Abstract
Ribose-5-phosphate isomerase (RPI) belongs to the non-oxidative branch of the pentose phosphate pathway, catalysing the inter-conversion of D-ribose-5-phosphate and D-ribulose-5-phosphate. Trypanosomatids encode a type B RPI, whereas humans have a structurally unrelated type A, making RPIB worthy of exploration as a potential drug target. Null mutant generation in Leishmania infantum was only possible when an episomal copy of RPIB gene was provided and the latter was retained both in vitro and in vivo in the absence of drug pressure. This suggests the gene is essential for parasite survival. Importantly, the inability to remove the second allele of RPIB gene in sKO mutants complemented with an episomal copy of RPIB carrying a mutation that abolishes isomerase activity suggests the essentiality is due to its metabolic function. In vitro, sKO promastigotes exhibited no defect in growth, metacyclogenesis or macrophage infection, however, an impairment in intracellular amastigotes’ replication was observed. Additionally, mice infected with sKO mutants rescued by RPIB complementation had a reduced parasite burden in the liver. Likewise, Trypanosoma brucei is resistant to complete RPIB gene removal and mice infected with sKO mutants showed prolonged survival upon infection. Taken together our results genetically validate RPIB as a potential drug target in trypanosomatids.
Similar content being viewed by others
Introduction
Some clinically relevant pathogens can survive and replicate in macrophages (MØ). However, only the protozoan Leishmania and Coxiella bacteria are known to thrive in fully mature phagolysosomes1,2. Mammals become infected with Leishmania through the bite of an infected sand fly, which injects non-replicative metacyclic promastigotes into the skin that are later phagocytosed by MØ2. Upon delivery to the phagolysosome, the internalized promastigotes differentiate into amastigotes3. The latter are able to rapidly re-infect other phagocytic cells (MØ or dendritic cells), as well as some non-phagocytic cells (fibroblasts), leading to several possible disease outcomes, such as acute disease (ranging from self-healing cutaneous infections to fatal, if untreated, visceral forms), as well as chronic or latent infections3.
Leishmaniasis is an important worldwide human health problem, affecting approximately 12 million people, with 1.3 million new cases every year4. Visceral leishmaniasis (VL), the most severe form of the disease, is mainly associated with Leishmania donovani or infantum infections. Currently, control of VL relies mainly on chemotherapy and vector control, both presenting several limitations that hinder disease eradication in endemic areas5. Traditional chemotherapy is often associated with high cost, toxicity, complex administration regimes and the emergence of resistance5. Consequently, approximately 20,000 to 30,000 people die every year4, rendering the search for novel chemotherapeutic options a priority.
The Pentose Phosphate Pathway (PPP) is a key metabolic pathway that relies on the use of glucose and is classically divided into two branches: an oxidative and a non-oxidative branch. In the trypanosomatids, enzymes from the oxidative branch, namely glucose-6-phosphate dehydrogenase (G6PD), 6-phosphogluconolactonase (6PGL) and 6-phosphogluconate dehydrogenase (6PGDH) play an important housekeeping role and are related to their cyanobacteria and plant orthologues6,7. The non-oxidative PPP is responsible for the interconversion of phosphorylated saccharides, giving rise to products (ribose-5-phosphate - R5P), intermediates (glyceraldehyde-3-phosphate - G3P, fructose-6-phosphate - F6P) and cofactors (NADPH) used to synthesize nucleic acids and lipids and to maintain redox homeostasis8. Curiously, the enzymes involved in the non-oxidative branch, such as ribose-5-phosphate isomerase B (RPIB), ribose-5-phosphate-3-epimerase (RPE), transketolase (TKT) and transaldolase (TAL), constitute a more heterogeneous group, including a member that lacks any mammalian orthologue (RPIB9) and others (RPE and TKT) that are developmentally regulated and dispensable in a species-specific manner10,11,12,13,14.
RPI enzymes catalyse the interconversion of ribose 5-phosphate (R5P) and ribulose 5-phosphate (Ru5P), depending on the substrate and product concentrations15. Two types of RPI enzymes can be found: type A (RPIA) is represented in all plant and animal kingdoms, contrasting with the type B (RPIB) that is restricted to some bacteria and protozoans15. Trypanosomatids possess a RPI type B which has no mammalian homologue9,16,17,18. Indeed, we have recently demonstrated that in Trypanosoma brucei bloodstream forms, RPIB knockdown induces a dramatic impairment in parasite infectivity18.
Taken together, RPIB appeared suitable for investigation as a drug target candidate in Leishmania. Thus, to evaluate the importance of this protein for Leishmania survival and infectivity, we have performed target gene replacement studies in L. infantum and characterized the mutants generated. Target gene replacement studies were also undertaken in T. brucei.
Results
LiRPIB and LmRPIB sequence alignment
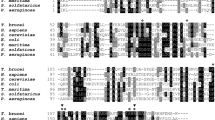
The open reading frames (ORFs) encoding a putative RPIB enzyme were identified in the genomes of L. infantum JPCM5 (LinJ.28.2100) and L. major Friedlin (LmjF.28.1970)19,20. The RPIB amplified sequences from these strains matched 100% with the annotated sequences. RPIB sequences of L. infantum (LiRPIB), L. major (LmRPIB), T. brucei (TbRPIB) and T. cruzi (TcRPIB) generate polypeptides containing 172, 172, 155 and 159 amino acids, respectively (Fig. 1a). LiRPIB displays a 93% sequence identity with LmRPIB and around 50% to RPIB from trypanosomes. However, LiRPIB possesses s only approximately 12% similarity to human RPIA. The amino acids involved in isomerisation (Cys69), phosphate stabilization (Arg113, Arg137, Arg141) and ring opening (His102, His138) have been identified in TcRPIB16 and are strictly conserved in the LiRPIB, LmRPIB and TbRPIB sequences (Fig. 1a).
Multiple-sequence alignment of RPIB proteins from trypanosomatids and analysis of recombinant LiRPIB and LmRPIB.
(a) Alignment of LiRPIB (NCBI-Gene ID: 5070424/LinJ.28.2100), LmRPIB (NCBI-Gene ID: 5653408/LmjF.28.1970), TbRPIB (NCBI-Gene ID: 3664062/Tb927.11.8970) and TcRPIB (NCBI-Gene ID: 3542840/TcCLB.508601.119). The colour pattern according to ALSCRIPT Calcons (Aline version 011208) indicates: red, identical residues; orange to blue, scale of conservation of amino acid properties in each column alignment; white, dissimilar residues). Secondary structure components of TcRPIB crystal structure (black) are represented above the alignment (arrows correspond to beta-sheets and cylinders correspond to alpha-helices). In all sequences, residues involved in ligand binding, isomerisation and the architecture of the catalytic pocket were represented with black circles above the alignment: Purple, light green and light blue circles below the alignment indicate the residues involved in the isomerisation, ring opening and phosphate stabilisation, respectively. (b) Coomassie blue stained SDS-PAGE gel of 10 μg of recombinant LiRPIB and LmRPIB post affinity chromatography purification. (c) Western-blot analysis of recombinant LiRPIB or whole L. infantum promastigote extract using a rabbit polyclonal anti-LiRPIB (1:1000). MW, molecular weight marker.
Enzymatic characterization of LiRPIB and LmRPIB
LiRPIB and LmRPIB recombinant proteins, containing a 6-histidine N-terminal tag, were expressed in Escherichia coli and purified by affinity chromatography under native conditions. Coomassie blue is represented in Fig. 1b, showing that both proteins have the predicted MW for the monomer (LiRPIB ~20.83, LmRPIB ~20.78 kDa). Coomassie staining also demonstrates that the purified recombinant proteins possess a suitable purity level to proceed to enzymatic studies. Additionally, recombinant LiRPIB was used to immunise rabbits to produce polyclonal anti-LiRPIB antibodies. Those antibodies recognise the recombinant protein and a single band in an extract of L. infantum promastigotes with the expected MW (~18.6 kDa, [web.expasy.org/protparam]) (Fig. 1c). Both LiRPIB and LmRPIB catalyse the conversion of R5P into Ru5P (direct reaction) or Ru5P into R5P (reverse reaction) in vitro, with similar kinetic constants, as shown in Supplementary Table S1. The reverse reaction, which generates R5P, seems to be favoured, with both enzymes presenting lower Km values and higher vmax and kcat.
RPIB expression and localisation in L. infantum promastigote and amastigote forms
The rabbit anti-LiRPIB polyclonal antibody was also validated by IFA, as the fluorescence intensity of WT promastigotes versus LiRPIB mutants (single knockout (sKO; clone 15) and overexpressing line (OE)) was measured and a positive correlation between protein levels and labelling intensity was found (Supplementary Fig. S1).
Using α-tubulin as a loading control and Western-blot analysis, we compared RPIB expression in different stages of the parasite’s life cycle. RPIB was found to be more highly expressed in amastigotes (5 fold increase) followed by logarithmic promastigotes (3 fold increase) when compared to late stationary phase promastigotes (Fig. 2a). LiAS-A (asparagine synthetase A21), which is equally expressed in the different stages and LimTXNPx (mitochondrial tryparedoxin22), which is more expressed in amastigotes, were used as controls (Fig. 2a). This result was also supported by immunofluorescence analysis, as the intensity of the RPIB signal, normalized for the parasite area, was higher in amastigotes, whereas in the case of AS-A no significant difference was observed (Supplementary Fig. S2).
