu | Sangri, Xizang, China; Liu1137 | AY914819 | — | — |
| Echinops przewalskii Iljin | Kunming, Yunnan, China; Liu2161 | AY914820 | AY914861 | AY914841 |
| Frolovia frolovii (Ledeb.) Raab-Straube | Altai, Xinjiang, China; A1194 | AY914822 | AY914862 | AY914842 |
| Hemistepta lyrata (Bunge) Bunge | Beijing, China; Wang040601 | AY914824 | AY914864 | AY914844 |
| Himalaiella deltoidea (DC.) Raab-Straube | Gonggashan, Sichuan, China; Liu2072 | AY914825 | AY914865 | AY914845 |
| Jurinea multiflora (L.) B. Fedtch. | Tacheng, Xinjiang, China; Guan4505 | AY914826 | — | AY914846 |
| Onopordum acanthium L. | Urumchi, Xinjiang, China; 79–149 | AY914827 | AY914866 | AY914847 |
| Saussurea alpina (L.) DC. | Upsaliensis, Suesica; 060802 | AY914829 | AY914867 | AY914848 |
| Saussurea costu s (Falc.) Raab-Straube | Weixi, Yunnan, China; Wang040602 | AY914821 | DQ874335 | DQ874336 |
| Saussurea forestii Diels | Cuona, Xizang, China; Qinghai-Tibet expedition, 3200 | DQ874337 | DQ874338 | DQ874339 |
| Serratula strangulata Iljin | Datong, Qinghai, China; Liu1608 | AY914830 | AY914868 | AY914850 |
| Silybum marianum (L.) Gaertn. | Wuding, Yunnan, China; Liu2143 | AY914831 | AY914869 | AY914849 |
| Xanthopappus subacaulis C. Winkl. | Henan, Qinghai, China; Liu050 (1) | AY914832 | AY914870 | AY914851 |
| Qilian, Qinghai, China; Liu1521 (2) | AY914833 | AY914871 | AY914852 |
| Species . | Origins . | ITS . | trnL-F . | psbA-trnH . |
|---|---|---|---|---|
| Ancathia igniaria (Spreng.) DC. | Xinjiang, Altai, China; Zhu6945 | AY914811 | — | — |
| Arctium lappa L. | Jiuzhi, Qinghai, China; Liu1834 | AY914812 | AY914854 | AY914834 |
| Carduus crispus L. | Tongren, Qinghai, China; Liu079 | AY914813 | AY914855 | AY914835 |
| Cirsium lidjiangense Petrak ex Hand.-Mazz. | Muli, Sichuan, China; Liu2137 | AY914828 | AY914856 | AY914836 |
| Diplazoptilon cooperi (Anthi.) Shih | Yadong, Xizang, China; 74–2338 | AY914814 | AY914857 | AY914837 |
| Dolomiaea calophylla Ling | Lasa, Xizang, China; Liu2565 | AY914816 | AY914859 | AY914839 |
| Dolomiaea edulis (Franch.) Shih | Hongyuan, Sichuan, China; Liu2191 | AY914817 | AY914860 | AY914840 |
| Dolomiaea scabrida (Shih et S. Y. Jin) Shih | Xizang, China; X102 | AY914818 | — | — |
| Dolomiaea souliei (Franch.) Shih | Lixian, Sichuan, China; Liu1942 | AY914815 | AY914858 | AY914838 |
| Dolomiaea tibetica S. W. Liu et J. Q. Liu | Sangri, Xizang, China; Liu1137 | AY914819 | — | — |
| Echinops przewalskii Iljin | Kunming, Yunnan, China; Liu2161 | AY914820 | AY914861 | AY914841 |
| Frolovia frolovii (Ledeb.) Raab-Straube | Altai, Xinjiang, China; A1194 | AY914822 | AY914862 | AY914842 |
| Hemistepta lyrata (Bunge) Bunge | Beijing, China; Wang040601 | AY914824 | AY914864 | AY914844 |
| Himalaiella deltoidea (DC.) Raab-Straube | Gonggashan, Sichuan, China; Liu2072 | AY914825 | AY914865 | AY914845 |
| Jurinea multiflora (L.) B. Fedtch. | Tacheng, Xinjiang, China; Guan4505 | AY914826 | — | AY914846 |
| Onopordum acanthium L. | Urumchi, Xinjiang, China; 79–149 | AY914827 | AY914866 | AY914847 |
| Saussurea alpina (L.) DC. | Upsaliensis, Suesica; 060802 | AY914829 | AY914867 | AY914848 |
| Saussurea costu s (Falc.) Raab-Straube | Weixi, Yunnan, China; Wang040602 | AY914821 | DQ874335 | DQ874336 |
| Saussurea forestii Diels | Cuona, Xizang, China; Qinghai-Tibet expedition, 3200 | DQ874337 | DQ874338 | DQ874339 |
