Abstract
Extracellular vesicles (EVs) isolated from mesenchymal stem/stromal cells (MSCs) contribute to recovery of damaged tissue. We have previously shown that porcine MSC-derived EVs transport mRNA and miRNA capable of modulating cellular pathways in recipient cells. To identify candidate factors that contribute to the therapeutic effects of porcine MSC-derived EVs, we characterized their protein cargo using proteomics. Porcine MSCs were cultured from abdominal fat, and EVs characterized for expression of typical MSC and EV markers. LC-MS/MS proteomic analysis was performed and proteins classified. Functional pathway analysis was performed and five candidate proteins were validated by western blot. Proteomics analysis identified 5,469 distinct proteins in MSCs and 4,937 in EVs. The average protein expression was higher in MSCs vs. EVs. Differential expression analysis revealed 128 proteins that are selectively enriched in EVs versus MSCs, whereas 563 proteins were excluded from EVs. Proteins enriched in EVs are linked to a broad range of biological functions, including angiogenesis, blood coagulation, apoptosis, extracellular matrix remodeling, and regulation of inflammation. Excluded are mostly nuclear proteins, like proteins involved in nucleotide binding and RNA splicing. EVs have a selectively-enriched protein cargo with a specific biological signature that MSCs may employ for intercellular communication to facilitate tissue repair.
Similar content being viewed by others
Introduction
Mesenchymal stem/stromal cells (MSCs) are principal components of an important endogenous repair system that allows constant self-renewal of adult tissues in many organs. MSCs were first described in 1970 as colony forming units fibroblasts1 and can be isolated from various tissues, including bone marrow, adipose tissue, umbilical cord blood, and peripheral blood. Although MSC populations have been characterized by different sets of markers, specific consensus criteria have been defined to identify MSCs, including adherence to plastic in standard culture conditions, expression of common MSC cell surface markers (e.g. CD73, CD90, and CD105), and capacity for trilineage differentiation into osteoblasts, adipocytes, and chondrocytes in cell culture2. In addition to their ability to migrate and differentiate into multiple cell linages, MSCs exert unique pro-angiogenic, anti-inflammatory, and immuno-modulatory effects that promote tissue repair and regeneration. These characteristics of MSCs are attractive for their application in cell-based therapies for tissue regeneration.
Over the last decade, MSCs have been employed to ameliorate tissue injury and accelerate repair in a wide variety of disorders. One particularly attractive application is in cardiovascular disease, the leading cause of mortality in countries with developed and emerging economies3. Indeed, a progressively increasing number of studies has focused on the ability of MSCs to repair the damaged myocardium in murine models of hypertension and heart failure4,5. Furthermore, in recent years, we and others have tested the efficacy of MSCs for attenuating cardiac and renal injury in large animal models of cardiovascular disease6. For example, intra-myocardial delivery of MSCs safely reduces infarct size and improves cardiac function in porcine ischaemic heart failure7. Furthermore, we have shown that intra-renal adjunctive MSC-based therapy with or without restoration of renal vascular patency improves renal and cardiac function in porcine renovascular disease, confirming the safety and efficacy of this intervention8,9,10,11.
Accumulating data indicate that MSCs release large amounts of extracellular vesicles (EVs) that mediate their paracrine effects by shuttling different types of RNAs (e.g., mRNA and miRNA) and proteins to recipient cells12,13. Consistent with this concept, we recently found that porcine MSC-derived EVs contain a selective combination of mRNAs and miRNAs capable of regulating transcription of genetic information and modulating angiogenesis, adipogenesis, and other pathways in recipient cells14. To support the potential utility of EVs in augmenting MSC-related therapies, the protein expression profile of porcine MSC-derived EVs needs to be characterized. In this study, we elucidated the protein content of porcine MSC-derived EVs to gain important insight into the molecular mechanisms by which they exert paracrine actions and contribute to the reparative potency of MSCs. Using a quantitative proteomic strategy to study the protein cargo of porcine MSC-derived EVs, we identified candidate proteins and biological signatures that are consistent with the postulated therapeutic effects of MSCs.
Materials and Methods
MSC and EV characterization and culture
All experiments were performed in accordance with the guidelines and approval of the Mayo Clinic Institutional Animal Care and Use Committee. MSCs were isolated from subcutaneous abdominal fat (5–10 g) of 3 female domestic pigs, as previously described14. Cells were cultured for 3 weeks in advanced MEM medium (Gibco/Invitrogen) supplemented with 5% platelet lysate (PLTmax, Mill Creek Life Sciences, Rochester, MN) in 37°/5% CO2, and the third passage collected for in vitro phenotypic and functional analyses. MSCs expressed CD44, CD90, and CD105 markers, and differentiated into osteocytes, chondrocytes, and adipocytes, as previously described8,9,15, consistent with our experience with human MSCs16.