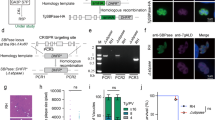
RPIB expression and localization in L. infantum.
(a) RPIB expression in different stages of the L. infantum life cycle. Promastigote forms: logarithmic phase (Log), early stationary phase (ES), late stationary phase (LS); axenic amastigote forms (Ama). Twenty μg of total extracts were analysed by Western-blot and probed with rabbit polyclonal anti-LiRPIB, anti-LiAS-A or LimTXNPx. α-tubulin, probed using a mouse monoclonal antibodywas used as loading control. Quantification, expressed in test protein/tubulin ratio, is presented in the right panel. Two-tailed unpaired t-test was performed: statistical significance p < 0.05. (b–e) Immunofluorescence analysis showing LiRPIB (red in b and d; green in c and e) localization in L. infantum promastigote (b,c) and amastigote (d,e) forms. Nucleus and kinetoplast DNA, cytosol and glycosomes were stained with DAPI (blue), sheep anti-LiTDR1 (thiol-dependent reductase 1, green) and rabbit anti-LiHGPRT (red), respectively. On panels b and d, red arrows in the zoomed areas correspond to sites of exclusive LiRPIB staining. On panels b–e, yellow arrows in the zoomed areas point to colocalisation sites. Images are maximal Z-projections of 30 to 35 contiguous stacks separated by 0.1 μm and were acquired with a 63x objective, using a LEICA SP5II confocal microscope. The scale bar corresponds to 5 μm. Data displayed in a–e are representative of three independent experiments.
To determine LiRPIB subcellular localization, we resorted to in silico prediction and performed digitonin fractionation and immunofluorescence analysis. Bioinformatics tools, Wolf Psort and CELLO, predicted a cytosolic localization for RPIB. Digitonin fractionation followed by Western-blot analysis was performed in promastigotes. The following antibodies were used to detect the fractioning pattern of proteins located in different subcellular compartments, namely, anti-TbEnolase (LiEnolase versus TbEnolase 79% identity, LiEnolase 39.6 kDa) as a cytosolic marker23 and anti-LdHGPRT (hypoxanthine guanine phosphoribosyl transferase, LiHGPRT 23.6 kDa) as a glycosomal marker24. Enolase (Supplementary Fig. S3a) was found in the supernatant at digitonin concentrations as low as 12.5 μg/ml. HGPRT, in turn, was retained longer in the pellet fractions (detected up to 1000 μg/ml), being significantly released in the supernatant only at 100 μg/ml. RPIB is released in the supernatant and retained in the pellet fractions similarly to Enolase and HGPRT, respectively. To further elucidate RPIB localization, we have also performed immunofluorescence in promastigotes and axenic amastigotes, followed by confocal microscopy. TDR1 (thiol-dependent reductase 1)25 and HGPRT were labelled as cytosolic and glycosomal markers, respectively. Representative images of the labelling with these two markers are shown in Supplementary Figs S3b and S4a. The RPIB distribution pattern and the co-localization with the cytosolic marker TDR1, with exception of few areas (highlighted with red arrows in Fig. 2b,d and Supplementary Figs S3c and S4b), indicate that the protein is mainly localized in the cytosol. However, some co-localization with the glycosomal marker, HGPRT, was observed (Fig. 2c,e and Supplementary Figs S3d and S4c), suggesting this protein might have a dual localization. Indeed, in all the parasites analysed, we always observed partial co-localization of RPIB with TDR1 and HGPRT. The images provided are representative of these observations.
In conclusion, RPIB despite having mainly cytosolic localisation in both promastigotes and axenic amastigotes can also be found in association with glycosomes.
LiRPIB facilitated null mutant generation by targeted gene replacement
A targeted gene replacement strategy was used for RPIB gene inactivation in L. infantum. The first transfection allowed the removal of one of the gene alleles generating sKO clones resistant to either neomycin (NEO) or hygromycin (HYG), depending on the cassette that was used. When attempting to remove the second gene allele, independently of using a HYG or NEO, no null mutants were obtained from a total of four independent experiments. Some of the mutants obtained following the dKO attempt were likely to have been aneuploid (Supplementary Table S2), as the parasites were shown by PCR and Southern-blot to carry both resistance cassettes and the gene at the target locus (data not shown). The repeated inability to remove the second allele, coupled with the constant aneuploidy generation was suggestive of gene essentiality. Therefore, a HYG construct has been transfected in parallel into parasites with the following genotypes: WT, sKONEO (sKO 15) and sKONEO complemented (sKO P15) with an episome carrying the RPIB gene (pSPαBLASTαRPIB). This approach enabled the generation of sKO mutants from the WT and dKO mutants only in the complemented sKO, therefore once more failing to generate mutants that were truly null in terms of RPIB. The WT parasites were transfected with the pSPαBLASTαRPIB plasmid to generate an overexpressing cell-line.
The genotype of the facilitated null mutants (dKO P15.3, dKO P15.14 and dKO P15.15) was confirmed by PCR (Fig. 3b) and Southern-blot (Fig. 3c), these respective strategies are illustrated in Fig. 3a. The successful integration of the selectable markers, the presence of RPIB in the episome and its absence from the chromosomic locus were verified by PCR. The genotypes were further confirmed by Southern-blot. A first hybridization was performed using 3′ UTR as a probe: the bands of ~7547 bp, ~3958 bp and ~4574 bp correspond to RPIB, NEO and HYG, respectively (Fig. 3c). The blot was then stripped and re-probed three additional times to confirm each of the bands. Re-probing with RPIB gene confirmed its presence in the chromosomes of WT, OE, sKO 15 and sKO P15 (~7547 bp) and in the episome in OE, sKO P15 and facilitated null mutants (~3606 bp) (Fig. 3c). All the clones in the promastigote stage were also analysed by Western-blot and showed RPIB reduction in sKO and overexpression in the sKO P15 and OE mutants (Fig. 3d). As expected, RPIB levels are also down regulated in the axenic amastigotes sKO (clone 15) when compared to WT or to the complemented line (sKO P15) (Fig. 3e).
Genetic and post-translational analysis of the LiRPIB mutants.
(a) LiRPIB locus (RPIB allele, targeted gene replacement cassettes, containing NEO and HYG) and pSPαBLASTαLiRPIB schematics. Horizontal arrows and numbers represent the primer pairs (sequences in Table S4) used to assess the genotype of the mutants: the black dashed line represents the expected PCR fragment. The colour of those arrows indicates the region of annealing: HYG, NEO, RPIB, UTRs used in the replacement cassettes, endogenous regions of the parasite or pSPαBLASTαLiRPIB backbone in blue, yellow, purple, green, black or grey, respectively. Southern-blot approach, upon digestion with EcoRI (vertical black arrows) and NaeI (vertical red arrows) is also represented: dashed red lines represent the expected digestion fragments. (b) PCR analysis of LiRPIB mutants to assess LiRPIB, NEO and HYG presence in chromosome 28, as well as pSPαBLASTαLiRPIB presence. B, blank. (c) Southern-blot analysis of 10 μg of LiRPIB mutants (versus WT) genomic DNA, previously digested with EcoRI and NaeI and probed using 3′ UTR. The blot was then stripped and reprobed using LiRPIB. The horizontal arrows highlight the bands that were obtained. (d,e) Western-blot analysis of RPIB expression in the LiRPIB mutants (versus WT), in both promastigotes (d) and axenic amastigotes (e). 1 × 107 parasites were used for total extract and LiCS (cysteine synthase) or LiAS-A (asparagine synthetase A) were used as loading control in d and e, respectively. (f) pSPαBLASTα quantification by qPCR using 10 ng of genomic DNA from LiRPIB OE and facilitated null mutants (fdKO) P15.3, P15.14 and P15.15 and from LiASA OE and complemented null mutants (cdKO) clones A and B. We used DNA from each of the mutants cultured for 3 and 6 months in complete RPMI in the absence of blasticidin and calibrated against the correspondent mutant maintained in culture in the presence of the drug. rRNA45 was used as the reference gene. The results correspond to means plus standard deviation of two independent experiments. WT, wild type; OE, overexpressor; sKO, single knockout (clones 13 and 15); sKO P, sKO (clones 13 and 15) complemented with pSPαBLASTαLiRPIB; dKO P15, facilitated null mutants (clones 3, 14 and 15).
The facilitated dKO mutants were subcultured weekly for 6 months in the presence or absence of blasticidin drug pressure. In the absence of drug pressure, LiRPIB facilitated mutants (fdKO) did not lose the plasmid, as assessed by qPCR (Fig. 3f). On the other hand, LiRPIB OE, as well as an unrelated non-essential protein (LiAS-A21) OE and its complemented null mutants (also using pSPαBLASTα as a vector; cdKO), lost the plasmid. Importantly, LiRPIB facilitated null mutant promastigotes when cultured in RPMI or SDM enriched or not with D-ribose do not lose the plasmid suggesting D-ribose supplementation appears not to compensate for the need of RPIB (Supplementary Fig. S5).