| Serratula strangulata Iljin | Datong, Qinghai, China; Liu1608 | AY914830 | AY914868 | AY914850 |
| Silybum marianum (L.) Gaertn. | Wuding, Yunnan, China; Liu2143 | AY914831 | AY914869 | AY914849 |
| Xanthopappus subacaulis C. Winkl. | Henan, Qinghai, China; Liu050 (1) | AY914832 | AY914870 | AY914851 |
| Qilian, Qinghai, China; Liu1521 (2) | AY914833 | AY914871 | AY914852 |
Three data sets were used for phylogenetic analyses. The first is composed of 49 nuclear ITS sequences from 41 genera of Cardueae and Brachylaena (outgroup) of Tarchonantheae (the closely related tribe; Panero and Funk, 2002 ). This data set was designed to detect the systematic positions of all three genera in Cardueae. The sequences covered both ITS1 and ITS2, but excluded the 5.8S gene because of the absence of any mutations in this fragment in the newly sampled species, and no corresponding record of this fragment for those species downloaded from GenBank. The construction of the second data set was based on the plastid trnL-F and psbA-trnH sequences from 27 accessions of 22 genera. The third data set was composed of both nuclear and plastid sequences from 27 taxa of 22 genera for which ITS, trnL-F and psbA-trnH sequences were all available. For the second and third analyses, Echinops of the sub-tribe Echinopinae was selected as the outgroup following previous reports ( Susanna et al. , 2006 ) and the present ITS analyses.
DNA extraction, amplification and sequencing
Total genomic DNA was isolated from samples directly dried in the field using the 2× CTAB (cetyltrimethyl ammonium bromide) method of Doyle and Doyle (1987) , and for samples from herbarium specimens with the Plant DNA Isolation Kit (Casarray, Shanghai, China) according to the manufacturer's protocol. The nuclear ITS region and the plastid trnL-F and psbA-trnH regions were amplified, respectively, with primers ITS1 and ITS4 ( White et al. , 1990 ), c and f ( Taberlet et al. , 1991 ) and psbA and trnH ( Sang et al. , 1997 ). The 25 µL volume polymerase chain reactions (PCRs) contained 12–60 ng of plant DNA, 50 m m Tris–HCl, 1.5 m m MgCl 2 , 0.5 m m dNTPs, 2 µ m of each primer and 0.75 U of Taq polymerase (Casarray, Shanghai, China). The PCR amplification profiles were identical for the three fragments: one cycle at 94 °C for 4 min; 32 cycles at 94 °C for 1 min, 50 °C for 1 min, 72 °C for 1·5 min; and one cycle at 72 °C for 7 min. PCR products were purified using a CASpure PCR Purification Kit following the protocol recommended by the manufacturer (Casarray, Shanghai, China). Sequencing primers were the same as those used for amplifying the corresponding regions. Sequencing reactions were carried out in a Biometra thermocycler using the DYEnamic ET terminator Cycle Sequencing Kit (Amersham Biosciences), following the manufacturer's protocol. Sequencing products were separated and analysed on a MegaBACE 500 DNA Analysis System. Both DNA stands were sequenced using forward and reverse primers, giving an overlap of at least 70 %.
Sequence alignment, boundary determination and data analysis
Alignment of all sequences was conducted using ClustalX ( Thompson et al. , 1997 ) and refined manually. Sequence boundaries were determined by comparison with published sequences of other genera of Asteraceae downloaded from GenBank. Pairwise distances, corrected with the Kimura 2-parameter (K2P) model ( Kimura, 1980 ), were calculated with MEGA version 3.1 ( Kumar et al. , 2004 ).