EVs were obtained from supernatants of MSCs using the ultra-centrifugation method14. In brief, 10 × 106 MSCs (a dose used for in vivo injections)8,9,10,11 were cultured for 48 h in advanced MEM medium without supplements and centrifuged at 2,000 g. Cell-free supernatants were then subjected to ultra-centrifugation at 100,000 g for 1 h at 4 °C, washed in serum-free medium containing HEPES 25 mM, and submitted to a second step of ultracentrifugation.
Transmission electron microscopy (TEM) of MSC supernatant with 2% uranyl acetate negative staining was performed, and cup-shaped 40–1000 nm structures identified as EVs. Micrographs were taken on a digital electron microscopy (JEOL 1200 EXII). EVs were further characterized based on the expression of EV (CD9, CD29, and CD63) and MSC (CD105 and CD73) surface markers by western blotting. In addition, EV size distribution was assessed by nanoparticle tracking analysis (NTA) using NanoSight NS300. EVs were diluted with PBS and samples continuously run through a flow-cell top-plate at 25 μL/min. Three videos (120 seconds each) of Brownian motion of nanoparticles were recorded and 1,000 completed tracks analyzed using NTA 2.3.5.
Proteomic profiling and network pathway analysis
Liquid chromatography mass spectrometry (LC-MS/MS) proteomic analysis was performed at the Mayo Clinic Proteomics Core, as previously described17,18. All MSC and EV pellets were solubilized in Tris buffer with 1%SDS HALT protease inhibitor and Benzonase. Lysis was achieved with by rapid agitation (‘vortexing’) followed by three consecutive 5 min incubations on ice. Protein samples were then denatured by incubation at 85 °C for 10 min. Aliquots with a protein equivalent of 20 μg were dried down and resolubilized in reducing sample buffer. Samples were electrophoresed in 4–20% TGX Ready gels at 200 V for 30 min.
The gel lanes were divided into 6 evenly spaced horizontal regions using as guides protein bands that were common to all samples. Each sample lane was cut length-wise in the middle to obtain technical duplicates. The resulting individual gel sections were diced and transferred into PCR tubes for digestion and extraction. Gel sections were digested with trypsin following a previously described protocol18. In brief, each gel section was de-stained and the proteins present in the section were reduced with dithiothreitol and alkylated with iodoacetamide. Proteins were digested by overnight incubation at 37 °C with 140 ng of trypsin dissolved in 25 mM Tris (pH 8.2). Peptides were extracted from the gel piece with 30 μL of 50% acetonitrile in 4% trifluoroacetic acid, followed by two additional extractions with 50 μL aliquots of acetonitrile. The combined extracts were evaporated to dryness on a vacuum concentrator and stored at −80 °C until further analysis.
Peptide extracts from each gel section were reconstituted in 40 μL HPLC-grade water containing 0.2% formic acid, 0.1% trifluoroacetic acid, and 0.002% Zwittergent 3–16. Aliquots of the peptide extracts (typically 10 μL, but 15 μL for the two gel sections encompasses higher molecular weight proteins) were loaded onto a 0.25 μL bed OptiPak trap (Optimize Technologies, Oregon City, Oregon) custom-packed with 5 μm, 200 Å Magic C8 (Bruker-Michrom, Auburn, CA) in the stationary phase. The loaded trap was washed for 4 min with an aqueous loading buffer of 0.2% formic acid and 0.05% trifluoroacetic acid at a flow rate of 10 μL/min. Following the wash, peptides were transferred via a 10-port valve onto a 35 cm × 100 μm PicoFrit column 9 (NewObjective, Woburn, MA), self-packed with Agilent Poroshell 120S 2.7 μm EC-C18 stationary phase, using a Dionex UltiMate 3000 RSLC LC system (Thermo-Fisher Scientific, Waltham, MA). Peptides were separated using a 400 nL/min LC gradient comprised of 2–30%B in 0–70 min, 30–50%B from 70–100 min, 50–95%B from 100–104 min, held at 95%B for 8 min and re-equilibrated to 2%B. Mobile phase A was 2% acetonitrile in water with 0.2% formic acid and mobile phase B was acetonitrile /isopropanol/water (80/10/10 by volume) with 0.2% formic acid. Eluting peptides were analyzed using a QExactive mass spectrometer (Thermo-Fisher Scientific, Waltham, MA). The instrument was operated in data-dependent mode by collecting MS1 data at 70,000 resolving power (measured at m/z 200) with an AGC value of 3E6 over an m/z range of 350–2000, using lock masses from background polysiloxanes at m/z 371.10123 and 445.12002. Precursors were fragmented with normalized collision energy of 27, and fragments measured at 17,500 resolving power and a fixed first mass of 140. Tandem mass spectra (MS/MS) were collected for the top 15 precursor masses present in each MS1 using an AGC value of 1E5, maximal ion fill time of 100 ms, an isolation window of 3.0 Da, isolation offset of 0.5 Da, and a dynamic exclusion time of 60 s.
