Abstract
The vertebrate gut is host to large communities of bacteria, and one of the beneficial contributions of this commensal gut microbiota is the increased nutritional gain from feed components that the host cannot degrade on its own. Fish larvae of similar age and under the same rearing conditions often diverge with regards to growth. The underlying reasons for this could be differences in genetic background, feeding behavior or digestive capacity. Both feeding behavior and digestion can be influenced by differences in the microbiota. To investigate possible correlations between the size of fish larvae and their gut microbiota, we analyzed the microbiota small and large genetically homogenous killifish and genetically heterogeneous cod larvae by Bray-Curtis Similarity measures of 16S DNA DGGE patterns. A significant difference in richness (p = 0.037) was observed in the gut microbiota of small and large killifish, but the overall gut microbiota was not found to be significantly different (p = 0.13), indicating strong genetic host selection on microbiota composition at the time of sampling. The microbiota of small and large cod larvae was significantly different with regards to evenness and diversity (p = 0.0001), and a strong correlation between microbiota and growth was observed.
Similar content being viewed by others
Introduction
The vertebrate gut contains numerous bacteria interacting with each other as well as with their host. The commensal gut microbiota contributes to the “energy harvest” from the diet by breaking down complex carbohydrates into monomers for further microbial fermentation. The end products of this fermentation process, short chain fatty acids (SCFA), are released into the gut and adsorbed by host cells1,2,3. Dietary carbohydrates are less nutritionally significant for carnivorous/planktivorous fish such as cod and killifish, where the main energy yield comes from ingested fats and protein4,5. However, the gut microbiota may also have an effect on digestion of lipids and fat, as germ-free zebrafish show reduced differentiation of gut epithelia and reduced lipid uptake compared to conventional controls6,7.
In accordance with the idea of microbes increasing the yield of ingested food, specific members of gut microbiota appear to be positively correlated with obesity in humans and mice2,8. Obese mice and humans have altered ratios of bacteria belonging to Firmicutes/Bacteroidetes (F/B) compared to their lean counterparts, as well as lower microbial diversity. In a recent paper by Zhao et al.9, the F/B ratio and causality of obesity is reviewed. Zhao suggests that the F/B ratio is too simple, and that a closer look at the underlying classes of bacteria such as the Mollicutes may be more relevant for how bacteria may contribute to obesity. In addition, Proteobacteria, specifically Enterobacter cloaca has been shown to be a co-factor in inducing obesity in gnotobiotic mice, by inducing a low grade inflammatory response that alters the lipid metabolism of the host10. Recently, expression of bacterial bile salt hydrolase has been found to direct host gene-expression and influence weight gain in mice11.
Fish larvae of similar age and under the same rearing conditions often diverge with regards to size, reflecting differences in growth rate. The underlying reasons for this could be genetic differences, different feeding behavior or digestive capacity. Both feeding behavior and digestive capacity could be influenced by differences in microbiota12,13. Genetic differences leading to different growth rates may also be indirectly linked to altered microbiota, as recent studies on mice and stickleback (Gasterosteus aculeatus) has revealed host QTLs correlated with specific members of the gut microbiota14,15,16. While obesity is considered unwanted in humans, a commensal microbiota that could increase the energy harvest from of feed would be considered a big advantage in farmed animals –including fish. Fish larvae are actively growing, and enhanced energy provision from the feed due to the activity of the gut microbiota would influence the growth rate of the larvae positively. While several probiotic trials claim that addition of specific bacteria improves growth17,18,19,20, there is to our knowledge only one published study that investigates gut microbiota and growth rate in fish21. However, this study was based on culture-dependent characterization of microbiota.
The objective of this study was to investigate potential correlations between growth and microbiota composition in fish larvae, and to investigate the effects of genetic background on microbiota composition. The microbiota of genetically homogenous Mangrove killifish (Kryptolebias marmoratus, clonal lineage DAN) juveniles and genetically heterogeneous Atlantic cod larvae (Gadus morhua L.) was characterized by 16S DNA denaturing gel electrophoresis (DGGE). To our knowledge this is the first study investigating correlations between microbiota and growth in fish by the use of culture-independent characterization techniques.
