Abstract
Plasiatine (1), isolated from the seeds of Plantago asiatica, is an unprecedented indole analogue linked to a phenylpropanoid moiety via a carbon bond that builds up a novel heteromeric construction with a C19N2 scaffold. Its structure was determined by spectroscopic data and computational evidence. Notably, experimental assay demonstrated that 1 significantly enhanced the activity of the nonreceptor protein tyrosine phosphatase Shp2 in vitro in a concentration-dependent manner with an EC50 value of 0.97 μM, and activated phosphorylation of ERK, a known target of Shp2. Moreover, plasiatine (1) promoted hepatocellular HepG2 cells migration. Molecular docking suggested that plasiatine (1) binds to the catalytic cleft of Shp2. These results identified plasiatine (1) as the first small molecule Shp2 activator, and it warrants further investigation as a novel pharmaceutical tool to study the function of Shp2 in tumorigenesis.
Similar content being viewed by others
Introduction
The Src homology 2-containing phosphotyrosine phosphatase 2 (Shp2) encoded by PTPN11 gene is a nonreceptor protein tyrosine phosphatase1. This phosphatase is a pivotal transducer of growth factors and cytokines, and is a critical intracellular regulator in mediating cell proliferation, differentiation, adhesion, migration, and apoptosis2,3. Somatic activating mutations in Shp2 have been demonstrated to be linked to hematologic disorders (e.g. juvenile myelomonocytic leukemia, myelodysplastic syndromes, acute myeloid leukemia, and acute lymphoid leukemia)4, and several types of solid tumors (e.g. lung adenocarcinoma, colon cancer, neuroblastoma, glioblastoma, melanoma, and breast carcinoma)5. In the last ten years, enormous efforts directed toward the discovery of specific Shp2 inhibitors as potential therapeutic agents for hematologic malignancies and solid tumors have been continuing in several research groups6,7,8,9,10,11. However, a paradoxical function demonstrated recently is that the appearance of inflammatory signaling through the Stat3 pathway and hepatic inflammation/necrosis is promoted by hepatocyte-specific deletion or inactivation of Shp2, leading to regenerative hyperplasia and development of cancers12,13. Therefore, the function of Shp2 in tumorigensis may be tissue-specific, and it is also important to discover Shp2 activators to dissect Shp2-meidated signaling pathways. However, such explorations are still lacking.
Plantago asiatica, a species of the Plantago genus of the Plantaginaceae family distributed worldwide, is a famous traditional Chinese medicine (TCM)14. Its seed is used as diuretic, detoxification, expectorant, and laxative; and its whole plant is used for the treatment of fever, sputum, and inflammatory15,16,17. The secondary metabolites of P. asiatica are characterized by phenylethanoid glycosides18,19,20,21 and other phenolic compounds22. In order to obtain structurally interesting and biologically active secondary metabolites, our preliminary study for chemical constituents of the seeds of P. asiatica provided a series of simple indole alkaloids, plasiaticines A–D, (+)-(R)-3-cyanomethyl-3-hydroxyoxindole, and indolyl-3-carboxylic acid23, which was the first report regarding the occurrence of alkaloids in P. asiatica. As part of our search for biologically active natural products from TCMs, further investigation of the seeds of P. asiatica led to the discovery of plasiatine (1), an unprecedented heteromeric structure with a C19N2 skeleton that is composed of an indole analogue and a phenylpropanoid moiety (Fig. 1). Bioactive assay showed that 1 potently promoted the activity of Shp2. Herein, we reported the isolation, structural elucidation, and bioactivity evaluation of compound 1.