Each data set was subjected to maximum parsimony (MP) and maximum likelihood (ML) using PAUP* 4·0b10 ( Swofford, 2003 ) and Bayesian analyses using MrBayes 3·0 ( Huelsenbeck and Ronquist, 2001 ). Modeltest ( Posada and Crandall, 1998 ) was used to select parameters and assumptions for ML analyses. ML heuristic search parameters were simple addition of sequences of taxa with TBR branch swapping, MULTREES and COLLAPSE. MP analyses (equally weighted characters and nucleotide transformations) involved a heuristic search strategy with 100 replicates of random addition of sequences, in combination with ACCTRAN character optimization, MULPARS+TBR branch swapping and STEEPEST DESCENT options on. Gaps were treated as missing characters. The bootstrap values (BP) were calculated from 1000 replicates using a heuristic search with simple addition with TBR and MULPARS options on ( Felsenstein, 1985 ). For Bayesian analyses, four simultaneous Monte Carlo Markov chains (MCMCs) were run for 2 000 000 generations saving a tree every 100 generations. Because the fittest models selected for the analysed data sets were not implemented in MrBayes, a common model GTR+I+G was used for the different data sets in the Bayesian analyses. Base frequencies were empirically derived. A majority rule consensus tree was calculated with PAUP* from the last 16 001 out of the 20 001 trees sampled. The first 4000 trees (burn-in) were excluded to avoid trees that might have been sampled prior to convergence of the Markov chains. Posterior probability (shown as percentages, PP) of each topological bipartition was estimated by its frequency across all the 16 001 trees sampled. Congruence between different DNA data sets was evaluated by the incongruence-length-difference (ILD) test with 1000 replicates on parsimony-informative characters using the TBR branch swapping algorithm, with the number of trees retained for each replicate limited to 1000 ( Farris et al. , 1995 ).
Molecular calibration
Because the putative pseudogenes of ITS usually originate more recently than the functional copies ( Bailey et al. , 2003 ), their existence in the aligned data set could confound phylogenetic reconstruction and affect the dating of the divergence among the orthologous sequences ( Alvarez and Wendel, 2003 ). The strategies here included direct sequencing of PCR products in both forward and reverse orientations, and using only those sequences that were unambiguously resolved. In addition, sequences from multiple accessions of individual species were obtained whenever possible. It was also checked whether the newly available sequences are functional sequences or putative pseudogenes by examining nucleotide substitutions in a highly conserved region (5.8S gene), a relatively reliable indicator for discerning ITS orthologues ( Hershkovitz et al. , 1999 ). In addition, there was no suggestion in previously published and unpublished studies that indicated that the taxa targeted in this study have confounding paralogous sequences from the ITS region.
The hypothesis of rate constancy was evaluated with the likelihood ratio test (LRT) that is twice the difference in log likelihood (likelihoods) of branch lengths between a rate-constrained tree (forcing the molecular clock in PAUP*) and a tree that has no constraints on branch lengths ( Goldmann, 1993 ). The molecular clock was rejected because constrained and unconstrained analyses differed significantly, so semi-parametric rate smoothing with a penalized likelihood (PL) approach, based on the ML tree without a molecular clock enforced, was used to produce an ultrametric tree with the aid of the computer program r8s ( Sanderson, 2002 ). Penalized likelihood combines the likelihood term for a saturated model with a different rate on every branch and the non-parametric penalty function that keeps those rate estimates from varying excessively across the tree. The relative contribution of the two terms is controlled by a smoothing parameter which was determined based on a data-driven cross-validation procedure implemented in r8s ( Sanderson, 2002 ). To obtain standard deviations for estimated divergence times, the data set was bootstrapped 100 times using the SEQBOOT module from PHYLIP ( Felsenstein, 1989 ), and branch lengths were re-estimated for each node under the constrained initial topology in PAUP*. The dating analyses were then repeated for each tree, and node statistics were summarized using the PROFILE command of program r8s.