In conclusion, our results indicate RPIB is essential for the survival of L. infantum.
Impact of LiRPIB partial inactivation on in vitro growth and infection
Several experiments using the different mutants were conducted in vitro to assess whether LiRPIB has a role in promastigote growth and metacyclogenesis, MØ infection and/or intracellular amastigote replication.
All promastigote mutants grew in a similar fashion to the WT (Fig. 4a). The expression of metacyclogenesis markers (Histone H4, SHERP, MET-126) in stationary cultures was not statistically different from the WT (Fig. 4b). Moreover, all the clones successfully differentiated into axenic amastigotes and apparently enter bone marrow derived MØ normally, as observed at 4 hours post infection, either by FACS (Fig. 4c) or microscopic analysis. Indeed, no differences in the percentage of infected cells nor the numbers of parasites per infected cell were seen at 4 and 24 (Fig. 4d,e) hours post-infection between WT and mutant parasites. While the percentage of infected cells remained similar during experiments for all the mutants, cells infected with sKO presented a statistically significant decrease in the average number of amastigotes per cell at 72 hours post-infection (Fig. 4e). Indeed, cells infected with WT presented 2.19 ± 0.10 amastigotes per cell, while cells infected with sKO 1.66 ± 0.01. This reduction was due to a higher percentage of cells with one amastigote and a lower percentage of cells containing, at least, three amastigotes when compared with the WT (Supplementary Fig. S6b). Importantly, this phenotype is not observed in complemented sKO (sKO P15). Interestingly, no defects are seen in the growth of sKO 15 axenic amastigotes when compared to WT or sKO P15 (Supplementary Fig. S6c).
In vitro characterization of LiRPIB mutants: promastigotes growth, metacyclogenesis, MØ invasion and intracellular amastigote replication.
(a) L. infantum promastigotes growth curves of LiRPIB mutants (versus WT), cultured in cRPMI. Data correspond to mean values of duplicates ± standard deviation. (b) Histone H4, SHERP and MET-1 gene quantification of expression levels by qRT-PCR. Total RNA from WT and LiRPIB mutant stationary promastigotes was extracted and converted into cDNA. rRNA45 was used as reference gene. Calibration was performed against WT parasite. Data correspond to mean values of duplicates plus standard deviation. (c) Percentage of infected bone marrow derived MØ at 4 hours post infection, determined by FACS analysis using CFSE-labelled WT or LiRPIB mutant stationary promastigotes. Infections were performed using a ratio of 2, 5 or 10 parasites per cell. Data correspond to mean values of duplicates plus standard deviation. (d,e) Percentage of infected bone marrow derived MØ (d) and average of amastigotes per cell (e) at 24 and 72 hours post infection. WT or LiRPIB mutant stationary promastigotes were used at a ratio of 10 parasites per cell. At the appropriate time points, the cells were Giemsa stained and analysed microscopically. Data correspond to mean values of duplicates plus standard deviation. Two-tailed unpaired t-test was performed: statistical significance p < 0.05. The data displayed in a–e correspond to two independent experiments.
In conclusion, the in vitro results suggest that LiRPIB might be important for the replication of intracellular amastigotes.
Impact of LiRPIB partial inactivation on the establishment of infection in mice
To evaluate the role of RPIB on the infectivity of the parasites, BALB/c mice were infected with WT or LiRPIB mutant promastigotes and the parasite burden in the spleen and liver determined at two (acute phase) and eight weeks (chronic phase) post infection. At two weeks post infection, differences in the splenic parasite loads were detected only for sKO mutant clone 15, while in the liver, animals infected with sKO mutants (clones 13 and 15) showed reduced parasite loads (Fig. 5a,b). Importantly, these differences were not seen in animals infected with sKO lines complemented with episomal RPIB (clones sKO P13 and sKO P15). Western-blot analysis of promastigotes differentiated back from the spleen and the liver in the acute phase demonstrates RPIB downregulation in sKO and overexpression in the complemented sKO. A representative Western-blot and the respective quantification are shown in Fig. 5c,d. In the chronic phase, there was overall a decrease of the parasite burden in the liver to levels close to the detection limit. This parasite clearance has been described in BALB/C model due to a partial self-resolution of the infection27. In the spleen, the parasite burdens were maintained similarly to the acute phase and in the sKO mutants there was no parasite clearance or a more accentuated defect (data not shown). Overall both WT and mutant parasites persisted in both organs. Mutant parasites recovered from the spleen and the liver of chronically infected animals were analysed by qPCR for the presence of the episome carrying RPIB. While the OE had lost the episome over time, the same finding did not occur with the facilitated null mutant clone P15.14 (Fig. 5e), suggesting RPIB is also essential in amastigote form.
Infectivity of LiRPIB mutants in mice.
(a,b) 1 × 108 early stationary promastigotes were intra-peritoneally injected in BALB/c mice that were sacrificed 2 weeks post infection. The parasite burden in spleen (a) and liver (b) was assessed by the limiting dilution method. The values represent the mean of four animals ± standard deviation. Two-tailed unpaired t test was performed: statistical significance p < 0.05. (c,d) Western-blot analysis of RPIB levels in WT and LiRPIB mutants, which were recovered from the limiting dilution plates of the spleen (S) or liver (L). 1 × 107 promastigotes were used for total extract preparation and LiCS (cysteine synthase) was detected as loading control. The fold difference in of RPIB expression in LiRPIB mutants in comparison to WT recovered from the spleen and liver at 2 weeks post-infection is shown in d. The results correspond to means plus standard deviation of two independent experiments. In c, a representative Western-blot is shown. (e) pSPαBLASTα quantification by qPCR using 10 ng of genomic DNA from LiRPIB OE and facilitated null mutant P15.14 that were left in culture for 6 months in the absence of blasticidin or recovered from spleen, or liver 2 months post-infection relative to the respective mutant maintained in culture under drug pressure. rRNA45 was used as a reference gene. The results correspond to means plus standard deviation of two independent experiments.
These data suggest RPIB is important in establishing Leishmania infection in the mouse, particularly in the liver.
Assessment whether LiRPIB essentiality is dependent on the isomerase function
We consider that assessing whether protein essentiality is due to the annotated metabolic function is particularly important in the context of drug target validation. T. cruzi RPIB isomerase activity is known to be abrogated by replacing cysteine 69 by an alanine without compromising the overall 3D conformation16. Therefore, we tested whether a similar mutation would abrogate LiRPIB isomerase activity and whether the removal of a second RPIB allele in a sKO complemented with a plasmid carrying the mutated RPIB (sKO P15C69A) would be successful. This experimental approach would reveal whether protein essentiality is independent (Fig. 6a6) or dependent (Fig. 6a7) on RPIB isomerase function, respectively.
Assessing whether LiRPIB essentiality depends on its isomerase function.
(a) Schematic overview of the strategy used to determine if LiRPIB essentiality depends on its isomerase function. WT (1), sKO 15 provided with pSP72αBLASTαRPIBWT (sKO P15WT, 2) or pSP72αBLASTαRPIBC69A (sKO P15C69A, 3) were transfected with a HYG containing construct. The generation of sKOHYG mutants from the transfection of the WT parasites (4) and the generation of facilitated null mutants from the transfection of sKO P15WT parasites (5) were used as controls. The ability to remove the second RPIB allele or not when transfecting sKO P15C69A parasites would determine whether the protein essentiality was independent (6) or dependent (7) on isomerase function, respectively. (b) Coomassie blue stained SDS-PAGE gel of 10 μg of recombinant LiRPIBWT and LiRPIBC69A post affinity chromatography purification. (c) Measurement of LiRPIBWT and LiRPIBC69A activity, catalysing the direct reaction (R5P−> Ru5P), using 50 mM of R5P and 0.5 or 1 μg of enzyme and the direct spectrophotometric method at 290 nm. The data are representative of 2 independent experiments. (d) Western-blot analysis of LiRPIB expression in WT, sKO 15, sKO P15WT, sKO P15C69A and OE, before the transfection scheme represented in A. 1 × 107 parasites were used for total extract preparation and LiCS (cysteine synthase) was used as loading control. (e) PCR analysis of LiRPIB mutants to assess LiRPIB, NEO and HYG presence in chromosome 28, as well as pSPαBLASTαLiRPIB presence. See Table S4 for primer sequences, the expected fragments and annealing sites are shown in Fig. 3s. B, blank.
For this purpose, a mutated recombinant LiRPIBC69A protein was expressed in E. coli to confirm the inactivation of the isomerase function. The recombinant protein possessed the expected MW (~21.09 kDa, Fig. 6b) but failed to convert R5P into Ru5P (Fig. 6c) or Ru5P into R5P (data not shown). The sKO 15 was complemented with pSP72αBLASTαRPIBC69A, and the expression levels of RPIB in sKO P15WT and sKO P15C69A were found to be similar (Fig. 6d). Afterwards, WT (Fig. 6a1) and sKO 15 provided of pSP72αBLASTαRPIBWT (sKO P15WT, Fig. 6a2) or pSP72αBLASTαRPIBC69A (sKO P15C69A, Fig. 6a3) were transfected with a HYG construct (Fig. 6a). As controls, two series of mutants were successfully generated: sKOHYG mutants from WT transfection (Fig. 6a4) and facilitated null mutants from sKO P15WT transfection (Fig. 6a5). Eleven out of 12 (Supplementary Table S2) facilitated null mutants were obtained from sKO P15WT transfection, while no facilitated null mutants were obtained with sKO P15C69A. PCR analysis of representative mutants from each series is shown in Fig. 6e, following the same strategies depicted in Fig. 3a. The previously generated facilitated null mutant clone P15.14 was used as positive control.