Results and Discussion
Compound 1, a white amorphous powder, presented a pseudomolecular ion peak at m/z 405 [M+Na]+ in the positive ESIMS, and its HREIMS data further afforded a molecular formula of C20H18N2O6 (m/z 382.1160, cacld for C20H18N2O6 382.1165), requiring 13 degrees of unsaturation. The IR spectrum showed characteristic bands at 1720, 1629, 1518, and 1469 cm−1 for the carbonyl groups and aromatic rings. In the 1H NMR spectrum (Table 1), a methoxyl group (δH 3.78) and two methines (δH 3.45 and one oxygenated at δH 5.44) were observed. A pair of AB doublet appeared at δH 3.03 (d, J = 16.6 Hz) and 2.83 (d, J = 16.6 Hz), indicating the existence of an isolated methylene. Three typical olefinic proton resonances at δH 6.90 (d, J = 1.6 Hz), 6.78 (dd, J = 8.1, 1.6 Hz), and 6.72 (d, J = 8.1 Hz) could be ascribed to a 1,3,4-trisubstituted benzene ring, and the remaining two olefinic singlets at δH 7.03 and 6.86 might be assigned to one 1,2,4,5-tetrasubstituted or two 1,2,3,4,5-pentasubstituted aromatic rings. The 13C NMR and DEPT spectra (Table 1) showed 20 carbon signals, corresponding to one methoxy, two methylenes (including one oxygenated), seven methines (including five olefinic methines and one oxygenated), and 10 quaternary carbons (including eight olefinic, one carbonyl, and one oxygenated). By analysis of the HSQC spectrum, all proton signals were unambiguously assigned to their respective carbons.
Its 1D NMR data (Table 1) showed that the characteristic signals for an oxoindoline (δC 179.3, C-2; 74.3, C-3; 131.0, C-4; 107.1, C-5; 157.3, C-6; 131.0, C-7; 108.8, C-8; 135.8, C-9) and a cyanomethyl (δC 27.3, C-10; 117.3, C-11) could be distinguished, indicating that 1 containing a 2-(3-hydroxy-2-oxoindolin-3-yl) acetonitrile shared a substructural similarity with plasiaticines B–D23. The HMBC correlations (Fig. 2) from H-5 (δH 7.03, s) to C-3 (δC 74.3) and from H2-10 (δH 2.83, d, J = 16.6 Hz; 3.03, d, J = 16.6 Hz) to C-2 (δC 179.3), C-3, and C-4 (δC 131.0) permitted the location of a hydroxyl and a cyanomethyl group at C-3. Correlations from both of H-5 and H-8 (δH 6.86, s) to C-6 (δC 157.3) and C-7 (δC 131.0) showed that C-6 was oxygenated and C-7 was substituted. Thus, the east part of the compound 1 was established to be 2-(3-hydroxy-2-oxoindolin-3-yl)acetonitrile (Fig. 2).
The 1,3,4-trisubstituted benzene ring with a methoxy and a hydroxyl at C-3′ and C-4′, respectively, was supported by the HMBC correlations from OMe (δH 3.78) to C-3′ (δC 149.1) and from H-6′ (δH 6.78) to C-4′ (δC 147.5), together with the ROESY correlation of OMe with H-2′ (δH 6.90, d, J = 1.6 Hz). In addition, the 1H–1H COSY correlations of H-7′/H-8′/H-9′ along with the HMBC correlations from H-6′ to C-1′ (δC 134.6) and from H-7′ (δH 5.44, d, J = 6.1 Hz) to C-1′, C-2′ (δC 110.4), C-6′ (δC 119.6), and C-9′ (δC 64.8) demonstrated that the west part of the compound 1 was phenylpropanoid unit (Fig. 2).
The carbon-carbon connection of C-7 and C-8′ was evidenced by the key HMBC correlations from H-8′ (δH 3.45, m) to C-6, C-7, and C-8 (δC 108.8) and from H-8 to C-8′ (δC 55.1) (Fig. 2). Meanwhile, given the deductive formula, the remaining one degree of unsaturation was fulfilled by the establishment of a five-membered oxygen-containing ring flanking the oxoindoline, which was supported by the HMBC correlations from H-7′ to C-6 and C-7 (Fig. 2).