RESULTS
The analyses of the nuclear ITS data set
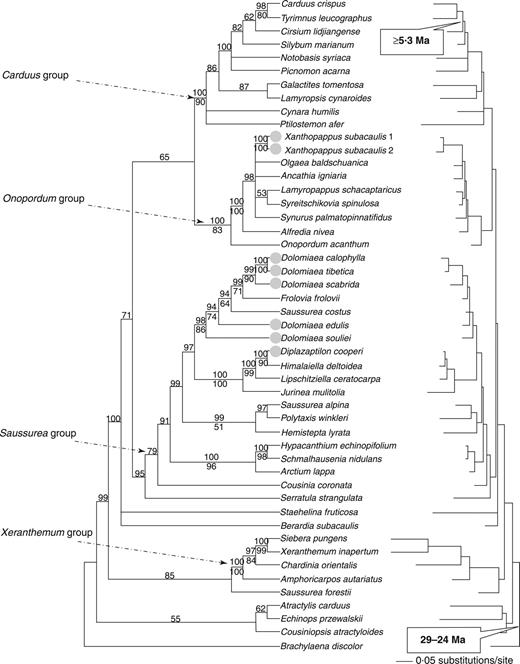
All the newly available sequences covered the 5·8S gene (164 bp), but no mutations were found within this segment. It was therefore assumed that all of these ITS sequences are functional orthologues rather than paralogues. Due to the unavailability of this fragment in the previously published species, it was omitted from any further analyses. The aligned ITS matrix is 498 bp long, 88 bp of which were variable but phylogenetically uninformative, and 271 bp that were variable and potentially phylogenetically informative when gaps were treated as missing characters. Parsimony analysis identified 142 trees with a length of 1512 steps, a consistence index (CI) of 0·388 and a retention index (RI) of 0·540. The consensus MP tree was mostly congruent with the ML tree (–lnL=7324·43926, the best-fit model GTR+I+G) in topology and the 50 % majority rule consensus tree derived from Bayesian analysis (Fig. 2 ). Of the excluded gaps, one indel of 6 bp is shared by all members of the Saussurea group. Two indels of 2 and 1 bp, respectively, support the clustering of Diplazoptilon and Himalaiella .
The 50 % majority rule consensus tree derived from Bayesian analysis of the ITS data set (left) and the maximum likelihood (ML) tree (right) constructed from the ITS data set with gaps excluded. Posterior probabilities are noted above branches, and bootstrap support values (>50 %) are given below branches. The circles mark the endemic genera on the Himalayan highlands. The shaded times indicate the calibrations used for the molecular dating.
Four major groups were recovered within Carduinae, here referred to as the Carduus , Saussurea , Onopordum and Xeranthemum groups, respectively. Two clades containing Diplazoptilon and Dolomiaea were closely related to each other within the Saussurea group. Diplazoptilon is sister to Himalaiella (BP=90 %; PP=100 %), a newly resurrected genus from Saussurea s.l. , and together they are sister to another newly recognized genus from Saussurea s.l. , Lipschitziella (BP=99 %; PP=100 %). The group comprising these three genera is sister to Jurinea (BP=100 %; PP=100 %). Frolovia frolovii and Saussurea costus were nested in a clade with five species of Dolomiaea (BP=86 %; PP=98 %), and this clade was resolved as sister to the lineage containing Diplazoptilon and three other genera. Xanthopappus fell in the Onopordum group (BP=83 %; PP=100 %).
Among the 41 genera examined within Cardueae, the average pairwise nucleotide distance was 17·85 %, with the smallest distance (0·8 %) occurring between Hypacanthium and Schmalhausenia and the largest (32·9 %) between Chardinia and Lamyropsis . Pairwise distances between Dolomiaea and its previously assumed closely related genera were 20.2–22 % ( Carduus ), 14·6–17·3 % ( Jurinea ) and 9·1–12·8 % ( Saussurea s.s. ). Within the Dolomiaea – Frolovia clade, the smallest distance (3·4 %) was detected between D. edulis and Saussurea costus. Diplazoptilon showed a pairwise distance variation of 1·8 % with Himalaiella . However, pairwise distances between Diplazoptilon and two genera previously assumed to be close relations were 13·9 % ( Saussurea s.s. ) and 11·4 % ( Dolomiaea ). Xanthopappus showed a pairwise distance variation of 7·1–7·3 % with Ancathia , but 19·9–21·2 % with Carduus .