We utilized a label-free peptide MS1 intensity-based method for finding differentially expressed proteins between experimental groups. The quality of the raw data was assessed using the quality control metrics in the Swift proteomic data processing pipeline. MaxQuant (version 1.5.1) software processed the raw data files to produce a list of protein groups and their corresponding intensities in each sample19. To accomplish this, MaxQuant was configured to use a composite porcine protein sequence database containing UniProt porcine reference proteome (downloaded on 08/12/2015) and sequences of common contaminants (trypsin, keratin, cotton, wool, etc.). Reversed protein sequences were appended to the database for estimating protein identification false discovery rates (FDRs). The software was configured to use 20 ppm m/z tolerance for precursors and fragments while performing peptide-spectrum matching. The software derived semi-tryptic peptides from the sequence database while looking for the following variable modifications: carbamidomethylation of cysteine (+57.023 Da.), oxidation of methionine (+15.994 Da.), formation of n-terminal pyroglutamic acid (−17.023 Da.) and protein n-terminal acetylation (+42.01 Da.). MaxQuant was instructed to align the runs and match features between multiple sample runs of the same gel region. The software filtered peptide and protein identifications at 1% FDR, assembled protein identifications into groups and reported protein group intensities.
An in-house R-programming script performed differential expression analysis using protein group intensities. First, protein group intensities of each sample were log2 transformed and normalized using a trimmed mean (5%) method. For each protein group, the normalized intensities observed in two groups of samples were modeled using a Gaussian-linked generalized linear model. ANOVA was used to detect the differentially expressed protein groups between pairs of experimental groups. Differential expression analysis was performed after the data was normalized by protein loading, and differential p-values FDR-corrected using the Benjamini-Hochberg-Yekutieli procedure20. Protein groups with an FDR < 0.05 and an absolute fold change (EVs/MSCs) > 10 were classified as enriched in EVs, whereas those with FDR < 0.05 and fold change <−10 were considered as excluded.
Proteins enriched in or excluded from EVs were classified by their molecular function, cellular localization, and class, using Protein Analysis Through Evolutionary Relationships (PANTHER)21. Functional pathway analysis was performed using the Database for Annotation and Integrated Discovery (DAVID 6.7)22.
Validation of proteomic analysis
Complement component (C2), vascular endothelial growth factor (VEGF), and von Willebrand factor (vWF) proteins, which were all enriched in EVs, were selected for validation, and their expression in EVs and MSCs measured by Western blot (Novus: Cat#NBP1-58985, Santa Cruz Biotech: cat# sc-152, and Abcam: cat#ab6994, respectively). Similarly, we validated the expression of RNA polymerase-associated protein RTF1 homolog (RTF1) and FOS-Like Antigen 1 (FOSL1) excluded from EVs (Abcam: cat#ab99362 and LifeSpan BioSiences, Inc. cat#:aa130-179, respectively).
Results
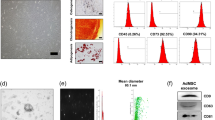
Cultured MSCs released significant amounts of EVs with a classic “cup-like” morphology (Fig. 1A), most of which were under 300 nm in size with a bi-phasic size-distribution (Fig. 1B) consistent with a composition of approximately 2/3 small microvesicles (~125 nm) and 1/3 exosomes (~55 nm). The EV expressed primarily EV (CD9, CD29, and CD63) and MSC surface markers (CD105 and CD73) (Fig. 1C).
(A) Transmission electron microscopy of MSC culture supernatants (negative staining) showing EV clusters (arrows) with the classic “cup-like” morphology. (B) Size distribution of isolated EVs revealed a composition of about 2/3 small microvesicles (~125 nm in size) and 1/3 exosomes (~55 nm). (C) MSC and EV protein expression of common MSC (CD105 and CD73) and EV (CD9, CD29, and CD63) markers.
Identification and classification of differentially expressed proteins
Proteomics analysis identified peptides derived from a total of 5,469 distinct protein groups in MSCs and 4,937 in EVs, with molecular weights ranging from 10–250 kDa (Fig. 2A). Average protein expression was higher in MSCs compared to EVs (Fig. 2B, p < 0.0001). EVs expressed 12 MSC and 77 EV markers (Tables 1 and 2). Differential expression analysis revealed 128 proteins upregulated in EVs vs. MSCs (Table S1, fold change EVs/MSCs >10, p < 0.05), whereas 573 proteins were excluded from EVs (fold change EVs/MSCs < −10, p < 0.05) (Fig. 2C).
(A) Master gel for LC-MS/MS proteomic analysis of 3 MSC and 3 EV samples. The gel shows the relative molecular weight on the left side. Slices were labeled (A–F) with A being at the highest molecular mass >250 kDa and F being the lowest (10–15 kDa). (B) Average protein expression was higher in MSCs compared to EVs. (C) Volcano plot of identified proteins in MSCs and EVs.