The configuration of C-3 in the oxoindoline could be readily elucidated on the basis of the experimental electronic circular dichroism (ECD) spectrum. Two negative Cotton effects (CEs) at 241 and 289 nm and two positive CEs at 211 and 271 nm in its experimental ECD spectrum, which were in accord with those of plasiaticines B–D23 and opposite to that of (+)-(S)-2-(3,4-dihydroxy-2-oxoindolin-3-yl)acetonitrile24, suggested that C-3 was R-configuration. In addition, the ROESY correlation of H-7′ with H2-9′ indicated that H-7′ and H-8′ were positioned on the opposite face of the molecule. However, the absolute configurations of C-7′ and C-8′ could not be established because the relative configuration of C-3 relative to those of C-7′ and C-8′ could not be determined by ROESY spectrum. After numerous unsuccessful attempts to obtain a suitable crystal for X-ray diffraction analysis, theoretical calculation studies for 13C NMR and ECD spectra of a pair of stereoisomers, (3R, 7′R, 8′S)-1 and (3R, 7′S, 8′R)-1, were perform using Gaussian 09 so as to provide further evidence to confirm the unprecedented skeleton and to determine the absolute configuration.
The 13C chemical shift predictions of both structures obtained by the gauge-independent atomic orbital method at MPW1PW91-SCRF/6-31 G(d,p) level with the polarizable continuum model (PCM) in MeOH solvent agreed well with the recorded shifts (Table 1 and Fig. 3), and the mean absolute error, the corrected mean absolute error, and R2 (Fig. 3) were 3.31 ppm, 1.46 ppm, and 0.9975 for (3R, 7′R, 8′S)-1, and 3.39 ppm, 1.44 ppm, and 0.9972 for (3R, 7′S, 8′R)-1, respectively. Thus, the 13C NMR calculation evidenced the proposed skeleton determined by our NMR analysis.
Subsequently, the absolute configurations of the stereogenic centers (C-7′ and C-8′) in the western hemisphere was elucidated by ECD calculation using time-dependent density-functional theory at B3LYP-SCRF/6-31 + G(d,p) level with PCM in MeOH solvent. The calculated ECD curves of two stereoisomers (3R, 7′R, 8′S)-1 and (3R, 7′S, 8′R)-1 were compared with the experimental spectrum. The wave troughs of the calculated curves of (3R, 7′R, 8′S)-1 and (3R, 7′S, 8′R)-1 around 240 nm were close to the experimental ECD band at 241 nm (Fig. 4). However, (3R, 7′R, 8′S)-1 had a wave trough around 205 nm, while (3R, 7′S, 8′R)-1 exhibited a wave crest at the same position which corresponded well with the experimental curve. Molecular orbital (MO) analysis (Figure S5) suggested that the negative CE at 241 nm in the experimental spectrum could be ascribed to the negative rotatory strength at 237.4 nm, which was caused by the electronic transitions from MO96 to MO101 in the 2-oxoindoline moiety; and the positive CE around 205 nm could be assigned to two positive rotatory strengths at 206.7 and 214.9 nm, which correlated with the electronic transitions from MO100 to MO114 and from MO99 to MO108, respectively. Therefore, the absolute configuration of compound 1 was established as 3R, 7′S, and 8′R.
Indole alkaloids, which may occupy a quarter of alkaloids from natural resources, represent a particular and vast group of natural products25. Generally, they are present as prenyl hybridsthat are biosynthetically derived from tryptophan or its precursors fused with isopentenyl pyrophosphate26, dimethylallyl pyrophosphate26, geranyl pyrophosphate27, or farnesyl pyrophosphate28,29, while other forms of indole hybrids are exceedingly rare. From a literature research, only one reference reports that ipobscurines B–D feature an indole analogue conjugated with two phenylpropanoid moieties via C–O and C–N bonds9; thus plasiatine (1) is the first heteromeric construction formed by a crucial carbon–carbon connection between an indole and a phenylpropanoid moiety, and its biogenetic pathway was also proposed (Fig. 5).