Results and Discussion
Growth of DAN killifish
Fourteen out of the 30 sampled killifish were selected for further analysis based on growth measurements. These two groups consisted of the 7 smallest and 7 largest killifish, referred to as S1-S7 and L1-L7, respectively. The regular length measurements of killifish individuals (Fig. 1) show a separation between the large and small group from about day 10 onwards (P = 0.09), with statistically significant differences from day 20 post hatch (dph) (p < 0.007). After 3 weeks the growth rate slowed down both for the large and small killifish. This is in accordance with previous observations of killifish development22,23 and coincides with the transition into the juvenile stage. The large differences in length and wet weight of the juveniles suggest a possible influence of microbiota on growth, as both the genetic background and rearing conditions of the killifish were uniform.
At sampling (40 dph) both the average length and weight of the small juveniles was significantly lower (p < 0.005) than for the large juveniles (Fig. 2) (for individual measurements see Supplementary table S1).
Microbiota associated with large and small mangrove DAN killifish juveniles
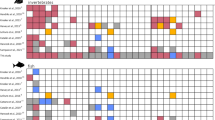
Forty-eight unique DGGE bands were detected, and the richness (k) varied between 23 and 34 bands among individuals (for gel image see supplementary Fig. S1). The average k was significantly lower for large killifish juveniles (p = 0.037). Neither the Shannon diversity index (H′) nor the Pielou’s evenness index (J′) showed significant differences (p > 0.05), between large and small killifish. The exploratory non metric multidimensional scaling (NMDS) of DGGE results showed a clear overlap in gut microbiota composition for most of the small and large killifish (Fig. 3). The DGGE profiles of three of the large killifish juveniles form a separate cluster on the NMDS, among these are the two largest individuals (L6 and L7). Hypothesis testing with One-Way PERMANOVA confirmed that there were no significant differences in the gastrointestinal microbiota between the two groups (p = 0.124). Average Bray Curtis similarities were calculated to compare fish, water and Artemia samples. Highest similarity (0.53 ± 0.02) was observed when comparing large and small killifish, whereas both water and Artemia samples showed low similarity to the killifish (average of 0.33 between killifish and water, and 0.32 between killifish and Artemia, see supplementary table S2 for all values and standard errors). Hypothesis testing with One-Way PERMANOVA confirmed that the Artemia microbiota was significantly different from the microbiota of both large and small killifish (p = 0.0031 and p = 0.0320, respectively).
As some separation in gut microbiota composition is suggested in the ordination in Fig. 3, we excluded the intermediate size fish (the largest of the small and the smallest of the large) from statistical analysis. One-way PERMANOVA confirmed significant differences in gut microbiota composition when analyzing only fish S1–S5 and L4–L7, P = 0.024.
SIMPER analysis showed subtle differences in abundance of 48 bands from the small and large killifish microbiota. Six bands from the DGGE gel were re-amplified and sequenced, among these were the top three bands identified in the SIMPER analysis. None of the sequenced bands were unique for small or large killifish, but were found in different amounts (Table 1). The Sequenced bands contribute to 27.7% of the variation between the two groups. Two opportunistic Vibrio species were among the identified bacteria. However, as there was no sign of infection in the sampled fish which exhibited normal growth and survival, and the highest abundance was found in the large fish, these bacteria were probably not harmful. The effects of the identified bacteria on growth were probably negligible, as the amounts found in the large and small killifish were not significantly different.
Statistically significant differences in gut microbiota composition between small and large killifish was only apparent after excluding the intermediate-size fish from the analysis, with this in mind follow-up studies should avoid sampling continuous sizes of fish, and rather focus on the very small and very large fish. Even though we did not observe significant differences in the GI microbiota composition when comparing the whole groups of large and small killifish at day 40, we cannot rule out the possibility of differences in microbiota at earlier developmental stages contributing to the differences in size. At sampling all killifish had entered the more developed juvenile stage. Conversely, the differences in size were observed from 20 days post hatch. Human microbiome studies have described an initial variable microbiota heavily influenced by environmental factors (such as mode of delivery, type of feed etc.), which subsequently stabilizes during early development, coinciding with increasing host selection24,25. A contrary trend has been shown in zebrafish and cod, where there seems to be strong host selection and negligible effects of environmental factors on the microbiota at the larval stage26,27. If genetic selection factors for gut microbiota develops with age it would seem to be expected that initial differences due to stochastic colonization of larvae will decrease with age in genetically homogenous populations. Reversely, if an initial deterministic (under strong host selection) community composition becomes more influenced by stochastic events as the killifish age, more dissimilar microbiotas would have been expected. The Artemia used to feed the killifish were obtained by standardized methods, resulting in similar microbial content in the feed administered to the fish (within Bray Curtis similarity was 0.52). The significant difference found between the Artemia microbiota and killifish microbiota also points towards a low influence of the feed microbiota, and strong host selection. Further characterization of the microbiota of early killifish larvae is needed to confirm whether gut microbiota variation is indeed higher in less developed individuals.