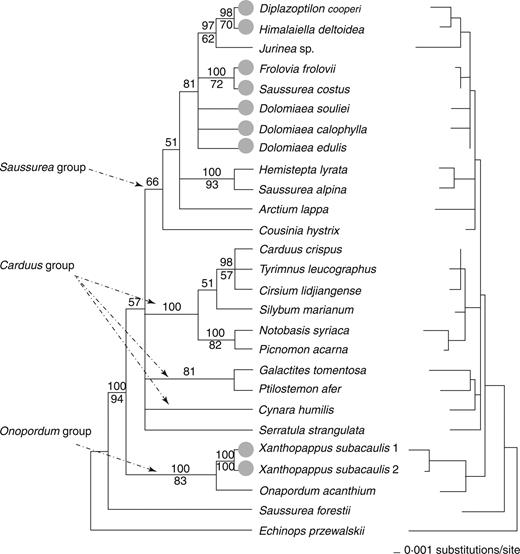
Combined analyses of plastid trnL-F and psbA-trnH regions
Both the trnL-F and psbA-trnH regions showed low variation, and partition homogeneity analyses gave no significant incongruence between them ( P =0·9). These two fragments were therefore combined together for phylogenetic analyses. The combined data set was composed of 27 accessions from 22 genera of Cardueae (20 genera from Carduinae, and one genus, respectively, from Centaureinae and Echinopsidinae). The aligned matrix consisted of 1350 positions, including 85 parsimony-uninformative variable sites and 39 potentially parsimony-informative variable sites. The MP analyses recovered 980 trees (length=155 steps; CI=0·832; RI=0·745). The strict consensus of these MP trees (not shown) is similar to the Bayesian majority consensus tree and the ML tree (Fig. 3 ).
The 50 % majority rule consensus tree derived from Bayesian analysis of two the combined plastid data sets (left) and the ML tree constructed from the same data set (right). Other details are the same as those in Fig. 2 .
The groupings of Dolomiaea and Frolovia , Diplazoptilon and Himalaiella , and Xanthopappus and Onopordum were again found. However, the relationships of these lineages and other clades and genera were less well resolved (Fig. 3 ). The pairwise distance between Dolomiaea and Frolovia is 0·26–0·6 %; in contrast, Dolomiaea showed a pairwise distance variation of 0·52–0·87 % with Carduus , 0·96–1·3 % with Jurinaea and 0·69–1·0 % with Saussurea s.s.. Diplazoptilon showed the smallest distance, 0·52 %, with Himalaiella , and the distances between it and its previously assumed closely related genera are 0·69–1·0 % ( Dolomiaea ) and 1·21 % ( Saussurea s.s. ). The pairwise distance between Xanthopappus and Onopordum was 1·0–1·3 %, and the distance between it and Carduus was 1·5–1·7 %.
Combined analyses of nuclear ITS and plastid trnL-F and psbA-trnH fragments
The ILD test ( P =1·0) showed that the two data sets, the nuclear rDNA ITS vs. the combination of the plastid trnL-F and psbA-trnH , were fully congruent, and it is therefore justifiable to combine them in a single data set for further analysis. The combined sequence matrix comprised 1848 characters after alignment, of which 189 sites were variable but uninformative, and 226 sites were variable and potentially parsimony-informative when gaps were treated as missing characters. The heuristic search resulted in 78 trees (length=1002 steps; CI=0·555; RI=0·553). Their strict consensus produced an identical topology to the majority consensus tree of the Bayesian inferences. The ML tree (TIM+I+G model, –lnL=8256·4629) also yielded a similar topology. The analyses of the combined data set confirmed all identified clades in both Figs 2 and 3 , and most of them received elevated support.
Divergence times
Because the ITS data matrix includes most representative genera of Cardueae, with a coverage of the wide morphological range of this tribe, only this data set was used to infer origin time scales of the Himalayan genera. LRT strongly rejected the assumption of a molecular clock for the nuclear ITS sequence matrix (–lnL=7324·43926 vs. 7439·92302; d.f.=47; P <0·001), so the ML phylogram was calculated in the absence of a molecular clock and then it was subjected to rate smoothing applying PL implemented in the software r8s ( Sanderson, 2002 ).