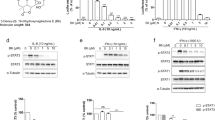
Our present study revealed that plasiatine (1) potently promoted the phosphatase activity of Shp2 in a concentration-dependent manner, with an EC50 value of 0.97 μM (Na3VO4 as a negative control, Fig. 6A,B). Further study found that plasiatine (1) enhanced the phosphorytion of Erk1/2 in the present or absent of EGF, a known downstream target of Shp26 (Fig. 6C). The wound-healing assay showed that plasiatine (1) at a concentration of 25 μM significantly promoted hepatocellular HepG2 cells migration (Fig. 6D,E). To map the binding site of plasiatine (1), we built complex structure of the compound with Shp2 by using computational docking method. As shown in Fig. 7, plasiatine (1) fitted well in the active site and the complex was stabilized by extensive hydrogen bonding and hydrophobic interaction. The model revealed that the oxoindoline moiety penetrated into the active site PTP signature motif (VHCSAGIGRTG)30. The oxoindoline carbonyl, acetonitrile, and hydroxyl group exhibited hydrogen bonding interactions with atoms of P-loop residues (Arg465 and Ser460) and Q-loop residues (Gln506 and Gln510). The dihydrofuran and hydroxymethyl group were located at the entrance of the active site and interacted with residues Lys364 and Tyr279. The methoxyl group formed hydrogen bonds with the side-chain NH2 group of Asn281 that is non-conserved PTP residues located adjacent to the phosphotyrosine recognition loop. The interaction between aromatic rings of the compound and the protein contributed to the binding through hydrophobic stabilization. The computational docking result was in good agreement with the bioactivity evaluation and obviously disclosed the detailed interactions between plasiatine (1) and Shp2. Compounds with oxindole scaffolds were identified to selectively inhibit Shp2 activity31. Plasiatine (1) and the previously reported oxindole compounds penetrate into the active site PTP signature motif through hydrogen bonding interactions with atoms of PTP loop residues (Arg465 and Ser460). In contrast to the oxindole compounds, plasiatine (1) also interacts with Gln506 and Tyr279, and mutations of these two residues are found to participate in the incidence of Noonan syndrome and juvenile myelomonocytic leukemia by increasing Shp2 activity32. Therefore, it is possible that plasiatine (1) promotes Shp2 activity through interaction with these two residues. Plasiatine (1) also forms hydrogen bonds with Asn282 adjacent to the phosphotyrosine recognition loop. It will be of interest to further investigate whether plasiatine (1) increases the combination with the substrate tyrosine to promote the Shp2 activity.
(A) The activity of Shp2 was determined after GST-Shp2 recombinant protein (0.1 μM) treated with Na3VO4 (50 μM) and different concentrations of plasiatine (1), respectively. (B) Presents the concentration-response curve of plasiatine (1) for activation of Shp2 activity using GST-Shp2 recombinant protein. (C) HEK293A cells were treated with 25 μM or 50 μM plasiatine (1) for 2 h with or without 75 ng/mL EGF pretreatment. The cell lysates were immunoblotted with an anti-pErk1/2 (P-ERK1/2), total Erk1/2 (T-ERK1/2) or anti-GAPDH antibody, respectively. (D,E) Effect of plasiatine (1) on hepatocelluar HepG2 cells migration. Panel E showed quantification analysis of wound healing assay from triplicate measurements ratio of the migration distance in each group divided by the migration distance in control (DMSO) cells. Bars represented the mean values ± SD of three experiments. Scale bar, 250 μm. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 vs control.
To our knowledge, this study represents the first identification of a small molecule Shp2 activator and opens a new way in Shp2-associated oncogenic mechanism research. This compound warrants further investigation as a novel pharmaceutical tool to study the function of Shp2 in tumorigenesis and facilitate the rational design of Shp2 activating agents.
Methods
General Experimental Procedures
Optical rotations were measured on a JASCO P-1020 digital polarimeter. UV data were obtained on a Shimadzu UV2401PC spectrophotometer. Experimental ECD spectra were measured on a Chirascan instrument. A Bruker Tensor-27 spectrophotometer was used for scanning IR spectroscopy with KBr pellets. 1D and 2D NMR spectra were recorded on Bruker DRX-500 spectrometer. Unless otherwise specified, chemical shifts (δ) were expressed in ppm with reference to the solvent signals. ESIMS were performed on Waters Xevo TQ-S. HREIMS were performed on Waters AutoSpec Premier P776. Column chromatography was performed with silica gel (200–300 mesh; Qingdao Marine Chemical, Inc., Qingdao, P. R. of China), MCI gel (75–150 μm, Mitsubishi Chemical Corporation, Tokyo, Japan), and Sephadex LH-20 gel (40–70 μm, Amersham Pharmacia Biotech AB, Uppsala, Sweden). Semipreparative HPLC was performed on an Agilent 1200 liquid chromatograph with a Zorbax SB-C18, 9.4 mm × 25 cm column. Fractions were monitored by TLC and spots were visualized by heating silica gel plates sprayed with 10% H2SO4 in EtOH. All solvents including petroleum ether (60–90 °C) were distilled prior to use.