Cod larvae size measurements
Twenty-three out of the 30 randomly sampled cod larvae (43 dph) were selected for analysis, excluding misshaped individuals. The larvae were divided in two groups, consisting of 11 small and 12 large larvae, referred to as S1–S11 and L1–L12 respectively. The 23 larvae ranged from 11 to 17 mm in length, and between 3 and 53 mg in weight (Fig. 4). Both average length and weight of the small larvae was significantly lower than for the large larvae (p < 0.005) (for individual measurements see supplementary Table S3).
Microbiota associated with large and small cod larvae
DGGE analysis detected a total of 47 unique bands, of which only one was present in all samples (for gel image see supplementary Fig. S2). Band richness (k) varied between 15 and 26, and the average k values were similar for large and small cod larvae (19.9 and 20.3 respectively). However, both the Shannon diversity index (H′) and Pielou’s evenness index (J′) were significantly different between the two groups (p < 0.005), the small larvae showing greater diversity and evenness than the large larvae. This indicates a more selective process of gut colonization in the large larvae. Recent shifts in niche availability caused by faster growth and maturation of the gut may (also) have led to a less diverse microbiota in larger larvae. The NMDS analysis clearly separates the large and small larvae (Fig. 5), and hypothesis testing with One-Way PERMANOVA confirmed a significant difference in microbiota composition between the two groups (p = 0.0002).
The differences in microbiota between large and small cod larvae cannot be explained by differences in feed or water microbiota. The feed supplied to all the cod larvae in this experiment came from the same batch cultivation system, and larvae were grown in tanks with the same water supply. The role of the feed and water microbiota is therefore assumed to be equal for all larvae, and analysis of feed/water microbiota was thus not performed. In general cod larvae microbiota is established under strong host selection, and tends to be more similar to the microbiota of the rearing water, and the effects of feed microbiota are negligible27,28.
A Cabfac factor analysis was performed to estimate the effect of cod larvae microbiota composition on larval size (Fig. 6). The Cabfac reconstruction confirms the strong correlation between size and microbiota composition in the cod larvae; 90% of the individual size variation can be explained by differences in microbiota composition across all individuals, and the slope of this relationship was not significantly different from 1 (0.95 and 0.98 for length and weight, respectively).
SIMPER analysis revealed that differences in abundance of 10 bands contributed to more than 70% of the dissimilarities between the small and large larvae. Out of these 10, taxonomy was assigned to 4 (Table 2). The sequenced bands contribute to 34% of the variation between the two samples. The amounts found in large and small larvae were significantly different for all four bands. The most dominating band found in all the large larvae was also present in all but one of the small larvae, but at significantly lower levels. Sequencing revealed that this band most likely represented Arcobacter (ɛ-Proteobacteria). Some members of the Arcobacter genus are considered foodborne pathogens, but they have also been isolated from oysters and the intestines of healthy livestock, as well as surface and groundwater29,30,31. For the cod larvae in this study this bacterial strain was not correlated with poorer growth, rather on the contrary as intensity of the band was significantly higher in the samples from the large larvae. Simper analysis showed that the difference in abundance of this strain between the large and small larvae contributed to ~18% of the total difference in microbiota composition, with mean abundances of 37.1 vs 21.1%. In a recent study by Bakke et al.27, pyrosequencing of cod larvae at different developmental stages revealed that Arcobacter dominated the microbiota of larvae at 17 and 32 dph, constituting 17–77% of the larval microbiota. The high abundance of this bacterium in seemingly healthy larvae in both studies may indicate that this species is a member of the commensal microbiota of cod larvae. Another band with significantly higher abundance in the large larvae was a Flavobacterium strain, contributing to 8.3% of the differences in the microbiota, with mean abundances of 10.4% and 3.6% for the large and small larvae, respectively. Flavobacterium was also found in larval microbiota at 8 and 32 dph in the study by Bakke et al.27, at comparable levels.