Proteins enriched in EVs
The majority of the proteins enriched in EVs showed binding, catalytic, and receptor activity, and were equally distributed between intracellular and extracellular compartment categories (Fig. 3A,B). A number of different classes of proteins were enriched in EVs, with receptors, signaling molecules, and enzyme modulators representing the most frequently occurring protein categories types (Fig. 3C). Functional classification showed a remarkable diversity of biological roles. A total of 52 clusters were identified, among which glycoproteins, extracellular space, and signaling proteins comprised the most relevant category (enrichment score = 15.4). From a biological perspective, these proteins are functionally involved in angiogenesis, blood coagulation, extracellular matrix remodeling, inflammatory response, and apoptosis (Fig. 4). These main categories were followed by closely related categories that encompass proteins involved in wound healing (enrichment score = 8.04), extracellular matrix (5.03), and adaptive immune response (4.46). The remaining categories were approximately equally distributed (average enrichment score = 1.2 ± 0.8).
Functional annotation clustering (using DAVID 6.7) of proteins enriched in EVs (top) and histograms showing distribution by fold change of EV proteins in the Glycoproteins, extracellular space, and signaling category (bottom).
1: Glycoproteins, extracellular space and signaling; 2: Wound healing; 3: Extracellular matrix; 4: Adaptive immune response; 5: Heparin binding; 6: Protein interaction; 7: Regulation of inflammatory response; 8: Peptidase activity; 9: Vesicle proteins; 10: Vesicle-mediated transport; 11: Response to steroid hormone stimulus; 12: Complement pathway; 13: Peptidase activity; 14: Cell adhesion; 15: Lipid binding; 16: Epidermal growth factor pathway; 17: Response to extracellular stimulus; 18: Lipoprotein; 19: Respiratory development; 20: Exocytosis; 21: TGF-beta signaling pathway; 22: Enzyme inhibitor activity; 23: Cell proliferation; 24: Cell surface; 25: Cell differentiation; 26: Vesicle-mediated transport; 27: Cell adhesion; 28: Protein complex assembly; 29: Positive regulation of transport; 30: Cytokine-cytokine receptor interaction; 31: Intracellular signaling; 32: Angiogenesis; 33: Response to carbohydrate stimulus; 34: Basement membrane; 35: Phospholipidic metabolic process; 36: Plasma membrane; 37: Regulation of cell size; 38: Protein transport; 39: Nucleotide binding; 40: Response to hypoxia; 41: Membrane proteins; 42: Regulation of kinase activity; 43: Intracellular signaling; 44: Cell migration; 45: ATP binding; 46: Calcium binding; 47: Lysosome; 48: Chemical homeostasis; 49: Microtubule; 50: Apoptosis; 51: Immunoglobulin; 52: Cytoskeleton.
Proteins excluded from EVs
Proteins excluded from EVs possessed binding and catalytic activity, and were mostly detected in key cell functions and organelles (Fig. 5A,B). The major functional class for these excluded proteins was the nucleic acid binding proteins, followed by transcription factors and enzyme modulators (Fig. 5C). Functional pathway analysis revealed their participation in a broad spectrum of cellular processes and subcellular locations (56 clusters). The most notable categories are nuclear proteins (enrichment score = 10.08), nucleoplasm (5.78), RNA splicing (4.9), nucleotide binding (4.67), and nucleolus (4.21) (Fig. 6A). The remaining 51 categories were equally distributed (average enrichment score = 0.7 ± 0.5).
(A) Functional annotation clustering (using DAVID 6.7) of proteins excluded from EVs. 1: Nucleolus; 2: Nucleoplasm; 3: RNA splicing; 4: Nucleotide binding; 5: Nucleolus; 6: Histone modification; 7: DNA binding; 8: RNA binding; 9: Apoptosis; 10: FHA regulatory proteins; 11: Isopeptide bond; 12: N-acetyltransferase activity; 13: GTPase regulator activity; 14: ATP binding; 15: RNA transport; 16: DNA replication; 17: Mitochondria; 18: Ubiquitin protein activity; 19: Transcription factors; 20: Macromolecular complex assembly; 21: Lipid synthesis; 22: Vesicle-mediated transport; 23: Transcription cofactor activity; 24: Helicase activity; 25: Regulation of cell death; 26: Cell division; 27: Regulation of transferase activity; 28: DNA repair; 29: Respiratory development; 30: Eye development; 31: Mitochondrial membrane; 32: Protein transport; 33: Transcription regulator activity; 34: Signaling proteins; 35: Transcription repressor activity; 36: Zinc fingers; 37: Organelle membrane; 38: Protein kinase cascade; 39: Vasculature development; 40: Immune system development; 41: MAPkinase signaling pathway; 42: Endoplasmic reticulum; 43: WD40 repeat region; 44: Zinc finger; 45: GTP binding; 46: Magnesium ion binding; 47: Cytoskeleton; 48: Enzyme receptor signaling; 49: Calcium ion binding; 50: Spermatogenesis; 51: Response to hormone stimulus; 52: Cell migration; 53: Cytoplasmic vesicle; 54: Protein dimerization activity; 55: Peptidase activity; 56: Cell fraction. (B) Expression of the candidate proteins complement component (C2), vascular endothelial growth factor (VEGF), von Willebrand factor (vWF), RNA polymerase-associated protein RTF1 homolog (RTF1), and FOS-Like Antigen 1 (FOSL1) was in accordance to the proteomics findings.