Plant Material
The seeds of P. asiatica were purchased from Juhuacun Traditional Chinese Medicine Market, Kunming, Yunnan Province, People’s Republic of China, in August 2011. A voucher specimen (No. KIB 2011-08-11) was identified by Mr. Yu Chen and was deposited at the State Key Laboratory of Phytochemistry and Plant Resources in West China, Kunming Institute of Botany, Chinese Academy of Sciences.
Extraction, Isolation, and Purification
The air-dried powders seeds of P. asiatica (10 kg) were extracted with 70% aqueous acetone (3 × 30 L) at room temperature and concentrated in vacuo to give a crude extract which was partitioned between H2O and EtOAc. The EtOAc extract (285 g) was chromatographed on a silica gel column eluted with gradient CHCl3–Me2CO (1:0 to 1:0) to afford fractions A–E. Fraction D (9.3 g) was chromatographed on MCI gel CHP 20 P eluted with gradient MeOH–H2O (1:4 to 1:0) to yield seven fractions, D1–7. Subfraction D3 (316 mg) was purified by Sephadex LH-20 (CHCl3–MeOH, 1:1) and semipreparative HPLC (MeCN–H2O, 15:85) to afford compound 1 (7 mg).
13C NMR and ECD Calculations and Molecular Orbital Analysis
The theoretical calculations of compound 1 were carried out using Gaussian 0933. Conformational analysis was initially performed using Maestro 9.0 with the OPLS_2005 force field. The conformers were optimized at B3LYP/6-31 G(d) level. Room-temperature equilibrium populations were calculated according to Boltzmann distribution law. The optimized conformation geometries, thermodynamic parameters, and populations of all conformations were provided in Figures S11 and S12 and Tables S1, S2, S5 and S6 in the Supplementary Information.
13C NMR shielding constants of compound 1 were calculated with the GIAO method34 at MPW1PW91-SCRF/6-31 G(d,p) level with the polarizable continuum model (PCM) in MeOH solvent. The shielding constants so obtained were converted into chemical shifts by referencing to TMS at 0 ppm (δcal = σTMS − σcal), where the σTMS was the shielding constant of TMS calculated at the same level. For each stereoisomer, the parameters a and b of the linear regression δcal = aδexp + b; the correlation coefficient, R2; the mean absolute error (MAE) defined as Σn |δcal − δexp|/n; the corrected mean absolute error (CMAE), defined as Σn |δcorr − δexp|/n, where δcorr = (δcal − b)/a and therefore corrects for systematic errors were presented.
The theoretical calculations of ECD were performed using TDDFT35,36 at B3LYP/6-31 + G(d,p) level in MeOH with PCM. The ECD spectra of compound 1 were obtained by weighing the Boltzmann distribution rate of each geometric conformation. The ECD spectra are simulated by overlapping Gaussian functions for each transition according to

where σ represents the width of the band at 1/e height, and ΔEi and Ri are the excitation energies and rotational strengths for transition i, respectively. σ = 0.20 eV and Rvelocity were used in this work.
The orbital information (NBO plot files) was generated by NBO program of Gaussian 0937. The predominantly populated conformers were selected for molecular orbital (MO) analysis. NBO plot files were used to generate corresponding Gaussian-type grid file by Multiwfn 2.438. After that, the isosurface of generated grid date was afforded by VMD software39.
Shp2 Activity Assay
Plasmids for expression of glutathione S-transferase (GST)-PTP fusion proteins of human Shp2 (residues 205–597) were constructed in pGEX-4T1 by PCR subcloning techniques. All constructs were verified by DNA sequencing. GST- Shp2 fusion proteins were expressed in Escherichia coli BL21 and affinity purified with glutathione Sepharose. After elution from glutathione affinity columns, GST-fusion proteins were dialyzed with dialysis buffer (12.5 mM Tris-Cl, pH 7.5, 25 mM NaCl, 1 mM dithiothreitol, and 0.1% β-mercaptoethanol) at 4 °C over night and then stored in dialysis buffer plus 20% glycerol at −80 °C.