Two DGGE bands representing Aliivibrio logei and Aliivibrio wodanis were found to be unique for the small larvae (B39 and B44, Table 2). These two strains are often found in association with known pathogenic strains causing diseases in salmon, but are to our knowledge not connected with any specific disease in cod32,33. The bands corresponding to these two vibrios were not present in all the small larvae, but were consistently found in the 6 smallest larvae in the “small larvae group”. This may be taken as an indication of an unfavorable colonization that affects larval growth negatively. Similarly, in a culture-based characterization study, fast growing grouper (Epinephelus coioides) was found to contain lower levels of vibrios than slow growing grouper21.
Possible role of genetic background on gut microbiota composition
The killifish in this study are expected to have very similar host selection for individuals at the same developmental stage, as they are clonal and therefore genetically homogenous. Any observed variation in microbiota would therefore most likely be due to stochastic events. Stochastic events during the early colonization of fish larvae may lead to establishment of a stable commensal microbiota, or to unstable or detrimental interactions with opportunistic bacteria34. Only by excluding the intermediate sized fish from analysis did we find significant differences in the gut microbiota of large and small killifish. A strong genetically encoded host selection may explain why we only observed subtle variation in microbiota composition. As the growth of the large and small killifish started to differ well before metamorphosis into the juvenile stage was complete, it may be that more dissimilar microbiotas initially colonized the larvae. In previous studies however, strong host selection has been indicated at early developmental stages for both cod and zebrafish larvae26,27. More studies are needed to investigate if there is strong host-selection in the earlier stages of killifish development.
The cod larvae were spawned from mixed parents, and thus have a heterogeneous genetic background. The large differences in microbiota between small and large larvae may be due to differences in host selection factors or subtle differences in development of the larvae, both of which could be influenced by genetic factors. In humans monozygotic twins has been shown to have higher gut microbiota similarity than dizygotic twins35, and in mice several quantitative trait loci which control abundances of different taxa have been identified by the use of advanced intercross breeding lines14,16. Also for other fish species there seems to be higher similarities in microbiota composition in closely related individuals36,37. Further studies on full sibling groups are needed to investigate whether growth and microbiota composition is more even in a less heterogeneous group of cod larvae.
Concluding remarks
To conclude, whereas the genetically homogenous killifish only had subtle differences in gut microbiota composition and were significantly different only when comparing the largest and smallest of the sampled fish, the genetically heterogeneous cod larvae had distinct differences in the microbiota of large and small larvae. For both species a strong host selection and a connection between the gut microbiota and the growth of the fish was indicated. Whether the large fish grew better as a result of the different microbiota, or acquired different microbiota as a result of differences in growth, needs further studies. Such studies should have a design which can disentangle the implications of host genetics on selection of gut microbiota. As 16S DGGE has limitations with regards to detecting low abundant species and does not supply information on taxonomy, future studies should use high throughput deep sequencing. The correlation between growth and microbiota is very interesting from a microbial management perspective, where a rewarding concept would be to steer the microbial communities in the rearing water (and thus in the fish gut) towards a microbiota that promotes rapid growth28,38,39.
Materials and Methods
Biological material and rearing conditions
All prevailing local, national and international regulations and conventions, and normal scientific ethical practices have been respected during the fish experiments. The cod experiment was approved by the Norwegian Animal Research Authority, and the killifish experiment followed the guidelines of the Animal Care and Use Committee of Nagasaki University, Japan.
Mangrove killifish
The DAN clonal strain of mangrove killifish was used in this study (obtained from W.P Davies, US Environmental Protection Agency, Florida USA). Larvae from this strain are descendants of one fish collected in Belize, and has been reared for over 10 generations at the Aquaculture Biology lab, Nagasaki University, Japan23.