These data indicate LiRPIB essentiality is due to its isomerase function.
TbRPIB null mutant generation attempt by targeted gene replacement
Taking into account RPIB essentiality in L. infantum and a previous report of its importance for T. brucei bloodstream forms infectivity18, we investigated whether essentiality would extend to this latter organism. To obtain TbRPIB null mutants, a target gene replacement strategy was also applied. One TbRPIB allele was replaced by HYG giving rise to sKO mutants at the first attempt. PCR and Southern blot, as shown in Fig. 7a, confirmed the correct insertion of the cassette (Fig. 7b,c). For the generation of TbRPIB dKO mutants, sKO parasites were transfected either with a BLEO construct or with a NEO construct, with no success. The NEO construct reliability was assessed by simultaneous transfection of WT and sKO mutant. Resistant parasites were only obtained for the WT but not for the sKO (Fig. 7d), enabling the generation of sKO but not null mutants. Therefore, we characterized the generated sKO cell lines, which showed a reduction in TbRPIB protein levels of approximately 50% (semi-quantification by densitometry) in sKO cell lines in comparison with the WT, as assessed by Western-blot (Fig. 7e). No significant differences were found on the in vitro growth of TbRPIB sKO bloodstream forms compared with WT (Fig. 7f). In vivo, TbRPIB sKO showed a reduced infectivity, evaluated by the parasitaemia (Fig. 7g), consequently, the infected mice had an increased life span (Fig. 7h) when compared with WT parasites.
Generation of TbRPIB mutants and their in vitro and in vivo characterization.
(a) TbRPIB locus (RPIB allele and targeted gene replacement cassettes, containing NEO and HYG) schematics. Horizontal arrows and numbers represent the primer pairs (sequences in Table S4) used to assess the genotype of the mutants: the black dashed line represents the expected PCR fragment. The colour of those arrows indicates the region of annealing: HYG, NEO or UTRs in blue, yellow or black, respectively. Southern-blot approach, upon digestion with BspHI (vertical red arrows) is also represented: dashed red lines represent the expected digestion fragments. (b) PCR analysis of TbRPIB sKO mutants to assess HYG 5′ integration. B, blank. (c) Southern-blot analysis of 10 μg of TbRPIB mutants (versus WT) genomic DNA, previously digested with BspHI and probed using 3′ UTR. (d) PCR analysis of TbRPIB sKO mutant to assess NEO 5′ integration. B, blank. (e) Western-blot analysis of RPIB expression in bloodstream mutants (versus WT) using aldolase as loading control. (f) Growth curve of WT versus a representative sKO cell line. Black and grey squares represent WT and sKO growth, respectively. Cumulative cell numbers (product of cell number and total dilution) are plotted. Values represent averages from two independent experiments using one representative sKO clone and ± error bars indicate standard deviation. (g) Groups of mice (n = 3) were infected intraperitoneally with 1 × 104 control WT (black) or a representative sKO clone (grey). Parasitemias of each animal are shown from 4th to 6th day post-infection. Values are means. 5 × 104 trypanosomes/ml of blood is the detection limit, which is represented by the black dashed line. For TbRPIB sKO, the mean of parasitemia in two animals at day 6 was below the detection limit. Mice were culled when parasitemia reached 1 × 108 cells/ml. (h) Kaplan–Meier survival analysis of mice infected with WT versus a representative sKO clone (black and grey line, respectively). Parasitemias and survival curve are representative of two independent experiments using two different sKO clones.
Our data strongly suggest that a functional copy of TbRPIB gene is essential for the parasite’s survival and that 50% decrease in RPIB levels is sufficient to compromise its infectivity.
Discussion
The genomes of all the trypanosomatid parasites contain sequences that encode for a putative RPIB. Recombinant enzymes from L. donovani17, T. brucei18 and T. cruzi9 have been formally demonstrated to have in vitro isomerase activity, catalyzing the interconversion of R5P and Ru5P. In this paper we have demonstrated that the same applies to L. infantum and L. major homologues. Leishmania enzymes share over 90% identity with each other but only ~50% with the RPIB from trypanosomes. Nevertheless, the protein residues so far associated with isomerization, ring opening and phosphate charge stabilization are strictly conserved. The Km values for both R5P and Ru5P were similar between Leishmania and trypanosomes, however, kcat values are considerably higher for Leishmania, in both direct and reverse reactions (Supplementary Table 1 versus9,17,18). A decrease in Km and an increase in kcat were consistently observed for Ru5P in comparison to R5P (Supplementary Table S1), suggesting that the conversion of Ru5P into R5P is favoured, which might be explained by the important role of R5P as a building block for nucleic acid synthesis.
Trypanosomatids have unique organelles to respond to the specific needs of their life cycles. Amongst these are glycosomes, peroxisome-related organelles that comprise enzymes of important metabolic pathways such glycolysis, PPP, among others28. This compartmentalisation of metabolic pathways may prevent the accumulation of toxic intermediates29 or enable fast metabolic adaptation to environmental changes28. LmRPIB possesses a peroxisome targeting sequence 2 (PTS-2), a glycosomal signal peptide sequence (–RVALGCDHA–30), that is conserved in both L. infantum and L. donovani. Our study suggests a dual localisation, despite most RPIB being detected in the cytosol of L. infantum promastigotes and amastigotes, it also localises to a certain extent in the glycosomes, similarly to what we have already described for TbRPIB18. Moreover, other enzymes of the same pathway have been shown to display the same pattern, for example, those enzymes immediately upstream, or downstream, of RPIB, 6PGDH and TKL, respectively11,12,30,31,32. However, recent proteomic analysis of the L. donovani glycosomes failed to detect RPIB33. This same analysis detected HGPRT (PTS-1), aldolase (PTS-2), as well as some enzymes of the PPP, namely: putative G6PD, TKL, putative RPE (PTS-1), putative 6PGDH and putative TAL (non-identified signal peptide), or other related proteins such as putative ribokinase (PTS-2). We wonder whether this is due to the amount of RPIB in the glycosomes, or whether it is not present in all of them (Supplementary Fig. S3d).
To assess the importance of RPIB to L. infantum survival and infectivity, we attempted to generate null mutants. Successive failures to obtain mutants null to RPIB, coupled with constant aneuploidy generation were suggestive of gene essentiality34,35. We used a classical approach to demonstrate this36,37, by providing the sKO parasites with an episomal copy of RPIB and removing the second copy of the gene, we generated facilitated null mutants. Moreover, the plasmid was not lost even in the long-term absence of drug pressure both in vitro (Fig. 3f) and in vivo (Fig. 5e) further supporting the essentiality of RPIB for L. infantum survival in both promastigote and amastigote forms.
Due to the impossibility of generating mutants truly null to RPIB, we have performed phenotypic studies using the sKO lines. In vitro, sKO promastigotes grow, undergo metacyclogenesis, enter macrophages and differentiate normally. However, our data point to a defective replication of the intracellular amastigotes. Indeed, under these experimental conditions, amastigote replication can be evaluated as the number of parasites per cell significantly increases from 24 to 72 hours post-infection with the WT parasite. Importantly, MØ infected with sKO mutants showed a reduced number of amastigotes per cell 72h post-infection and this phenotype was rescued by complementation with episomal RPIB. It is noteworthy, that the remaining amount of protein probably suffices the parasite needs for minimal intracellular replication and therefore the overall effect is very modest. In fact, in vivo, an equally mild defect was observed, as expected considering the sKO lines present only a 50% downregulation in the protein levels (semi-quantification by densitometry). Moreover, there was no parasite clearance, not even at later stages of infection (data not shown), meaning that the parasites can persist. This type of phenotype is frequently described in mutants of Leishmania for enzymes involved in energy metabolism, e.g., glucose transporter knockout in L. mexicana38 and gluconeogenic enzyme fructose-1,6-bisphosphatase (FBPase) knockout in L. major39, as promastigotes are able to infect and differentiate, but the resulting amastigotes do not replicate failing to generate the normal lesions seen in the mouse model.
The higher expression of RPIB in amastigotes (Fig. 2a) and its importance for intracellular amastigote replication (Fig. 4e and Supplementary Fig. S6a,b) suggests a major role in this stage of the parasite’s life cycle.