Validation of proteomic analysis
Expression of the candidate proteins followed the same patterns as the proteomics findings. Specifically, C2, VEGF, and vWF were higher in EVs compared to their parent MSCs, whereas RTF1 and FOSL1 were higher in MSCs (Fig. 6B).
Discussion
In the current study, we describe the biological signature of porcine MSC-derived EVs from a proteomics perspective. Our analysis identified a large number of proteins enriched in EVs, bearing the potential to act as modulators of angiogenesis, blood coagulation, extracellular matrix remodeling, apoptosis, and inflammation. Moroever, we identified several nuclear proteins and proteins involved in nucleotide binding and RNA splicing that are selectively excluded from EVs. These observations provide important insights into the molecular mechanisms underpinning the reparative capacity of MSC-derived EVs and underscore the broad spectrum of factors present in EVs as mediators of MSC-related intercellular communication. Moreover, our findings might help develop novel regenerative therapies using MSC-derived EVs for tissue repair.
Accumulating evidence indicates that EVs are major effectors of the paracrine actions of MSCs, and mediate their cell-to-cell comunication. Bruno and colleagues demonstrated that EVs derived from human bone marrow MSCs stimulate proliferation in vitro and confer resistance to apoptosis in renal tubular epithelial cells by activating a proliferative program via horizontal transfer of mRNA23. MSC-derived EVs have also been shown to reduce infarct size in mice with myocardial ischemia/reperfusion injury24 and promote angiogenesis in rats with myocardial infarction25. These results clearly indicate that EVs contribute to the principal trophic effects of MSC transplantation.
We have recently characterized the mRNA and miRNA cargo of porcine MSC-derived EVs using high-throughput RNA sequencing analysis to elucidate nucleic acid-based mechanisms by which they exert their tissue protective properties14. We found that MSC-derived EVs contain a combination of mRNAs and miRNAs capable of regulating transcription of genetic information and modulating angiogenesis, adipogenesis, extracellular matrix turnover, TGF-β signaling, and other pathways in recipient cells. The current study extends our previous observations and demonstrates that MSC-derived EVs contain proteins capable of modulating several signaling pathways and gene networks, providing molecular evidence useful to elucidate the paracrine effects of MSCs. Of particular interest for the use of EVs in supporting cardiovascular repair, our current proteomic findings combined with our previous transcriptomic findings indicate that EVs are enriched for proteins, mRNAs and miRNAs that individually, and perhaps together, support angiogenesis.
LC-MS/MS analysis offers the possibility to quantitatively characterize multiple protein groups by combining the physical separation capabilities of LC with the mass analysis capabilities of MS. Our proteomic analysis show that EVs derived from porcine adipose tissue MSC express several prototypical MSC markers (e.g. CD44, CD73, and CD105), as well as 79 of the top 100 EV markers identified in the Exocarta database (http://www.exocarta.org/, e.g. CD29 and CD63). Notably, we identified 128 proteins enriched in EVs compared to MSCs. Funtional enrichment analysis of these proteins indicates high representation of glycoprotein, extracelular space, and signaling proteins involved in angiogeneis, inflammation, matrix remodeling, blood coagulation, and apoptosis.
Experimental studies have demonstrated the pro-angiogenic potential of MSCs both in vivo and in vitro, providing a mechanistic rationale for their therapeutic use in stimulating angiogenesis. We have previously shown that a single intra-renal delivery of MSC substantialy improved the porcine renal microvasculature, associated with increased renal expression of VEGF, a key angiogenic factor that promotes endothelial cell survival and vascular sprouting8. The current study shows that VEGF is enriched in EVs, associated with upregulation of Ephrin-B2, a transmembrane ligand for Eph receptor tyrosine kinases that promotes VEGF-induced angiogesis26. Therefore, EVs may not only deliver the VEGF protein, but also upregulate its production in recipient cells. Furthermore, the EV proteome also includes several other key proteins involved in angiogenesis, such as angiopoietin-related protein-4, platelet-derived growth factor-C (PDGFC), and Wingless-Type (wnt) MMTV Integration Site Family, Member 7B (WNT7B). Our observations are underscored by recent findings showing up-regulated angiogenic associated pathways in exosomes derived from human bone marrow MSCs27, illustrating the conservation of the nature of EV cargo across species. Indeed, using a Venn diagram analysis, we compared our findings with those reported by Anderson et al., and identified 1,362 common EV proteins, among which several are involved in vascular development and angiogenesis including angiopoietin-1, Wnt Family Member-5A, and Notch-1 (Figure S1). Overall, there incomplete (~70%) overlap of EV proteins between the studies may be due to differences in MSC origin, interspecies variability, culture medium, and EV isolation methods. Importantly, our study extends previous findings, as our LC-MS/MS analysis identified a larger number of EV proteins.