Mangrove killifish eggs were manually dechorionated following the protocol by Koenig and Chasar40, using fine forceps to release the larvae. Thirty larvae were reared individually in 100 mL plastic cups, containing 60 mL artificial seawater (Marine Art High, Senju, Saiaku Co., Ltd, Osaka) dissolved to 17 ppt in ozonated tap water. The fish were held at 25 ± 1°C with a photoperiod of 14 h light and 10 h darkness. Water was exchanged every 10 days, coinciding with larval length measurements. Larval length was determined using a digital microscope (Keyence VH-6300). Total length was converted into standard length by the use of the following equations; y = −0.21 + 0.84x. Wet weight was recorded after drying the fish with KimWipes®.
Cysts of Artemia (Great Salt Lake, Aquafauna Biomarine Inc., CA, USA) were decapsulated with hypochlorite according to the protocol of Sorgeloos et al.41. Decapsulated Artemia (1–2 day old naupli) were added as feed for the killifish, ad libitum every 2–3 days. Rearing water, as well as Artemia from 4 different feed “batches” (13, 17, 21 and 27 dph) were sampled for microbiological analysis. When all the fish had reached the juvenile stage at 40 dph, length was measured and the fish were put to death by triacaine methane sulphonate (MS 222). Intestines from all 30 juveniles were sampled for microbiological analysis, using forceps and fine needles.
Cod larvae
Cod larvae were reared at SINTEF Fisheries and Aquaculture, Trondheim. The cultivation regime included addition of rotifers (fed Reed Mariculture Rotifer diet® and Pavlova 1800 paste) from 2 dph, and MarolE (Sintef, Trondheim, Norway) enriched A. fransiscana from 18 dph (for details see Supplementary Table S4.). At 43 dph 30 larvae were randomly sampled from identically treated rearing tanks, and put to death using MS 222. Total length of individual larvae was measured by the use of a stereo microscope. Rinsed larvae were dried with KimWipes® before wet weight was determined. Larvae were stored at −80°C before microbiological analysis. As a result of the small size of cod larvae, the guts were not dissected out. As whole cod larvae were used in the microbiota analysis, a contribution from skin-associated microbiota cannot be ruled out. However, the number of skin-associated bacteria have been reported to be low compared to that of the gut microbiota42.
DNA extraction
Killifish intestines and whole cod larvae were homogenized in 200 μl autoclaved seawater using a sterile plastic rod and a syringe. DNA was extracted from the fish, water and Artemia samples using the QIAGEN DNeasy blood and tissue kit with some modifications from the manufacturer’s protocol28. DNA concentration was measured with a NanoDrop spectrophotometer (Thermo Fisher Scientific).
PCR-DGGE
To avoid co-amplification of eukaryote DNA, a nested PCR protocol was used to amplify the V3 region of the bacterial 16S rDNA43. The quantity and size of PCR products were investigated on 1% agarose gels casted with GelRedTM. DGGE was performed as described by Muyzer et al.44, with the INGENY phorU DGGE system. A denaturing gradient of 35–55% (where 100% corresponds to 7 M urea and 40% formamide) was used in 8% acrylamide gels. Depending on the template concentration, 2.5–15 μl of sample was loaded into the wells, to obtain comparable amounts of PCR product. The gels were run with 0,5 × TAE buffer at 100 V for 17 hours at 60 °C. After electrophoresis, the gels were stained with SYBR Gold (1:10 000 dilution, InvitrogenTM) for 1 hour, rinsed with MilliQ water and photographed under UV light (GenBox geldoc system, Syngene).
Sequence analysis
Selected bands from the DGGE gels were stamped out using a micropipette, and left to dissolve in MilliQ water for 24 hours at 4 °C. After re-amplification as described in Bakke et al.28, PCR products were purified using QIAquick PCR Purification kit (Qiagen) and DNA sequencing was performed by Eurofins MWG operon using M13R (5′-caggaaacagctatgac-3′) as a sequencing primer. Sequences were then analyzed through the Ribosomal Database Project (RDP) Classifier45.