Likewise, RPIB knockdown in T. brucei bloodstream forms, has been demonstrated in vivo to reduce infectivity, with an extension of survival for the murine host18. The defective phenotype observed in vivo for the TbRPIB sKO mutants (Fig. 7g,h) was less dramatic when compared to the one obtained with RNAi18. This might be explained by the more pronounced RPIB downregulation achieved with the RNAi system. Three independent attempts to generate dKO mutants have failed, suggesting that RPIB is essential for T. brucei survival. A TbRPIB conditional knockout would ultimately prove gene essentiality40. Nevertheless, our results support the hypothesis that RPIB is a potential therapeutic target in the fight against both Leishmania and T. brucei infections. However, critically analysing the in vivo data for both parasites, it appears that a high inhibition of the enzyme would be required to have a substantial impact on the infection, considering that for 50% downregulation the defects are modest, especially in the case of Leishmania.
Several enzymes have been reported to exhibit protein moonlighting in trypanosomatids22,41,42,43, some involved in carbohydrate metabolism40. In this sense, from a drug discovery perspective, it is critical to assess if the expected metabolic function is one that is detrimental to parasite survival and/or infectivity and therefore the one to be targeted by inhibitory molecules. We have unequivocally demonstrated that RPIB isomerase function is obligatory for the survival of L. infantum (Fig. 6).
We can only speculate why an enzyme involved in the non-oxidative branch of PPP, in which substrate interconversion takes place, should be essential. Amastigotes are known to have complex nutritional requirements, which may have precluded their establishment in early endosomal or non-hydrolytic vacuoles of MØ43, where the levels of amino acids and sugars are not sufficient for the parasite’s demands44,45. Incidentally, the variable nutritional composition of host cell phagolysosomes may be one of the reasons why the Leishmania promastigotes fail to differentiate and the ensuing amastigotes, to replicate in neutrophils2, but are successful in MØs43. This may also explain why sKO mutants exhibit a more pronounced infectivity defect in the liver compared with the spleen.
Many Leishmania auxotrophs do not exhibit any loss of virulence and manage to persist in animal hosts, suggesting that the apparently hostile environment of the phagolysosome is somehow a permissive niche43. Importantly, amastigotes undergo a “stringent metabolic response”, characterized by a sharp decrease in glucose uptake that allows for more efficient energy metabolism, associated with a decrease of energy expenditure in anabolic processes and redirecting carbon to intracellular carbohydrate reserves46. It has been proposed that it facilitates long-term amastigote survival in the nutrient-limited environment of the phagolysosome, but simultaneously and paradoxically increasing dependency on carbohydrate uptake and metabolism46 Ribose can be imported47 and metabolized48 by Leishmania promastigotes and apart from its importance for nucleic acid synthesis it can also be an important source of energy for intracellular amastigotes49. The fact that RPIB sKO axenic amastigotes do not have any intrinsic defect in multiplication (Supplementary Fig. S6c), suggests glucose/ribose uptake from the phagolysosome might be a limiting factor. Interestingly in promastigotes, even upon D-ribose supplementation, the facilitated null mutants did not lose the plasmid carrying RPIB (Supplementary Fig. S5) suggesting that their ability to import ribose and/or convert it into R5P via ribokinase may not be sufficient to compensate RPIB ablation.
Moreover, organisms such as yeast exhibit an alternative NADP-independent pathway for R5P synthesis designated the riboneogenesis pathway50. Although not conclusively shown to be operational in trypanosomes, Leishmania lacks the sequence encoding for the putative key enzyme of the process13. The decrease in glucose and ribose importation along with the absence of an alternative pathway for RPIB generation would render the parasites more dependent on other enzymes that ultimately generate this metabolite.
Apart from the effects from the reduction in the R5P pool, absence of RPIB may lead to the potential accumulation of Ru5P. Indeed, if the upstream enzymes are regulated by the concentration of the end products, then the oxidative branch would become less operational leading to a decrease in NADPH production, rendering the parasites more susceptible to reactive oxygen species. Additionally, Ru5P accumulation could trigger another outcome, if we consider observations made in hepatocarcinoma cells, upon human RPIA knockdown51. In this study, the accumulated Ru5P was converted into xylulose-5P by RPE. This metabolite can activate PP2A activity52, which negatively regulates ERK signalling for cell proliferation. Whether this sort of regulation takes place in Leishmania has not been described, however, the genome does encode for putative RPE19,20, PP2A53 and several MAP kinases54.
In summary, RPIB, which lacks a human homologue, is essential for L. infantum survival and infectivity. We have indications that this may also be the case in T. brucei, thus taking a broader perspective RPIB may be considered a potential drug target in trypanosomatids. The fact that a simple in vitro activity assay could be used for the screening of inhibitory molecules coupled to the existence of structural data16, reinforce this protein as a good candidate. However, further studies concerning its druggability will need to be carried out.
Materials and Methods
Ethics statement
All experiments were approved by the IBMC. INEB Animal Ethics Committees and the Portuguese National Authorities for Animal Health guidelines, according to the statements on the directive 2010/63/EU of the European Parliament and of the Council.
Chemicals and reagents
D-ribose-5-phosphate disodium salt hydrate, D-ribulose-5-phosphate disodium salt, D-ribose, EDTA, cysteinium chloride, tetracycline, carbazole, sulphuric acid, dNTPs, Tween-20, Tris-base, urea, thiourea, DTT, 2-mercaptoethanol, Triton X-100 and IPTG (isopropyl-β-D- thiogalactopyranoside) were purchased from Sigma. Oligonucleotide primers were obtained from STAB VIDA. Restriction endonucleases were from New England Biolabs. Polyclonal antibodies against LiRPIB and TbRPIB were obtained in rabbits inoculated with purified recombinant His-tagged LiRPIB and TbRPIB, respectively.
Parasites
L. infantum (MHOM/MA/67/ITMAP-263) promastigote forms were cultured in complete RPMI 1640 or SDM media with or without supplementation with 100 μM of D-ribose at 26 °C. Axenic amastigote forms were cultured in complete MAA medium at 37 °C, 5% CO2, as described previously26. For in vitro and in vivo characterization, different cell lines were firstly recovered from the spleen of infected BALB/c to restore virulence and posteriorly maintained in culture for no longer than 10 passages26. T. brucei brucei Lister 427 bloodstream forms were cultivated in HMI-9 medium, as previously described55. Depending on the analysis, protein extracts were prepared as follows: 1) 1 × 107 late-stationary L. infantum promastigotes or T. b. brucei bloodstream forms were resuspended in T8 lysis buffer (Tris-base 0.6%, urea 42%, thiourea 15%, DTT 0.3%, Triton X-100 1%); or 2) 1 × 108 promastigotes or axenic amastigotes were resuspended in 100 μL of PBS containing protease inhibitor (Roche) and following 6 freeze/thaw cycles, the parasite suspension supernatants were recovered and then quantified using Bio-Rad DC Protein Assay (Biorad). All the samples were stored at −80 °C.
RPIB protein alignments
LiRPIB, LmRPIB, TbRPIB and TcRPIB protein alignments were performed using the ClustalW program56. Aline program, Version 01120857 was used for editing protein sequence alignments.
RPIB in silico localization prediction
LiRPIB subcellular localisation prediction was performed using WoLF PSORT58 and CELLO59.
Cloning RPIB genes
Ribose 5-phosphate isomerase B from L. infantum (LiRPIB) and L. major (LmRPIB) was obtained by performing PCR on genomic DNA, extracted using DNAzol (Invitrogen) from L. infantum (MHOM/MA/67/ITMAP-263) and L. major strain Friedlin. Fragments of the open reading frames of LiRPIB (LinJ.28.2100; chromosome LinJ.28; 781,581–782,099) and LmRPIB (LmjF.28.1970; chromosome LmjF.28; 766,756–767,274)19,20 were PCR-amplified, using primers 1 + 2 and 3 + 4 (Supplementary Table S3), respectively. PCR conditions were as follows: initial denaturation (2 min at 94 °C), 35 cycles of denaturation (30 s at 94 °C), annealing (30 s at 45 °C,) elongation (2 min at 68 °C) and a final extension step (10 min at 68 °C). In the case of LiRPIB, another restriction strategy was required to clone the gene into a Leishmania overexpression vector – pSPαBLASTα (primers 7 + 8, Supplementary Table S3). PCR conditions were as follows: initial denaturation (2 min at 94 °C), 30 cycles of denaturation (15 s at 94 °C), annealing (30 s at 62 °C,) elongation (1 min at 72 °C) and a final extension step (10 min at 72 °C). In order to obtain LiRPIB with a point mutation with Cys69 (replaced by an alanine, C69A) – LiRPIBC69A, primers 5 + 6 and 7 + 6 (Supplementary Table S3) were used to amplify fragments of the ORF containing the restriction sites; XbaI/AgeI or NheI/AgeI and the desired point mutation, that were then cloned into pGEM-T in order to ultimately clone them in the pET28a(+) (Novagen) and pSPαBLASTα vectors, respectively. PCR conditions were as follows: initial denaturation (2 min at 94 °C), 35 cycles of denaturation (15 s at 94 °C), annealing (30 s at 62 °C,) elongation (30 s at 72 °C) and a final extension step (10 min at 72 °C). All the PCR products were obtained using a Taq DNA polymerase with proofreading activity (Roche), isolated, cloned into a pGEM-T Easy vector (Promega) and sequenced.