Shp2 activity was measured as described previously6. Using the 8-difluoro-4-methylumbelliferyl phosphate (DiFMUP; Invitrogen, Carlsbad, CA) as the substrate, reaction buffer contained 50 mM Hepes (pH 7.0), 150 mM NaCl, 0.05% Tween 20, 2 mM dithiothreitol, 1 mM EDTA, 20 μM DiFMUP, 0.1 μM GST-Shp2, and different concentrations of test compound or dimethyl sulfoxide (solvent) in a total reaction volume of 100 μL in black 96-well plates. Reaction was initiated by addition of DiFMUP, and the incubation time was 30 min at 37 °C. DiFMUP fluorescence signal was measured at an excitation of 355 nm and an emission of 455 nm with a plate reader (Thermo Scientific Verioskan Flash).
Western Blotting
Cells cultured in 60-mm dishes were lysed with RIPA Lysis Buffer (Beyotime, Haimen, China) supplemented with protease inhibitor cocktail (Sigma). Equal protein content of total cell lysates were mixed in loading buffer, boiled for 5 min, and then subjected to SDS-PAGE gel (10%) electrophoresis along with EasySee western marker (TransGen Biotech, Beijing, China). After electrophoresis, proteins were blotted onto nitrocellulose membranes and blocked with 5% bovine serum albumin, probed with a primary antibody at 4 °C overnight, followed by a horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), then enhanced chemiluminesence detection (Amersham Bioscience, Piscataway, NJ).
In vitro Wound-healing Assay
Wound healing experiments were performed as described previously40. In brief, 1 × 105 HepG2 cells per well were plated in 6-well plates.The confluent HepG2 cells were scratched with a 200 μL disposable plastic pipette tip and were allowed to migrate towards the wound. To examine migration in HepG2 cells, the medium was replaced with that supplemented with 2% FBS either in the presence or absence of plasiatine (1), followed by scratching. Wound closure rates were expressed as a percentage of the migratory distance in control cells (100%).
Preparation of Protein and Molecular Docking
The 3D crystal structure of Shp2 was retrieved from the Protein Data Bank (PDB entry: 4PVG). The program Maestro 9.0 (Schrödinger, Inc.) was used for this assessment. All water molecules were removed from the structure of the complex. Hydrogen atoms and charges were added during a brief relaxation that was performed using the “Protein Preparation Wizard” workflow. After optimizing the hydrogen bond network, the crystal structure was minimized using the OPLS_2005 force field with the maximum root mean square deviation (RMSD) value of 0.3 Å. The grid-enclosing box was centered on the ligand L88N79 in the refined crystal structure as described above. This domain has been identified as the PTP catalytic domain. The three-dimensional structure of plasiatine (1) was generated with the Ligprep module. Docking process was performed using Glide with default docking parameter settings with standard precision (SP) mode.
Plasiatine (1) White amorphous powder;  −37.1 (c 0.12, MeOH); UV (MeOH) λmax (log ε) 203 (3.91), 277 (3.33), 320 (2.63) nm; ECD (c 0.09 MeOH) λmax (Δε) 211 (+21.36), 241 (−17.70), 271 (+1.44), 289 (−2.34) nm; IR (KBr) νmax 3429, 1720, 1629, 1469, 1274 cm−1; 1H and 13C NMR data, see Table 1; positive ESIMS m/z 405 (100) [M+Na]+; HREIMS m/z 382.1160 [M]+ (calcd for C20H18N2O6, 382.1165).
−37.1 (c 0.12, MeOH); UV (MeOH) λmax (log ε) 203 (3.91), 277 (3.33), 320 (2.63) nm; ECD (c 0.09 MeOH) λmax (Δε) 211 (+21.36), 241 (−17.70), 271 (+1.44), 289 (−2.34) nm; IR (KBr) νmax 3429, 1720, 1629, 1469, 1274 cm−1; 1H and 13C NMR data, see Table 1; positive ESIMS m/z 405 (100) [M+Na]+; HREIMS m/z 382.1160 [M]+ (calcd for C20H18N2O6, 382.1165).