MSCs also possess potent immunosuppressive and anti-inflammatory properties, partly via non-specific anti-proliferative actions on a wide range of immune cells28. Interestingly, we found increased EV expression of several inflammatory mediators, particularly members of the complement system, which modulate adaptive immune responses by interacting with receptors on dendritic cells and lymphocytes29. Furthermore, the complement system mediates the migration of MSCs to sites of inflammation and tissue injury, enhancing its immuno-modulatory actions30.
Additionally, proteins involved in extracellular matrix remodeling were enriched in EVs compared to MSCs. These included matrix metalloproteinase 9 (MMP9), which is involved in the breakdown of extracellular matrix, but also secreted by MSC and downregulates the cytotoxicity of natural killer cells31,32. Thus, delivery of this enzyme in EVs may supplement MSC immuno-modulatory actions. Furthermore, TGF-β1 is enriched in EVs. We have previously shown that EVs contain high levels of several mRNAs that encode for protein ligands within the TGFβ family, including TGFB1, TGFB3, FURIN, and ENG14. TGFβ proteins are involved in tissue remodeling and fibrosis, but also induce CD4+/CD25+ regulatory T-cells, dampering the inflammatory response33. In addition, TGFβ induces release of cytochrome-c from mitochondria triggering apoptosis34. Interestingly, we observed upregulation in other proteins that modulate apoptosis, including netrin-1 and secreted frizzled-related protein (SFRP)-535,36. These observations may therefore indicate a pro-apoptotic function for EVs, possibly to facilitate removal of damaged cells.
We also found robust EV expression of proteins involved in blood coagulation, such as vWF, coagulation factor X, and plasma kallikrein, which may reflect the innate procoagulant activity of MSCs. Consistent with this concept, a recent study showed that intracoronary MSC delivered during reperfusion in porcine may manifest pro-coagulant properties, emphasizing the need for caution when applying cell-based therapy37. Interestingly, our EVs express several members of the secretory component of platelets, including vWF, PDGFC, and TGFβ, possibly because MSCs were cultured in platelet lysate38. However, our MSCs do not express endothelial markers like CD31 and CD348,9, arguing against contamination with endothelial cells. Further studies are needed to explore the mechanisms by which vWF is enriched in MSC-derived EVs.
Apart from enrichment, our results revealed selective exclusion of a number of proteins from EVs, such as nuclear proteins and proteins involved in nucleotide binding and RNA splicing, including nucleolar complex associated-4 homolog, cleavage and polyadenylation specific factor-3, and CWC15 spliceosome-associated protein homolog. The mechanisms that regulate packaging these molecules into EVs remain unclear, but evidently cells are definitely selective in determining what proteins emerge as cargo in EVs.
Our study is limited by the relatively low number of samples, given the labor and cost of obtaining EVs from porcine disease models and of proteomic analysis. Nevertheless, a similar number of samples is commonly used in comparable studies reporting cell/tissue proteomic profiles39,40. Furthermore, post-translational changes in MSCs and EVs proteins unrevealed by proteomic analysis were not be assessed here, although LC-MS/MS has high sensitivity and robust precision for detecting MSC and EV protein groups. Indeed, follow-up studies using western blots demonstrated the efficacy of our proteomic analysis to detect changes in representative protein expression between MSCs and the EVs they produce. Future studies are needed to confirm these findings and test whether the protein content of in-vivo MSCs correlates with our observations in cultured MSCs. In addition, additional studies may ascribe specific cargo in our EV to microvesicles versus exosomes.
In summary, having examined the proteome of porcine MSCs and EVs, this study identified a number of proteins that may contribute to the molecular mechanisms involved in MSC-mediated tissue repair. We identified 128 proteins enriched and 563 excluded from EVs. Importantly, proteins enriched in EVs can potentially participate in tissue repair and regeneration by modulating angiogenesis, blood coagulation, apoptosis, extracellular matrix remodeling, and inflammation, whereas those actively depleted from EVs were mostly nuclear proteins or involved in nucleotide binding and RNA splicing. Therefore, our findings provide the foundation for future studies that focus on tissue protective effects of MSC-derived EVs and may guide the development of novel EV-based therapies for several diseases.
Additional Information
How to cite this article: Eirin, A. et al. Comparative proteomic analysis of extracellular vesicles isolated from porcine adipose tissue-derived mesenchymal stem/stromal cells. Sci. Rep. 6, 36120; doi: 10.1038/srep36120 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Friedenstein, A. J., Chailakhjan, R. K. & Lalykina, K. S. The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell Tissue Kinet. 3, 393–403 (1970).
Dominici, M. et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8, 315–317, Q2183N8UT042W62H (2006).
Mozaffarian, D. et al. Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association. Circulation 133, e38–e360, 10.1161/CIR.0000000000000350 (2016).
Tano, N. et al. Allogeneic Mesenchymal Stromal Cells Transplanted Onto the Heart Surface Achieve Therapeutic Myocardial Repair Despite Immunologic Responses in Rats. J Am Heart Assoc 5, e002815/JAHA.115.002815 (2016).