Expression and purification of recombinant LiRPIB (WT and C69A) and LmRPIB
The LiRPIBWT, LiRPIBC69A and LmRPIB genes were excised from the pGEM-T Easy vector (using NdeI/EcoRI, NheI/SacI and NdeI/EcoRI, respectively), gel purified and subcloned into pET28a(+) expression vector. The resulting constructs presented a 6-histidine tag at the N-terminal and were transformed into E. coli BL21DE3 cells. Proteins were purified by affinity chromatography as previously described18. Concentration was determined by measuring absorbance at 280 nm using the theoretical molar extinction coefficients of 12950, 12950 and 7450 M−1.cm−1 for LiRPIBWT, LiRPIBC69A and LmRPIB, respectively, making use of NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies). Ten μg of purified recombinant proteins were resolved in SDS/PAGE stained with Coomassie Brilliant Blue G-250 (Biorad).
Western-blot analysis
Five hundred ng of LiRPIBWT recombinant protein, 20 μg of total soluble extracts from both promastigote and amastigote forms, or 1 × 107 parasites were resolved in SDS-PAGE and transferred onto a nitrocellulose membrane (TransBlot Turbo, Bio-Rad), which was blocked, probed, washed and developed as previously described18. The following primary antibodies were used: mouse anti-α-tubulin (clone DM1A, Neomarkers, 1:1000), rabbit anti-LiRPIB (1:500 or 1:1000), rabbit anti-TbRPIB (1:1000), rabbit anti-LiCS (cysteine synthase, 1:2000), rabbit anti-LdHGPRT (hypoxanthine guanine phosphoribosyl transferase, 1:2000), rabbit anti-TbEnolase (1:5000), abbit anti-TbAldolase (1:5000), rabbit anti-LimTXNPx (mitochondrial peroxiredoxin, 1:1000) and rabbit anti-LiAS-A (asparagine synthetase A, 1:1000). Horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse IgG (Amersham) (1:5000 for 1 h, at RT) were used as the secondary antibody. ImageJ software (version 1.47) was used for protein semi-quantification.
Enzymatic Assay
To determine the Km for R5P and for Ru5P, a direct spectrophotometric method at 290 nm or a modification of Dische’s Cysteine-Carbazole method were used, respectively9. The experimental set up was performed as previously18.
Generation of LiRPIB overexpressed (OE) and facilitated null mutants
A targeted gene replacement strategy was used for L. infantum RPIB gene knockout. All the primer sequences for gene replacement cassettes and confirmation of the mutants’ genotypes are given in Supplementary Table S4. Briefly, RPIB flanking regions were amplified from L. infantum genomic DNA and were linked to neomycin phosphotransferase (NEO) or hygromycin phosphotransferase (HYG) genes using a fusion PCR approach. The 5′ and 3′ UTR were amplified using primers 1 + 2 and 3 + 4, respectively. NEO and HYG were amplified from pSP72αNEOα and pGL345HYG templates, using primers 5 + 6 and 7 + 8 respectively, which possess around 30 nucleotides of the 5′ UTR in the sense primer and the first 30 nucleotides of the 3′ UTR in the antisense primer. 5′ UTR_NEO_3′ UTR and 5′ UTR_HYG_3′ UTR constructs were obtained using primers 9 + 10. To obtain an episomal copy of RPIB, LiRPIBWT or LiRPIBC69A, genes were excised from the pGEM-T Easy vector (using XbaI/NdeI), gel purified and subcloned into the vector pSP72αBLASTα. Approximately 10 μg of either linear fragments obtained by fusion PCR or plasmid were purified, concentrated and transfected into 5 × 107 mid-log promastigotes, using an Amaxa Nucleofector II device with human T-cell nucleofector kit (Lonza). The day after transfection, drug selection was carried out at 20 μg/mL of G418 (Invitrogen), 50 μg/mL of hygromycin B (InVivoGen) and/or 30 μg/mL blasticidin (InVivoGen). Parasite cloning, except for transfections with episomes was performed by diluting the parasite suspension to a concentration of 0.5 cells per well in 96-well plates using SDM culture medium. Mutants overexpressing LiRPIB (OE) were obtained by transfecting WT parasites with pSP72αBLASTα LiRPIBWT. The facilitated null mutants were generated by complementing sKO mutants with pSP72αBLASTαLiRPIBWT, followed transfection with 5′ UTR_HYG_3′ UTR.
Generation of TbRPIB mutants
Three different DNA cassettes were generated. The DNA fragments consisted ORFs of the resistance genes (hygromycin, bleomycin and neomycin), flanked by the 5′ and 3′ UTRs of the TbRPIB gene. For the generation of sKO mutants, the plasmid used was pGL345-HYG modified using the 5′ (924 bp, primers 19 + 20, Supplementary Table S4) and 3′ (997 bp, primers 21 + 22, Supplementary Table S4) RPIB flanks obtained through PCR from T. brucei genomic DNA template. The primers contained HindIII/SalI and SmaI/BgIII restriction sites were used for cloning into the appropriate pre-digested pGL345HYG. To remove the second allele, pGL345BLEO or pGL345NEO plasmids were constructed by replacing the HYG with BLEO or NEO, respectively, in the plasmid used to obtain the sKOs, employing SpeI/BamHI restriction enzymes. Thereafter the final plasmids were digested with HindIII and BglII to obtain the final DNA fragment for transfection. Transfection was performed as described for LiRPIB mutants, using 10 μg of DNA. Selection was undertaken using 7.5 μg/ml, 0.2 μg/ml and 5 μg/ml of hygromycin, bleomycin or neomycin, respectively.
PCR and Southern-blot analysis of RPIB mutants
LiRPIB mutants were analysed by PCR (NZYTech or Invitrogen Taq Polymerase) for the presence of the following events: LiRPIB and selectable markers, LiRPIB, LiRPIB 5′ integration, NEO 5′ integration, HYG 5′ integration and pSP72αBLASTαLiRPIB, using primers pairs 9 + 10, 11 + 12, 1 + 12 and 9 + 12, 13 + 14, 15 + 16 and 17 + 18, respectively (Supplementary Table S4). TbRPIB mutants were analysed by PCR for the following events: HYG and NEO 5′ integration, using primers pairs 23 + 24, 25 + 26, respectively (Supplementary Table S4). For Southern blot (SB) analysis, total genomic DNA was extracted. Ten μg of genomic DNA were digested O/N with a 5 fold excess of EcoRI and NaeI (LiRPIB mutants) or BspHI (TbRPIB mutants) at 37 °C and samples were run O/N in a 0.8% agarose gel. The transfer into a nylan membrane, nucleic acid fixation, hybridisation and revelation were performed as previously described21. In the case of LiRPIB mutants, the blots were probed sequentially with 3′ UTR, LiRPIB (Fig. 3c), NEO and HYG, which were PCR amplified, using primers 3 + 4, 11 + 12, 5 + 6 and 7 + 8 (Supplementary Table S4), respectively. In the case of TbRPIB mutants, the blots were probed with 3′ UTR (Fig. 7d), amplified using primers 21 + 22 (Supplementary Table S4).
Real-Time quantitative PCR (qPCR) analysis of LiRPIB mutants
qPCR analysis was used to assess the expression of metacyclogenesis markers in LiRPIB promastigote mutants, as well as pSP72αBLASTαLiRPIBWT copy number. Concerning the expression of the markers for metacyclogenesis (MET1, Histone H4, SHERP (small hydrophilic endoplasmic reticulum associated protein)), total RNA extraction, reverse transcription and qPCR were performed as shown previously26. To assess pSP72αBLASTαLiRPIBWT copy number, 10 ng of genomic DNA, primers 17 + 18 (Supplementary Table S4) and purified plasmid (positive control) were used. In both cases, rRNA45 was used as a reference gene and qPCR reactions were run in duplicate for each sample on a Bio-Rad My Cycler iQ5 (BioRad).
In vitro growth of RPIB mutants
Growth curves of LiRPIB mutants and WT were seeded at 1 × 106 parasites/ml in complete RPMI or complete MAA for promastigote and axenic amastigote forms, respectively. Before launching growth curves, the parasites were maintained in log phase for 2–3 passages in the absence of selection. For TbRPIB mutants and WT in vitro growth curves, cell lines were seeded at 1 × 105 parasites/ml of complete HMI-9 medium, after 48h in the absence of selective drugs. Every 24 h, until day 7 or day 10 in the case of L. infantum or T brucei, respectively, cell growth was monitored microscopically.
In vitro bone marrow derived macrophages (BMMø) infection by LiRPIB mutants
Cell suspension of bone marrow was obtained by flushing the femurs of susceptible BALB/c mice and culturing in 96-well plates (1 × 105 cells per well) or in 24-well plates containing cover glasses (3.5 × 105 cells per well) in LCCM supplemented DMEM, as previously26. For FACS analysis, CFSE labelled promastigotes26 were incubated with the BMMø at ratios of 2:1, 5:1 and 10:1 during 4 hours and then cells were washed to remove non-internalized parasites. The infection rates were determined at 4, 24 and 48 hours post-infection using the BD FACS Canto II cytometer and analysed by FlowJo software. For microscopic analysis, at day 7 of culture, promastigotes were incubated with the BMMø at a 10:1 ratio and again after 4 hours, infection was stopped. Infection ratios and number of parasites per cell were assessed at 4, 24 and 72 hours post infection. At these time points, cells were fixed with 3% p-formaldehyde (PFA), Giemsa stained and cover glasses were mounted on slides, using Vectashield (Vector Labs). Microscopic analysis was performed using a Nikon eclipse 80i (Nikon).