Fossil records for Cardueae are rare, and the earliest of the known records is pollen of Cirsium from the early Pliocene ( Menke, 1976 ; Graham, 1996 ). It was hypothesized that the divergence of Cirsium and its sister Carduus occurred before the early Pliocene, and constrained 5·3 Ma as a minimum age to the stem node of Cirsium . At the same time, the stem age of Cardueae was estimated as 29–24 Ma in a recent dating of Asteraceae ( Kim et al. , 2005 ) and therefore this dating was used to calibrate the divergence of Cardueae from Tarchonantheae (Fig. 2 ). The DIVTIME command of the r8s program with the cross-validation function enforced identified the smoothing parameter value of 1·6 as the optimal and it was then selected to execute the PL estimates. The results indicated that the divergences of Dolomiaea , Diplazoptilon and Xanthopappus from their sisters probably occurred between 13·6 and 12·2 Ma, 2·0 and 1·6 Ma or 5·7 and 4·7 Ma, respectively.
DISCUSSION
Systematic position of Xanthopappus
The close relationship between Xanthopappus and Carduus has never previously been questioned because of their close similarity in spiny morphology and their sympatric distributions ( Smith, 1917 ; Kazmi, 1963 , 1964 ; Dittrich, 1977 ; Shi, 1987 ; Bremer, 1994 ; Wu et al. , 2003 ). These two genera commonly share hairy receptacles, entire pericarpal crowns, straight carpopodia and multiserial, scabrid, basally connate pappus bristles. Xanthopappus differs from Carduus in having yellow florets and smooth filaments ( Smith, 1917 ; Shi, 1987 ). However, the molecular data indicated that Xanthopappus nested within the Onopordum group together with Alfredia , Ancathia , Lamyropappus , Olgaea , Synurus and Syreitschikovia whereas Carduus fell in a separate Carduus group with Cirsium and eight other genera.
The exclusion of Xanthopappus from the Carduus group further supports natural circumscription of this group, as suggested by Häffner and Hellwig (1999) , García-Jacas et al. (2002) and Susanna et al. (2006) . This ‘thistle’ group was considered to share common rose-coloured flowers, but the flowers of the Onopordum group are similarly pink or purple ( Shi, 1987 ). However, floral colour appears to have developed as a result of parallel evolution within Cardueae. All genera in this group have hairy or papillate anther filaments. In addition, their habit, habitat and distribution are similar, according to Häffner (2000) . In contrast, members of the Onopordum group, including Xanthopappus , are characterized by glabrous filaments and yellow and highly fragile pappus bristles ( Shi, 1987 ; Bremer, 1994 ; Petit et al. , 1996 ; Petit, 1997 ; Häffner, 2000 ). Xanthopappus is endemic to the QTP, and the remaining genera in this group are centred in central Asia ( Shi, 1987 ; Bremer, 1994 ; Häffner, 2000 ).
The generic delimitation and the phylogenetic relationships of Dolomiaea and Diplazoptilon
The nuclear ITS analyses indicate that all sampled species of Dolomiaea and Frolovia frolovii and Saussurea costus form a well supported clade (Fig. 2 ). The genetic distances, 2·8–8·9 % for the ITS data within the Dolomiaea – Frolovia clade fall well within the infrageneric variation reported for Asteraceae ( Fernández et al. , 2001 ). The combined plastid analyses also revealed a close relationship between these species (Fig. 3 ). Therefore, Frolovia and S. costus should be placed in the broadly recircumscribed Dolomiaea . Raab-Straube (2003) resurrected Frolovia from Saussurea sub-genus Frolovia , and pointed out its close relationship with Jurinea . However, he did not include S. costus within Frolovia , which was formerly placed in Saussurea subgen. Frolovia , although the major reason for its exclusion from Frolovia might be that it differs from the other species in this genus in having entire leaves. The present results suggest that S. costus is more closely related to Frolovia and Dolomiaea than to species of Saussurea s.s. , and that it should similarly be placed in the expanded Dolomiaea . These findings are basically consistent with the recent investigation conducted by Susanna et al. (2006) . Possible morphological evidence for this lineage is that all the species have apical pericarp rim projections. The chromosome number, pollen type and cypsela anatomy remain unknown for all species of Dolomiaea and Frolovia and S. costus examined here.