Additional Information
How to cite this article: Gao, Z.-H. et al. Plasiatine, an Unprecedented Indole–Phenylpropanoid Hybrid from Plantago asiatica as a Potent Activator of the Nonreceptor Protein Tyrosine Phosphatase Shp2. Sci. Rep. 6, 24945; doi: 10.1038/srep24945 (2016).
References
Tartaglia, M. et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 29, 465–468 (2001).
Chan, R. J. & Feng, G. S. PTPN11 is the first identified proto-oncogene that encodes a tyrosine phosphatase. Blood 109, 862–867 (2007).
Qu, C. K. The SHP-2 tyrosine phosphatase: signaling mechanisms and biological functions. Cell Res. 10, 279–288 (2000).
Tartaglia, M. et al. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat. Genet. 34, 148–150 (2003).
Bentires-Alj, M. et al. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 64, 8816–8820 (2004).
Chen, L. et al. Discovery of a novel Shp2 protein tyrosine phosphatase inhibitor. Mol. Pharmacol. 70, 562–570 (2006).
Hellmuth, K. et al. Specific inhibitors of the protein tyrosine phosphatase Shp2 identified by high-throughput docking. Proc. Natl. Acad. Sci. USA 105, 7275–7280 (2008).
Lawrence, H. R. et al. Inhibitors of Src homology-2 domain containing protein tyrosine phosphatase-2 (Shp2) based on oxindole scaffolds. J. Med. Chem. 51, 4948–4956 (2008).
Zhang, X. et al. Salicylic acid based small molecule inhibitor for the oncogenic Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2). J. Med. Chem. 53, 2482–2493 (2010).
Zeng, L. F. et al. Therapeutic potential of targeting the oncogenic SHP2 phosphatase. J. Med. Chem. 57, 6594–6609 (2014).
Liu, W. et al. Identification of cryptotanshinone as an inhibitor of oncogenic protein tyrosine phosphatase SHP2 (PTPN11). J. Med. Chem. 56, 7212–7221 (2013).
Bard-Chapeau, A. E. et al. Ptpn11/Shp2 acts as a tumor suppressor in hepatocellular carcinogenesis. Cancer Cell 19, 629–639 (2011).
Feng, G. S. Conflicting roles of molecules in hepatocarcinogenesis: paradigm or paradox. Cancer Cell 21, 150–154 (2012).
Li, Z. Y., Wei, L. & Hoggard, K. R. Plantaginaceae. Flora of China 19, 495–503 (2011).
Pharmacopoeia of People’s Republic of China. Vol. 1, 63–64 (China Medical Science Press, 2010).
Samuelsen, A. B. The traditional uses, chemical constituents and biological activities of Plantago major L. A review. J. Ethnopharmacol. 71, 1–21 (2000).
Mitsuhashi, H. Illustrated Medicinal Plants of the World in Colour. (Tokyo: Hokuryukan, 1988).
Nishibe, S., Tamayama, Y., Sasahara, M. & Andary, C. A phenylethanoid glycoside from Plantago asiatica . Phytochemistry 38, 741–743 (1995).
Huang, D. F. et al. Eur. J. Pharmacol. 620, 105–111 (2009).
Miyase, T. et al. Phenylethanoid glycosides from Plantago asiatica . Phytochemistry 30, 2015–2018 (1991).
Li, L., Liu, C. M., Liu, Z. Q. & Wang, J. Isolation and purification of phenylethanoid glycosides from plant extract of Plantago asiatica by high performance centrifugal partition chromatography. Chin. Chem. Lett. 19, 1349–1352 (2008).
Ravn, H., Nishibe, S., Sasahara, M. & Li, X. Phenolic compounds from Plantago asiatica . Phytochemistry 29, 3627–3631 (1990).
Gao, Z. H. et al. Four new indole alkaloids from Plantago asiatica . Nat. Prod. Bioprospect. 2, 249–254 (2012).
Chen, M. H. et al. Alkaloids from the root of Isatis indigotica . J. Nat. Prod. 75, 1167–1176 (2012).