In vivo infectivity of RPIB mutants
To characterize LiRPIB mutants, five to six weeks old female BALB/c mice were obtained from Charles River. For mouse infections, promastigotes from 4 days old stationary cultures were collected, washed, resuspended in PBS to a final amount of 1 × 108 parasites/animal and injected intraperitoneally. Mice were sacrificed at 2 or 8 weeks post-infection. The parasite burden in the spleen and liver was determined by limiting dilution as previously described60. To characterize TbRPIB mutants, after 48h in the absence of selective drugs, 1 × 104 WT and sKO parasites were inoculated intraperitoneally into 6–8 week old BALB/c mice. Parasitaemia was measured on the fourth, fifth and sixth day post-infection through tail blood extraction.
Digitonin Fractionation
This procedure has been performed as previously described21. All fractions were analysed by Western-blot.
Immunofluorescence
L. infantum mid-log or late stationary promastigotes and axenic amastigotes were fixed, permeabilised and stained as previously described61. Parasites were spread on 8 well-IF slides (Polysciences) or 18 well-IF slides (Ibidi) for wide field and confocal microscopy, respectively. The following primary antibodies were used: rabbit anti-LiRPIB (1:500), sheep anti-LiTDR1 (1:500) and rabbit anti-LiHGPRT (1:500). The following secondary antibodies were used: goat anti-rabbit Alexa Fluor 488 or Alexa Fluor 568 and donkey anti-sheep Alexa Fluor 488 (Molecular probes, Life Technologies). In the case of RPIB and HGPRT colocalisation studies, parasites were sequentially incubated with rabbit anti-LiRPIB O/N at 4 °C, unconjugated sheep anti-rabbit for 60 min at RT, donkey anti-sheep Alexa Fluor 488 for 60 min at RT, rabbit anti-LiHGPRT for 2 hours at RT and goat anti-rabbit Alexa Fluor 568 for 60 min at RT (sequential stainings lacking only anti-LiRPIB or anti-LiHGPRT as well as single staining of each of the proteins were performed simultaneously as controls). Images were captured using wide field fluorescence microscope AxioImager Z1 (Carl Zeiss) and confocal Leica TCS SP5II microscope (Leica) for antibody validation and colocalisation studies, respectively. Images were analysed using ImageJ (version 1.47) or Fiji (version 1.45) software’s.
Statistical Analysis
For statistical analysis, two-tailed unpaired t-test was used. Statistical analysis was performed using GraphPad Prism Software (version 5.0) and significance was found when p < 0.05.
Additional Information
How to cite this article: Faria, J. et al. Disclosing the essentiality of ribose-5-phosphate isomerase B in Trypanosomatids. Sci. Rep. 6, 26937; doi: 10.1038/srep26937 (2016).
References
Rabinovitch, M. & Veras, P. S. Cohabitation of Leishmania amazonensis and Coxiella burnetii. Trends Microbiol. 4, 158–161 (1996).
van Zandbergen, G. et al. Cutting edge: neutrophil granulocyte serves as a vector for Leishmania entry into macrophages. J. Immunol. 173, 6521–6525 (2004).
Antoine, J. C., Prina, E., Courret, N. & Lang, T. Leishmania spp. on the interactions they establish with antigen-presenting cells of their mammalian hosts. Adv. Parasitol. 58, 1–68 (2004).
WHO Leishmaniasis, Fact sheet No 375 (2014).
Maltezou, H. C. Drug resistance in visceral leishmaniasis. J. Biomed. Biotechnol. 617521 (2010).
Krepinsky, K., Plaumann, M., Martin, W. & Schnarrenberger, C. Purification and cloning of choroplast 6-phsophogluconate dehydrogenase from spinach. Cyanobacterial genes for chloroplast and cytosolic isoenzymes encoded in eukaryotic chromosomes. Eur. J. Biochem. 268, 2678–2686 (2001).
Hannaert, V., Bringaud, F., Opperdoes, F. R. & Michels, P. A. Evolution of energy metabolism and its compartmentation in Kinetoplastida. J. Mol. Biol. 331, 653–665 (2003).
Stryer, L. Biochemistry, 4th edition, Freeman, New York, 559–565 (1999).
Stern, A. L., Burgos, E., Salmon, L. & Cazzulo, J. J. Ribose-5-phosphate isomerase type B from Trypanosoma cruzi: kinetic properties and site directed mutagenesis reveal information about the reaction mechanism. Biochem. J. 401, 279–285 (2007).
Maugeri, D. A., Cazzulo, J. J., Burchmore, R. J., Barrett, M. P. & Ogbunude, P. O. Pentose phosphate metabolism in Leishmania mexicana. Mol. Biochem. Parasitol. 130, 117–125 (2003).
Maugeri, D. A. & Cazullo, J. J. The pentose phosphate pathway in Trypanosoma cruzi. FEMS Microbiol. Lett. 234, 117–123 (2004).
Veitch, N. J., Maugeri, D. A., Cazzulo, J. J., Lindqvist, Y. & Barrett, M. P. Transketolase from Leishmania mexicana has a dual subcellular localisation. Biochem. J. 382, 759–767 (2004).
Opperdoes, F. R. & Coombs, G. H. Metabolism of Leishmania: proven and predicted. Trends Parasitol. 23, 149–158 (2007).
Stoffel, S. A. et al. Transketolase in Trypanosoma brucei. Mol. Biochem. Parasitol. 179, 1–7 (2011).
Sorensen, K. I. & Hove-Jensen, B. Ribose catabolism of Escherichia coli: characterization of RpiB gene encoding ribose-5-phosphate isomerase Band of the RpiR gene, which is involved in the regulation of RpiB expression. J. Bacteriol. 178, 1003–1011 (1996).
Stern, A. L., Naworyta, A., Cazzulo, J. J. & Mowbray, S. L. Structures of type B ribose-5-phosphate isomerase from Trypanosoma cruzi shed light on the determinants of sugar specificity in the structural family. FEBS J. 278, 793–808 (2011).
Kaur, P. K., Dinesh, N., Soumya, N., Babu, N. K. & Singh, S. Identification and characterization of a novel ribose-5-phosphate isomerase B from Leishmania donovani. Biochem. Biophys. Res. Commun. 421, 51–56 (2012).
Loureiro, I. et al. Ribose-5-phosphate isomerase B knockdown compromises Trypanosoma brucei bloodstream form infectivity. PLoS Negl Trop Dis 9, e3430 (2015).
El-Sayed, N. M. et al. Comparative genomics of trypanosomatid parasitic protozoa. Science 309, 404–409 (2005).
Ivens, A. C. et al. The genome of the kinetoplastid parasite, Leishmania major. Science 309, 436–442 (2005).
Faria, J. et al. Leishmania infantum asparagine synthetase A is dispensable for parasites survival and infectivity. PLoS Negl Trop Dis 10, e0004365 (2016).
Teixeira, F. et al. Mitochondrial peroxiredoxin functions as crucial chaperone reservoir in Leishmania infantum. Proc. Natl. Acad. Sci. USA 112, E616–24 (2015).
Gupta, R. et al. Characterization of glycolytic enzymes-rAldolase and rEnolase of Leishmania donovani, identified as Th1 stimulatory proteins, for their immunogenicity and immunoprophylactic efficacies against experimental visceral leishmaniasis. PLoS ONE 9, e86073 (2014).
Shih, S., Hwang, H. Y., Carter, D., Stenberg, P. & Ullman, B. Localization and targeting of the Leishmania donovani hypoxanthine-guanine phosphoribosyltransferase to the glycosome. J. Biol. Chem. 273, 1534–1541 (1998).
Silva, A. M. et al. Characterization of Leishmania infantum thiol-dependent reductase 1 and evaluation of its potential to induce immune protection. Parasite Immunol. 34, 345–350 (2012).
Moreira, D. et al. Impact of continuous axenic cultivation in Leishmania infantum virulence. PLoS Negl Trop Dis 6, e1469 (2012).
Beattie, L. et al. Dynamic imaging of experimental Leishmania donovani-induced hepatic granulomas detects Kupffer cell-restricted antigen presentation to antigen-specific CD8+ T cells. PLoS Pathog. 6, e1000805 (2010).
Michels, P. A. M., Bringaud, F., Herman, M. & Hannaert, V. Metabolic functions of glycosomes in trypanosomatids. Biochimica et Biophysica Acta (BBA) – Mol. Cell Res. 1763, 1463–1477 (2006).
Haanstra, J. R. et al. Compartmentation prevents a lethal turbo-explosion of glycolysis in trypanosomes. Proc. Natl. Acad. Sci. USA 105, 17718–17723 (2008).
Opperdoes, F. R. & Szikora, J. P. In silico prediction of the glycosomal enzymes of Leishmania major and trypanosomes. Mol. Biochem. Parasitol. 147, 193–206 (2006).