Ling (1965) treated Vladimiria as a separate genus and established two sections, section Vladimiria and section Sorocephalos , according to the number of capitula on the inflorescence. Shi (1986) , however, did not adopt such a classification and recognized only section Vladimiria and section Dolomiaea when he merged these two genera. The differences proposed for these two sections are the same as those for generic delimitation listed by Ling (1965) : long and acute vs. short and round styles. Both D. calophylla and D. tibetica with short and round styles in section Dolomiaea form a clade, nested within the other species, but the three species D. souliei , D. edulis and D. scabrida , with long and acute styles, do not comprise a corresponding clade (Fig. 2 ). Similarly, the topology (Fig. 2 ) indicates that two species, D. souliei and D. scabrida , with more than one capitulum on the inflorescence do not form a clade. These results support a combination of two genera as a single genus, and further suggest that the infrageneric classification of the broadly circumscribed genus needs further revision. The long and acute styles and the inflorescence with more than one capitulum are likely to represent plesiomorphic states within Dolomiaea as assessed from the molecular trees. The inclusion of Frolovia and S. costus within the broadly circumscribed Dolomiaea makes the morphological variation in this genus more complex. Further examinations of morphological variation in Dolomiaea and the correlation of this with molecular data are needed before a final infrageneric system can be established and taxonomic treatments can be made.
According to both individual and combined ITS and plastid analyses, Diplazoptilon is sister to the newly established genus Himalaiella . These results reject the relationships of Diplazoptilon with Saussurea , Dolomiaea or Jurinea , as suggested by various authors (e.g. Ling, 1965 ; Shi, 1986 ). Diplazoptilon and Saussurea are similar in having two rows of pappus bristles ( Ling, 1965 ), but Diplazoptilon differs from Saussurea s.s. (as defined by Raab-Straube, 2003 ) by its pericarp crown and alveolate receptacles. These two traits are also found in Dolomiaea as broadly defined here, but the anther bases of Diplazoptilon are hairy, unlike those of Dolomiaea . A combination of granulate and echinate pollen in Diplazoptilon is distinctly different from the echinate pollen form of Jurinea. Diplazoptilon differs from Himalaiella by the double layer of plump pappus bristles ( Shi, 1987 ; Raab-Straube, 2003 ). Susanna et al. (2006) suggested that Diplazoptilon , Himalaiella , Lipschitziella and Jurinea should be taxonomically treated together as a single genus. However, up to now, due to the lack of morphological investigation, it is still difficult to find distinct characters to unite this lineage as a broadly circumscribed genus.
Origins of the Himalayan endemic genera and alpine flora
Within the Saussurea group, the crude dating based on ITS sequence divergence revealed that Dolomiaea , including F. frolovii and S. costus , diverged from its sister clade probably around 13·6–12·2 Ma. The divergence times of Diplazoptilon from Himalaiella were calculated to be around 2·0–1·6 Ma. Xanthopappus probably diverged from its close relatives some 5·7–4·7 Ma. These crude estimates indicate that the origins of the three sampled genera distributed in the high elevations of the Himalayan region occurred during different periods, probably from 13·6 to 1·6 Ma, and therefore suggest a continuous origin hypothesis for endemic genera in this area since the middle Miocene. Molecular calibration of branching time in phylogenetic trees is controversial, and should be treated with caution. However, when palaeontological data are lacking, molecular estimates provide the only means of inferring the age of lineages ( Li, 1997 ; Bromham and Penny, 2003 ). Despite these concerns, we are reasonably confident that these genera originated through gradual allopatric speciation, for two reasons. First, a discernible clustering of genetic mutations in some endemics within phylogenetic trees was obvious, and these endemic genera were revealed to have different genetic distances with their sister lineages. These findings suggest gradual and continuous origins of these endemics, notwithstanding the unavailability of precise dating. Secondly, geological evidence indicates that recent extensive uplifting of the QTP occurred during at least four different periods since the early Miocene, i.e. 22 Ma, 15–13 Ma, 8–7 Ma and 3·5–1·6 Ma ( Harrison et al. , 1992 ; Li et al. , 1995 ; Shi et al. , 1998 ; Spicer et al. , 2003 ). It is feasible, therefore, that these uplifts created continuous fragmentation of habitats, and continuously produced endemic genera through allopatric speciation in the QTP and the adjacent Himalayan region.