Gel analysis and statistical analysis
DGGE gel images were analyzed with the Gel2k software46 (provided by Svein Norland, Dept. Biology, Univ. Bergen, Norway) to convert band profiles to histograms, resulting in sample-peak area matrices. Individual peak areas for DGGE bands were normalized by dividing by the sum of all peak areas from each respective DGGE profile. The Shannon index (H′)47, and the relative diversity, J′ (evenness) were used as measures of diversity in the DGGE profiles, in addition to band richness (k). Student’s t-test (unpaired) was used to investigate significance in differences in Shannon indices, abundance of individual DGGE bands and larval growth measurements. One-way PERMANOVA (non-parametric MANOVA48) based on Bray-Curtis similarity was used to test for differences in DGGE profiles between groups of samples. Ordination by Non-metric multidimensional scaling (NMDS) based on Bray-Curtis similarity was used to visualize the similarity/dissimilarity in bacterial community profiles among the samples49. SIMPER analysis was performed to identify which DGGE bands contributed most to dissimilarities between samples. CABFAC analysis (a factor analysis of abundance data with associated environmental data) was used to reconstruct larval weight/length based on microbial community DGGE profiles). The multivariate analyses were performed with the PAST software package50.
Additional Information
How to cite this article: Forberg, T. et al. Correlation between microbiota and growth in Mangrove Killifish (Kryptolebias marmoratus) and Atlantic cod (Gadus morhua). Sci. Rep. 6, 21192; doi: 10.1038/srep21192 (2016).
References
Hooper, L. V., Midtvedt, T. & Gordon, J. I. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22, 283–307, 10.1146/annurev.nutr.22.011602.092259 (2002).
Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031, 10.1038/nature05414 (2006).
Flint, H. J., Scott, K. P., Louis, P. & Duncan, S. H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 9, 577–589, 10.1038/nrgastro.2012.156 (2012).
Taylor, D. S. Diet of the killifish Rivulus marmoratus collected from land crab burrows, with further ecological notes. Environ. Biol. Fish. 33, 389–393, 10.1007/BF00010951 (1992).
Hamre, K. Nutrition in cod (Gadus morhua) larvae and juveniles. ICES J. Mar. Sci. 63, 267–274, 10.1016/j.icesjms.2005.11.011 (2006).
Bates, J. M. et al. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Develop. Biol. 297, 374–386, 10.1016/j.ydbio.2006.05.006 (2006).
Semova, I. et al. Microbiota Regulate Intestinal Absorption and Metabolism of Fatty Acids in the Zebrafish. Cell Host Microbe 12, 277–288, 10.1016/j.chom.2012.08.003 (2012).
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023, 10.1038/4441022a (2006).
Zhao, L. The gut microbiota and obesity: from correlation to causality. Nat. Rev. Microbiol. 11, 639–647, 10.1038/nrmicro3089 (2013).
Fei, N. & Zhao, L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. Isme J. 7, 880–884, 10.1038/ismej.2012.153 (2013).
Joyce, S. A. et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. 111, 7421–7426, 10.1073/pnas.1323599111 (2014).
Cani, P. D. et al. Gut microbiota fermentation of prebiotics increases satietogenic and incretin gut peptide production with consequences for appetite sensation and glucose response after a meal. Am. J. Clin. Nutr. 90, 1236–1243, 10.3945/ajcn.2009.28095 (2009).
Tremaroli, V. & Backhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249, 10.1038/nature11552 (2012).
McKnite, A. M. et al. Murine Gut Microbiota Is Defined by Host Genetics and Modulates Variation of Metabolic Traits. Plos One 7, e39191, 10.1371/journal.pone.0039191 (2012).
Stagaman, K., Guillemin, K. & Milligan-Myhre, K. Tending a complex microbiota requires major immune complexity. Mol Ecol 23, 4679–4681, 10.1111/mec.12895 (2014).
Benson, A. K. et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. 107, 18933–18938, 10.1073/pnas.1007028107 (2010).
Sun, Y.-Z., Yang, H.-L., Ma, R.-L. & Lin, W.-Y. Probiotic applications of two dominant gut Bacillus strains with antagonistic activity improved the growth performance and immune responses of grouper Epinephelus coioides . Fish Shellfish Immun. 29, 803–809 (2010).
El‐Haroun, E., Goda, A. S. & Chowdhury, K. Effect of dietary probiotic Biogen® supplementation as a growth promoter on growth performance and feed utilization of Nile tilapia Oreochromis niloticus (L.). Aquac. Res. 37, 1473–1480 (2006).
Suzer, C. et al. Lactobacillus spp. bacteria as probiotics in gilthead sea bream (Sparus aurata, L.) larvae: Effects on growth performance and digestive enzyme activities. Aquaculture 280, 140–145 (2008).