Ammar, H. I. et al. Comparison of adipose tissue- and bone marrow- derived mesenchymal stem cells for alleviating doxorubicin-induced cardiac dysfunction in diabetic rats. Stem Cell Res Ther 6, 148, 10.1186/s13287-015-0142-x (2015).
Lerman, L. O., Chade, A. R., Sica, V. & Napoli, C. Animal models of hypertension: an overview. J. Lab. Clin. Med. 146, 160–173, S0022-2143(05)00160-5/j.lab.2005.05.005 (2005).
Schuleri, K. H. et al. Autologous mesenchymal stem cells produce reverse remodelling in chronic ischaemic cardiomyopathy. Eur. Heart J. 30, 2722–2732, 10.1093/eurheartj/ehp265 (2009).
Eirin, A. et al. Adipose tissue-derived mesenchymal stem cells improve revascularization outcomes to restore renal function in swine atherosclerotic renal artery stenosis. Stem Cells 30, 1030–1041, 10.1002/stem.1047 (2012).
Zhu, X. Y. et al. Mesenchymal stem cells and endothelial progenitor cells decrease renal injury in experimental swine renal artery stenosis through different mechanisms. Stem Cells 31, 117–125, 10.1002/stem.1263 (2013).
Eirin, A. et al. Intra-renal delivery of mesenchymal stem cells attenuates myocardial injury after reversal of hypertension in porcine renovascular disease. Stem Cell Res Ther 6, 7, scrt541/scrt541 (2015).
Eirin, A. et al. Intra-renal delivery of mesenchymal stem cells and endothelial progenitor cells attenuates hypertensive cardiomyopathy in experimental renovascular hypertension. Cell Transplant, 10.3727/096368914X685582 (2014).
Lai, R. C., Chen, T. S. & Lim, S. K. Mesenchymal stem cell exosome: a novel stem cell-based therapy for cardiovascular disease. Regen Med 6, 481–492, 10.2217/rme.11.35 (2011).
Yeo, R. W. et al. Mesenchymal stem cell: an efficient mass producer of exosomes for drug delivery. Adv Drug Deliv Rev 65, 336–341, 10.1016/j.addr.2012.07.001S0169-409X(12)00212-8 (2013).
Eirin, A. et al. MicroRNA and mRNA cargo of extracellular vesicles from porcine adipose tissue-derived mesenchymal stem cells. Gene 551, 55–64, 10.1016/j.gene.2014.08.041S0378-1119(14)00980-9 (2014).
Ebrahimi, B. et al. Mesenchymal stem cells improve medullary inflammation and fibrosis after revascularization of swine atherosclerotic renal artery stenosis. PLoS One 8, e67474, 10.1371/journal.pone.0067474 (2013).
Dudakovic, A. et al. High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. J. Cell. Biochem. 115, 1816–1828, 10.1002/jcb.24852 (2014).
Hogan, M. C. et al. Subfractionation, characterization, and in-depth proteomic analysis of glomerular membrane vesicles in human urine. Kidney Int. 85, 1225–1237, 10.1038/ki.2013.422 (2014).
Hogan, M. C. et al. Identification of Biomarkers for PKD1 Using Urinary Exosomes. J. Am. Soc. Nephrol. 26, 1661–1670, 10.1681/ASN.2014040354 (2015).
Cox, J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13, 2513–2526, 10.1074/mcp.M113.031591 [pii] (2014).
Kim, K. I. & van de Wiel, M. A. Effects of dependence in high-dimensional multiple testing problems. BMC Bioinformatics 9, 114, 10.1186/1471-2105-9-114 (2008).
Mi, H. et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res 33, D284–D288, 33/suppl_1/D28410.1093/nar/gki078 (2005).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57, 10.1038/nprot.2008.211 (2009).
Bruno, S. et al. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J. Am. Soc. Nephrol. 20, 1053–1067, 10.1681/ASN.2008070798 (2009).
Lai, R. C. et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res 4, 214–222, 10.1016/j.scr.2009.12.003 (2010).
Bian, S. et al. Extracellular vesicles derived from human bone marrow mesenchymal stem cells promote angiogenesis in a rat myocardial infarction model. J Mol Med (Berl) 92, 387–397, 10.1007/s00109-013-1110-5 (2014).
Wang, Y. et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 465, 483–486, 10.1038/nature09002 (2010).
Anderson, J. D. et al. Comprehensive Proteomic Analysis of Mesenchymal Stem Cell Exosomes Reveals Modulation of Angiogenesis via Nuclear Factor-KappaB Signaling. Stem Cells 34, 601–613, 10.1002/stem.2298 (2016).
Shi, M., Liu, Z. W. & Wang, F. S. Immunomodulatory properties and therapeutic application of mesenchymal stem cells. Clin. Exp. Immunol. 164, 1–8, 10.1111/j.1365-2249.2011.04327.x (2011).
Carroll, M. C. The complement system in regulation of adaptive immunity. Nat Immunol 5, 981–986, 10.1038/ni1113 (2004).