Heise, N. & Opperdoes, F. R. Purification, localisation and characterization of glucose-6-phosphate dehydrogenase of Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 21–32 (1999).
Duffieux, F., Van Roy, J., Michels, P. A. & Opperdoes, F. R. Molecular characterization of the first two enzymes of the pentose phosphate pathway of Trypanosoma brucei. J. Biol. Chem. 275, 27,559–27,565 (2000).
Jamdhade, M. D. et al. Comprehensive proteomic analysis of glycosomes from Leishmania donovani. OMICS 19, 157–169 (2015).
Cruz, A. K., Titus, R. & Beverley, S. M. Plasticity in chromosome number and testing essential genes in Leishmania by targeting. Proc. Natl. Acad. Sci. USA 90, 1599–1603 (1993).
Mukherjee, A., Langston, L. D. & Ouellette, M. Intrachromosomal tandem duplication and repeat expansion during attempts to inactivate the subtelomeric essential gene GSH1 in Leishmania. Nucleic Acids Res. 39, 7499–7511 (2011).
Dacher, M. et al. Probing druggability and biological function of essential proteins in Leishmania combining facilitated null mutant and plasmid shuffle analyses. Mol. Microbiol. 93, 146–166 (2014).
Pedrosa, A. L. & Cruz, A. K. The effect of location and direction of an episomal gene on the restoration of a phenotype by functional complementation in Leishmania. Mol. Biochem. Parasitol. 122, 141–148 (2002).
Rodríguez-Contreras, D. & Landfear, S. M. Metabolic changes in glucose transporter-deficient Leishmania mexicana and parasite virulence. J. Biol. Chem. 281, 20,068–20,076 (2006).
Naderer, T. et al. Virulence of Leishmania major in macrophages and mice requires the gluconeogenic enzyme fructose-1,6-bisphosphatase. Proc. Natl. Acad. Sci. USA 103, 5502–5507 (2006).
Wyllie, S. et al. Dissecting the essentiality of the bifunctional trypanothione synthetase-amidase in Trypanosoma brucei using chemical and genetic methods. Mol. Microbiol. 74, 529–40 (2009).
Castro, H. et al. Leishmania mitochondrial peroxiredoxin plays a crucial peroxidase-unrelated role during infection: insight into its novel chaperone activity. PLoS Pathog. 7, e1002325 (2011).
Gómez-Arreaza, A. et al. Extracellular functions of glycolytic enzymes of parasites: unpredicted use of ancient proteins. Mol. Biochem. Parasitol. 193, 75–81 (2014).
McConville, M. J., de Souza, D., Saunders, E., Likic, V. A. & Naderer, T. Living in the phagolysosome: metabolism of Leishmania amastigotes. Trends Parasitol. 23, 368–375 (2007).
Lorenz, M. V., Bender, J. A. & Fink, J. R. Transcriptional response to Candida albicans upon internalization by macrophages. Eukaryotic Cell 3, 1076–1087 (2004).
Muñoz-Elías, E. J. & McKinney, J. D. Mycobacterium tuberculosis isocitrate lyases 1 and 2 jointly required for in vivo growth and virulence. Nat. Med. 11, 638–644 (2005).
Saunders, E. C. et al. Induction of a stringent metabolic response in intracellular stages of Leishmania mexicana leads to increased dependence on mitochondrial metabolism. PLoS Pathog. 10, e1003888 (2014).
Naula, C. M., Logan, F. M., Wong, P. E., Barrett, P. M. & Burchmore, R. J. A glucose transporter can mediate ribose uptake. J. Biol. Chem. 285, 29,721–29,728 (2010).
Berens, R. L., Deutsch-King, L. C. & Marr, J. J. Leishmania donovani and Leishmania braziliensis: hexokinase, glucose-6-phosphate dehydrogenase and pentose phosphate shunt activity. Exp. Parasitol. 49, 1–8 (1980).
Burchmore, R. J. & Barrett, M. P. Life in vacuoles – nutrient acquisition by Leishmania amastigotes. Int. J. Parasitol. 12, 1311–1320 (2001).
Clasquin, M. F. et al. Riboneogenesis in yeast. Cell 145, 969–980 (2011).
Ciou, S. C. et al. Ribose-5-phosphate isomerase A regulates hepatocarcinogenesis via PP2A and ERK signalling. Int. J. Cancer 137, 104–115 (2014).
Kabashima, T., Kawaguchi, T., Wadzinski, B. E. & Uyeda, K. Xylulose 5-phosphate mediates glucose-induced lipogenesis by xylulose 5-phosphate-activated protein phosphatase in rat liver. Proc. Natl. Acad. Sci. USA 100, 5107–5112 (2003).
Brenchley, R. et al. The TryTryp Phosphatome: analysis of the protein phosphatase catalytic domains. BMC Genomics 8, 434 (2007).
Parsons, M., Worthey, E. A., Ward, P. N. & Mottram, J. C. Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics 6, 127 (2005).
Schlecker, T. et al. Substrate specificity, localization and essential role of the glutathione peroxidase-type tryparedoxin peroxidases in Trypanosoma brucei. J. Biol. Chem. 280, 14,385–14,394 (2005).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Bond, C. S. & Schuttelkopf, A. W. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr. D. Biol. Crystallogr. 65, 510–512 (2009).
Horton, P. et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, 585–587 (2007).
Yu, C. S., Chen, Y. C., Lu, C. H. & Hwang, J. K. Prediction of protein subcellular localization. Proteins 64, 643–651 (2006).
Silvestre, R. et al. SIR2-deficient Leishmania infantum induces a defined IFN-gamma/IL-10 pattern that correlates with protection. J. Immunol. 179, 3161–3170 (2007).
Cull, B. et al. Glycosome turnover in Leishmania major is mediated by autophagy. Autophagy 12, 2143–2157 (2015).
Acknowledgements
We would like to thank Professor Ana Tomás from the Institute for Molecular and Cell Biology, University of Porto, Portugal, for providing LimTXNPx antibody; Dr. Paul Michels from Université Catholique de Louvain, Belgium, for providing Tbenolase antibody; Professor Graham Coombs, Strathclyde University, Glasgow, for LmCS antibody; Professor Buddy Ullman, School of Medicine, Oregan Health and Science University, USA, for LdHGPRT antibody; Dr. Christine Clayton, Zentrum fur Molekulare Biologie der Universitat Heidelberg, Germany, for TbAldolase antibody. We would also like to thank Professor Jeremy Mottram, University of Glasgow, for pGL345HYG and Professor Marc Ouellette, Centre de Recherche en Infectiologie, of Laval University, Canada, for pSPαNEOα and pSPαBLASTα. We would also like to thank Dr. Jane MacDougall from Photeomix, France, for proofreading the English of the manuscript. The research leading to these results has received funding from the European Community’s Seventh Framework Programme under grant agreement No. 602773 (Project KINDRED).’ The COST Action CM1307: Targeted chemotherapy towards diseases caused by endoparasites has also contributed for this work. We would like to acknowledge Fundação para a Ciência e Tecnologia (FTC) for supporting Joana Faria (SFRH/BD/79712/2011) and Inês Loureiro (SFRH/BD/64528/2009). Inês Loureiro was also supported by the European Community’s Seventh Framework Programme (KINDRED-PR300102-BD). JT is an Investigator FCT funded by National funds through FCT and co-funded through European Social Fund within the Human Potential Operating Programme. Nuno Santarem and Pedro Cecílio are supported by fellowships from the European Community’s Seventh Framework Programme under grant agreements No. 602773 (Project KINDRED) and No. 603181 (Project MuLeVaClin), respectively.
Author information
Authors and Affiliations
Contributions
J.F., I.L., J.T., N.S. and A.C.S. conceived and designed the experiments. J.F., I.L. and P.C. performed the experiments and analyzed the data. J.F., I.L., P.C., J.T., N.S., S.M.-R. and A.C.-d.-S. critically discussed the results. A.C.-d.-S. and S.M.R. contributed with reagents, materials and analysis tools. J.F., I.L., J.T., N.S., S.M.R. and A.C.-d.-S. wrote the paper. All the authors reviewed the results and approved the final version of the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Faria, J., Loureiro, I., Santarém, N. et al. Disclosing the essentiality of ribose-5-phosphate isomerase B in Trypanosomatids. Sci Rep 6, 26937 (2016). https://doi.org/10.1038/srep26937
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep26937
This article is cited by
-
Engineering ribose-5-phosphate isomerase B from a central carbon metabolic enzyme to a promising sugar biocatalyst
Applied Microbiology and Biotechnology (2021)
-
Ribose-5-phosphate isomerases: characteristics, structural features, and applications
Applied Microbiology and Biotechnology (2020)
-
Backbone assignment of ribose-5-phosphate isomerase of Mycobacterium tuberculosis (MtRpiB)
Biomolecular NMR Assignments (2020)
-
Murine infection with bioluminescent Leishmania infantum axenic amastigotes applied to drug discovery
Scientific Reports (2019)
-
The glyceraldehyde-3-phosphate dehydrogenase GapDH of Corynebacterium diphtheriae is redox-controlled by protein S-mycothiolation under oxidative stress
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.