Lobo, C. et al. Dietary probiotic supplementation (Shewanella putrefaciens Pdp11) modulates gut microbiota and promotes growth and condition in Senegalese sole larviculture. Fish Physiol. Biochem. 40, 295–309, 10.1007/s10695-013-9844-0 (2014).
Sun, Y., Yang, H., Ling, Z., Chang, J. & Ye, J. Gut microbiota of fast and slow growing grouper Epinephelus coioides. Afr. J. Microb. Res. 3, 637–640 (2009).
Grageda, C. M. V., Sakakura, Y. & Hagiwara, A. Early development of the self-fertilizing mangrove killifish Rivulus marmoratus reared in the laboratory. Ichthyol. Res. 51, 309–315, 10.1007/s10228-004-0235-5 (2004).
Grageda, M. V. C., Sakakura, Y., Minamimoto, M. & Hagiwara, A. Differences in life-history traits in two clonal strains of the self-fertilizing fish, Rivulus marmoratus. Environ. Biol. Fish. 73, 427–436, 10.1007/s10641-005-2196-6 (2005).
Palmer, C., Bik, E. M., DiGiulio, D. B., Relman, D. A. & Brown, P. O. Development of the human infant intestinal microbiota. PLoS biol. 5, e177, 10.1371/journal.pbio.0050177 (2007).
Wall, R. et al. Role of Gut Microbiota in Early Infant Development. Clin. Med. Pediatr. 3, 45–54 (2009).
Yan, Q., van der Gast, C. J. & Yu, Y. Bacterial Community Assembly and Turnover within the Intestines of Developing Zebrafish. Plos One 7, e30603, 10.1371/journal.pone.0030603 (2012).
Bakke, I., Coward, E., Andersen, T. & Vadstein, O. Selection in the host structures the microbiota associated with developing cod larvae (Gadus morhua). Environ. Microbiol. 17, 3914–3924, 10.1111/1462-2920.12888 (2015).
Bakke, I., Skjermo, J., Vo, T. A. & Vadstein, O. Live feed is not a major determinant of the microbiota associated with cod larvae (Gadus morhua). Env. Microbiol. Rep. 5, 537–548, 10.1111/1758-2229.12042 (2013).
Romero, J., García-Varela, M., Laclette, J. P. & Espejo, R. T. Bacterial 16S rRNA Gene Analysis Revealed That Bacteria Related to Arcobacter spp. Constitute an Abundant and Common Component of the Oyster Microbiota (Tiostrea chilensis). Microb. Ecol. 44, 365–371, 10.1007/s00248-002-1063-7 (2002).
Wesley, I. et al. Fecal Shedding of Campylobacter and Arcobacter spp. in Dairy Cattle. Appl. Environ. Microbiol. 66, 1994–2000 (2000).
Lehner, A., Tasara, T. & Stephan, R. Relevant aspects of Arcobacter spp. as potential foodborne pathogen. Int. J. Food. Microbiol. 102, 127–135, 10.1016/j.ijfoodmicro.2005.03.003 (2005).
Lunder, T. et al. Phenotypic and genotypic characterization of Vibrio viscosus sp. nov. and Vibrio wodanis sp. nov. isolated from Atlantic salmon (Salmo salar) with ‘winter ulcer’. Int. J. Syst. Evol. Micr. 50, 427–450, 10.1099/00207713-50-2-427 (2000).
Benediktsdóttir, H. & Sigurjónsdóttir . Vibrio spp. isolated from salmonids with shallow skin lesions and reared at low temperature. J. Fish Dis. 21, 19–28, 10.1046/j.1365-2761.1998.00065.x (1998).
Olafsen, J. A. Interactions between fish larvae and bacteria in marine aquaculture. Aquaculture 200, 223–247, 10.1016/S0044-8486(01)00702-5 (2001).
Goodrich, Julia K. et al. Human Genetics Shape the Gut Microbiome. Cell 159, 789–799, 10.1016/j.cell.2014.09.053 (2014).
Sullam, K. E. et al. Divergence across diet, time and populations rules out parallel evolution in the gut microbiomes of Trinidadian guppies. Isme J. 9, 1508–1522, 10.1038/ismej.2014.231 (2015).