The endemic genera occurring in the QTP and the adjacent Himalayan highlands are small, with fewer than 15 species, and many are monotypic ( Wu, 1987 ; Wu et al. , 1995 ). In contrast to morphological distinctness, some of these endemics showed low genetic divergence from their sister groups, e.g. Diplazoptilon , or even nest within other genera, e.g. Frolovia within Dolomiaea , as revealed here. Several recent molecular studies of the QTP endemic genera uncovered similar results, e.g. Sinadoxa (Adoxaceae) ( Liu et al. , 2000 ), Milula (Alliaceae) ( Friesen et al. , 2000 ), Lomatogoniopsis (Gentianaceae) ( Liu et al. , 2001 ) and Sinacalia (Asteraceae) ( Liu et al. , 2006 ). Undoubtedly these endemics originated in situ with the more recent uplifts of the plateau since the Pliocene (within apaprox. 5 Ma), and with rapid formation of aberrant morphology. However, a few genera could have greater genetic distances and more ancient divergence times with their sister lineages, e.g. broadly defined Dolomiaea (approx. 13·55–12·17 Ma) in the present study, Nannoglottis of Asteraceae (probably between 22 and 32 Ma; Liu et al. , 2002 ) and Pomatosace of Primulaceae (probably around 10 Ma; Wang et al. , 2004 ). Except for the endemic genera with aberrant morphology, another important characteristic of alpine flora in the Himalayan highlands is the high diversification of some lineages, especially some genera with >100 species endemic to the region, e.g. Gentiana (Gentianaceae) and Rhododendron (Ericaceae) ( Wu, 1987 ; Wu et al. , 1995 ). The species richness in these genera contributes to the majority of the alpine flora in this region ( Wu et al. , 1995 ). Molecular analyses of a few such large genera have revealed poorly resolved infrageneric phylogenetic relationships suggesting radiative diversification. Molecular calibrations suggested that the radiation of some genera (e.g. Pedicularis , Yang et al. , 2003 ; Saussurea , Wang and Liu, 2004 a ; Ligularia – Cremanthodium – Parasenecio complex, Liu et al. , 2006 ) was triggered by the earlier uplifts of the QTP, whereas the onset of diversification of others (e.g. Gentiana , Yuan and Kupfer, 1997 ; Rheum , Wang et al. , 2005 a ; Rhododendron sub-genus Hymenanthes , Milne, 2004 and pers. comm.) seem to have occurred more recently (e.g. radiation of Rhododendron sub-genus Hymenanthes , approx. 4–6 Ma; Milne, 2004 and pers. comm.).
In conclusion, this research has provided evidence for a continuous development of endemic genera, probably triggered by different uplifts of the QTP between the Miocene (approx. 22 Ma) and the Quaternary (approx. 2 Ma). These continuous uplifts might similarly have contributed to the radiation and diversification of species-rich genera occurring there and in adjacent regions, where the formation of the major habitats and geography can be mainly attributed to the uplift of the QTP. Apart from botanical examples, recent research on animal diversification in the QTP and adjacent Himalayan regions revealed a similar correlation with different uplift stages of the plateau from the Miocene to the Quaternary. Pikas, oriental voles and sand lizards were estimated to have initiated extensive diversification around or within the Pliocene ( Yu et al. , 2000 ; Pang et al. , 2003 ; Luo et al. , 2004 ). However, Chinese sisorid catfishes originated in the Oligocene–Miocene boundary (24–19 Ma), and radiated from Miocene to Pleistocene ( Guo et al. , 2005 ). Further studies of other organisms are now required to establish if the development of endemic genera and radiation of species-rich lineages triggered by different uplifts of the plateau are a common phenomenon and of major importance in generating the current high diversity of plants and other organisms within this region and adjacent areas.
ACKNOWLEDGEMENTS
We are grateful to Dr Mike Fay and two anonymous referees for their constructive comments on an earlier version of this paper. Support for this research was provided by the Key Innovation Plan KSCX2-SW-106, KSCX2-YW-Z, the Special Fund for an Outstanding PhD Dissertation, FANEDD 200327, the Specialized Research Fund for the Doctoral Program of Higher Education and a grant from the German Research Council (Mi 271/15-1).