Yang, L. Q. & Stockigt, J. Trends for diverse production strategies of plant medicinal alkaloids. Nat. Prod. Rep. 27, 1469–1479 (2010).
Li, S. M. Prenylated indole derivatives from fungi: structure diversity, biological activities, biosynthesis and chemoenzymatic synthesis. Nat. Prod. Rep. 27, 57–78 (2010).
O’Connor, S. E. & Maresh, J. J. Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat. Prod. Rep. 23, 532–547 (2006).
Marcos, I. S., Moro, R. F., Costales, I., Basabe, P. & Diez, D. Sesquiterpenyl indoles. Nat. Prod. Rep. 30, 1509–1526 (2013).
Jenett-Siems, K., Weigl, R., Kaloga, M., Schulz, J. & Eich, E. Ipobscurines C and D: macrolactam-type indole alkaloids from the seeds of Ipomoea obscura . Phytochemistry 62, 1257–1263 (2003).
Andersen, J. N. et al. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell Biol. 21, 7117–7136 (2001).
Lawrence, H. R. et al. Inhibitors of Src homology-2 domain containing protein tyrosine phosphatase-2 (Shp2) based on oxindole scaffolds. J. Med. Chem. 51, 4948–56 (2008).
Chan, G., Kalaitzidis, D. & Neel, B. G. The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metastasis Rev. 27, 179–192 (2008).
Frisch, M. J. et al. Gaussian 09 Revision C.01. [Gaussian, Inc. (ed.)] (Wallingford CT, USA, 2010).
Lodewyk, M. W., Siebert, M. R. & Tantillo, D. J. Computational prediction of 1H and 13C chemical shifts: a useful tool for natural product, mechanistic, and synthetic organic chemistry. Chem. Rev. 112, 1839–1862 (2012).
Bringmann, G., Bruhn, T., Maksimenka, K. & Hemberger, Y. The assignment of absolute stereostructures through quantum chemical circular dichroism calculations. Eur. J. Org. Chem. 2009, 2717–2727 (2009).
Berova, N., Di Bari, L. & Pescitelli, G. Application of electronic circular dichroism in configurational and conformational analysis of organic compounds. Chem. Soc. Rev. 36, 914–931 (2007).
Glendening, E. D., Reed, A. E., Carpenter, E. J. & Weinhold, F. NBO Version 3.1 [Gaussian, Inc. (ed.)] (Wallingford CT, USA, 2009).
Lu, T. & Chen, F. W. Multiwfn: a multifunctional wavefunction analyzer. J. Comput. Chem. 33, 580–592 (2012).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Ishida-Takagishi, M. et al. The Dishevelled-associating protein Daple controls the non-canonical Wnt/Rac pathway and cell motility. Nat. Commun. 29, 3:859 (2012).
Acknowledgements
This project was supported financially by the NSFC for Outstanding Young Scholar (81422046), the NSF of Yunnan Province (2012FB178), and sponsored by SRF for ROCS, SEM to Wei-Lie Xiao. The calculation sections were supported by the HPC Center of KIB, CAS.
Author information
Authors and Affiliations
Contributions
W.-L.X., H.L. and F.W. conceived the experiments. Z.-H.G., S.-Z.S. and Y.Y. conducted the phytochemical experiment. Y.-M.S conducted the quantum chemical calculations. Z.Q., B.-W.D. and H.-P.P. conducted the biological assay. X.W. conducted the molecular docking. W.-L.X., H.L., F.W., Y.-M.S. and X.J. analyzed the data. W.-L.X., H.L., F.W. and Y.-M.S. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Gao, ZH., Shi, YM., Qiang, Z. et al. Plasiatine, an Unprecedented Indole–Phenylpropanoid Hybrid from Plantago asiatica as a Potent Activator of the Nonreceptor Protein Tyrosine Phosphatase Shp2. Sci Rep 6, 24945 (2016). https://doi.org/10.1038/srep24945
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep24945
This article is cited by
-
Targeting protein phosphatases for the treatment of inflammation-related diseases: From signaling to therapy
Signal Transduction and Targeted Therapy (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.