Navarrete, P. et al. PCR-TTGE Analysis of 16S rRNA from Rainbow Trout (Oncorhynchus mykiss) Gut Microbiota Reveals Host-Specific Communities of Active Bacteria. Plos One 7, e31335, 10.1371/journal.pone.0031335 (2012).
Attramadal, K. J. et al. Recirculation as a possible microbial control strategy in the production of marine larvae. Aquacult. Eng. 46, 27–39, 10.1016/j.aquaeng.2011.10.003 (2012).
Vadstein, O. et al. Microbiology and immunology of fish larvae. Rev. Aquacult. 5, S1–S25, 10.1111/j.1753-5131.2012.01082.x (2013).
Koenig, C. C. & Chasar, M. P. Usefulness of the hermaphroditic marine fish, Rivulus marmoratus, in carcinogenicity testing. J. Natl. Canc. Inst. Monogr. 65, 15–33 (1984).
Sorgeloos, P., Bossuyt, E., Laviña, E., Baeza-Mesa, M. & Persoone, G. Decapsulation of Artemia cysts: A simple technique for the improvement of the use of brine shrimp in aquaculture. Aquaculture 12, 311–315, 10.1016/0044-8486(77)90209-5 (1977).
Austin, B. The Bacterial Microflora of Fish, Revised. Scientific World J. 6, 931–945, 10.1100/tsw.2006.181 (2006).
Bakke, I., De Schryver, P., Boon, N. & Vadstein, O. PCR-based community structure studies of Bacteria associated with eukaryotic organisms: A simple PCR strategy to avoid co-amplification of eukaryotic DNA. J. Microbiol. Meth. 84, 349–351, 10.1016/j.mimet.2010.12.015 (2011).
Muyzer, G., de Waal, E. C. & Uitterlinden, A. G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700 (1993).
Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267, 10.1128/AEM.00062-07 (2007).
Norland, S. Gel2K gel analysis software. University of Bergen, Norway, http://folk.uib.no/nimsn/gel2k (2004).
Shannon, C. E. A Mathematical Theory of Communication. At&T Tech. J. 27, 623–656 (1948).
Anderson, M. J. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 26, 32–46, 10.1111/j.1442-9993.2001.01070.pp.x (2001).
Taguchi, Y. H. & Oono, Y. Nonmetric Multidimensional Scaling as a Data-Mining Tool: New Algorithm and New Targets. Adv. Chem. Phys. 130, 315–351, 10.1002/0471712531.ch18 (2005).
Hammer, O., Harper, D. & Ryan, P. Paleontol. Electron. 4, http://palaeo-electronica.org/2001_1/past/issue1_01.htm (2001) (10/07/2015).
Acknowledgements
Thanks to the Laboratory of Aquaculture Biology Nagasaki University for hosting E.B.S. Matsuura Kana and Hori Kyoko reared the killifish. The cod start feeding experiment was a part of The European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement n°227197 Promicrobe “Microbes as positive actors for more sustainable aquaculture”. T.F. is financed through NRC grant 233865.
Author information
Authors and Affiliations
Contributions
O.V., Y.O., A.H. and Y.S. designed the research. E.B.S. in collaboration with A.H. and Y.S. carried out killifish experiments, E.B.S. sampled cod larvae, and performed DGGE-analysis and sequencing in collaboration with I.B. T.F. and I.B. processed DGGE data and interpreted the results. O.V. and T.F. performed statistical data analysis. T.F. wrote the manuscript with revision and contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Forberg, T., Sjulstad, E., Bakke, I. et al. Correlation between microbiota and growth in Mangrove Killifish (Kryptolebias marmoratus) and Atlantic cod (Gadus morhua). Sci Rep 6, 21192 (2016). https://doi.org/10.1038/srep21192
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep21192
This article is cited by
-
Opportunistic feeding habits of two African freshwater clupeid fishes: DNA metabarcoding unravels spatial differences in diet and microbiome, and identifies new prey taxa
Hydrobiologia (2023)
-
Correlations of age and growth rate with microbiota composition in Atlantic cod (Gadus morhua) larvae
Scientific Reports (2017)
-
The highly variable microbiota associated to intestinal mucosa correlates with growth and hypoxia resistance of sea bass, Dicentrarchus labrax, submitted to different nutritional histories
BMC Microbiology (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.