Qiu, Y., Marquez-Curtis, L. A. & Janowska-Wieczorek, A. Mesenchymal stromal cells derived from umbilical cord blood migrate in response to complement C1q. Cytotherapy 14, 285–295, 10.3109/14653249.2011.651532 (2012).
Lee, B. K. et al. A high concentration of MMP-2/gelatinase A and MMP-9/gelatinase B reduce NK cell-mediated cytotoxicity against an oral squamous cell carcinoma cell line. In Vivo 22, 593–597 (2008).
Ding, Y. et al. Mesenchymal stem cells prevent the rejection of fully allogenic islet grafts by the immunosuppressive activity of matrix metalloproteinase-2 and -9. Diabetes 58, 1797–1806, 10.2337/db09-0317 (2009).
Fu, S. et al. TGF-beta induces Foxp3 + T-regulatory cells from CD4 + CD25 - precursors. Am J Transplant 4, 1614–1627, 10.1111/j.1600-6143.2004.00566.xAJT566 (2004).
Jang, C. W. et al. TGF-beta induces apoptosis through Smad-mediated expression of DAP-kinase. Nat Cell Biol 4, 51–58, 10.1038/ncb731 (2002).
Castets, M. et al. Inhibition of endothelial cell apoptosis by netrin-1 during angiogenesis. Dev Cell 16, 614–620, 10.1016/j.devcel.2009.02.006 (2009).
Cho, S. W. et al. Differential effects of secreted frizzled-related proteins (sFRPs) on osteoblastic differentiation of mouse mesenchymal cells and apoptosis of osteoblasts. Biochem. Biophys. Res. Commun. 367, 399–405, 10.1016/j.bbrc.2007.12.128 (2008).
Gleeson, B. M. et al. Bone Marrow-Derived Mesenchymal Stem Cells Have Innate Procoagulant Activity and Cause Microvascular Obstruction Following Intracoronary Delivery: Amelioration by Antithrombin Therapy. Stem Cells 33, 2726–2737, 10.1002/stem.2050 (2015).
Crespo-Diaz, R. et al. Platelet lysate consisting of a natural repair proteome supports human mesenchymal stem cell proliferation and chromosomal stability. Cell Transplant 20, 797–811, 10.3727/096368910X543376 (2011).
Shu-Shan, Z., Jian-Jun, D., Cai-Feng, W., Ting-Yu, Z. & De-Fu, Z. Comparative proteomic analysis of hearts of adult SCNT Bama miniature pigs (Sus scrofa). Theriogenology 81, 901–905, 10.1016/j.theriogenology.2014.01.006 (2014).
Arce, C., Lucena, C., Moreno, A. & Garrido, J. J. Proteomic analysis of intestinal mucosa responses to Salmonella enterica serovar typhimurium in naturally infected pig. Comp. Immunol. Microbiol. Infect. Dis. 37, 59–67, 10.1016/j.cimid.2013.10.008 (2014).
Acknowledgements
This study was partly supported by the NIH grant numbers: DK73608, DK104273, HL123160, and DK102325, DK106427, the Mayo Clinic Center for Regenerative Medicine, and the Mayo Clinic Foundation: Mary Kathryn and Michael B. Panitch Career Development Award.
Author information
Authors and Affiliations
Contributions
A.E. conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, and final approval of manuscript; X.-Y.Z. collection and/or assembly of data, data analysis and interpretation, and manuscript writing; J.R.W., H.T., A.S.P. and S.D. collection and/or assembly of data and data analysis and interpretation; A.J.v.W. and A.L. manuscript writing and final approval of manuscript; L.O.L. conception and design, financial support, collection and/or assembly of data, data analysis and interpretation, manuscript writing, and final approval of manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Eirin, A., Zhu, XY., Puranik, A. et al. Comparative proteomic analysis of extracellular vesicles isolated from porcine adipose tissue-derived mesenchymal stem/stromal cells. Sci Rep 6, 36120 (2016). https://doi.org/10.1038/srep36120
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep36120
This article is cited by
-
Osteogenic human MSC-derived extracellular vesicles regulate MSC activity and osteogenic differentiation and promote bone regeneration in a rat calvarial defect model
Stem Cell Research & Therapy (2024)
-
Autologous Extracellular Vesicles Attenuate Cardiac Injury in Experimental Atherosclerotic Renovascular Disease More Effectively Than Their Parent Mesenchymal Stem/Stromal Cells
Stem Cell Reviews and Reports (2023)
-
Bone marrow mesenchymal stem cells’ osteogenic potential: superiority or non-superiority to other sources of mesenchymal stem cells?
Cell and Tissue Banking (2023)
-
Heterogeneity of mesenchymal stem cell-derived extracellular vesicles is highly impacted by the tissue/cell source and culture conditions
Cell & Bioscience (2022)
-
Extracellular vesicles for ischemia/reperfusion injury-induced acute kidney injury: a systematic review and meta-analysis of data from animal models
Systematic Reviews (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.