Abstract
Novel disulfide-containing polypeptide toxin was discovered in the venom of the Tibellus oblongus spider. We report on isolation, spatial structure determination and electrophysiological characterization of this 41-residue toxin, called ω-Tbo-IT1. It has an insect-toxic effect with LD50 19 μg/g in experiments on house fly Musca domestica larvae and with LD50 20 μg/g on juvenile Gromphadorhina portentosa cockroaches. Electrophysiological experiments revealed a reversible inhibition of evoked excitatory postsynaptic currents in blow fly Calliphora vicina neuromuscular junctions, while parameters of spontaneous ones were not affected. The inhibition was concentration dependent, with IC50 value 40 ± 10 nM and Hill coefficient 3.4 ± 0.3. The toxin did not affect frog neuromuscular junctions or glutamatergic and GABAergic transmission in rat brains. Ca2+ currents in Calliphora vicina muscle were not inhibited, whereas in Periplaneta americana cockroach neurons at least one type of voltage gated Ca2+ current was inhibited by ω-Tbo-IT1. Thus, the toxin apparently acts as an inhibitor of presynaptic insect Ca2+ channels. Spatial structure analysis of the recombinant ω-Tbo-IT1 by NMR spectroscopy in aqueous solution revealed that the toxin comprises the conventional ICK fold containing an extended β-hairpin loop and short β-hairpin loop which are capable of making “scissors-like mutual motions”.
Similar content being viewed by others
Introduction
For many years, natural venoms have been valuable sources of biologically active compounds. The development of different analysis methods made it possible to unravel the complexity of spider venoms1,2,3,4,5,6 and identify action modes of isolated compounds on various molecular targets7,8,9,10,11,12,13,14. These investigations have shown that, among other venomous creatures, spiders produce the largest variety of venom compounds15. Taking into account the fact that spiders are the most abundant terrestrial predators16, this huge library of bioactive compounds remains poorly investigated. According to ArachnoServer (http://www.arachnoserver.org), the number of individual spider toxins investigated to date is 1403 from 97 different spider species17. Today, the average number of polypeptide toxins in spider venom is estimated at about 170 per species18. And according to The World Spider Catalog Version 16.5 (http://www.wsc.nmbe.ch/), there are currently 45,670 species of spiders on our planet (belonging to 114 different families); therefore, the estimated number of individual spider toxins seems to be quite amazing. Consequently, scientists have described only a tiny part of all spider venom toxins to date.
Regarding long and delicate tuning of venom composition by evolution, the most predominant spider toxins are those serving two main aims: predation of prey and protection of the spider itself15. Because insects are the main source of spider nutrition, a lot of insect-acting toxins have been detected by spider venom studies. Many of these toxins are insect specific and can, at low concentrations, cause paralysis or death of insects, but are not toxic to mammals10,13,19,20,21. The vast majority of insect-specific toxins are disulfide-stabilized polypeptides less than 10 kDa that shared one common fold named inhibitor cystine knot (ICK). Such polypeptide toxins act generally, whether on insect sodium, on insect calcium ion channels and rarer on potassium ones10,11,13. The action selectivity may be explained by structural differences of insect ion channels and their mammalian forms. Therefore, spider venom polypeptides may be regarded as a candidate base for the development of new insecticides16,22.
Development of such compounds is still very important because insects are acting in a disturbing way to different human activities. An increasing human population could require a large amount of food in the future. In this light, large annual crop yield reductions by insects pose a threat16. Moreover, such infectious diseases as malaria are transmitted by mosquitoes. In spite of some recent success in combating malaria23,24, there still remain many obstacles; therefore, mosquito control is a very urgent issue. A critical factor is also extending pest resistance to existing insecticides.
Tibellus oblongus (Walckenaer, 1802) belongs to the Philodromidae family and is distributed throughout the Holarctic25. T. oblongus, as another species of the Tibellus genus, is an active hunter that does not spin webs but actively pursues its prey. Papers describing any venom polypeptides for this spider have not been published to date, but cytochrome oxidase subunit 1, together with its gene and some ribosomal RNA genes, are published26. In preliminary experiments, we have observed strong insect toxicity for crude T. oblongus venom. And now we report on isolation, structure determination and recombinant production as well as electrophysiological characterization of a novel insecticidal polypeptide–ω-Tbo-IT1, which inhibits insect calcium channels, presumably the Cav2 subtype.
Materials and Methods
Venom collection
The crude T. oblongus venom was purchased from Fauna Laboratories, Ltd. (Almaty, Republic of Kazakhstan). Female spiders were collected in the nearby Almaty region and crude venom for an investigation was obtained by electrostimulation of several species.
All experiments were approved by the Animal Care and Use Committees of the Shemyakin-Ovchinnikov Institute of the Russian Academy of Sciences and Sechenov Institute of Evolutionary Physiology and Biochemistry of the Russian Academy of Sciences. Experiments were carried out in accordance with the guidelines approved by national Animal Protection Law, which is fully compatible with European Community Council directives 86/609/EEC.
All solvents, salts and reagents were purchased in the best quality available from certified suppliers. All gels and buffer solutions were prepared according to manufacturers’ manuals.
Purification
ω-Tbo-IT1 toxin was fractionated from T. oblongus venom using a size-exclusion chromatography (SEC) and several steps of reverse-phase high pressure liquid chromatography (RP-HPLC). Soluble venom (2.7 mg in 5 ml) was applied to a Beckman TSK 2000SW column (7.5 × 600 mm) which was equilibrated with 20 mM sodium phosphate buffer (150 mM NaCl, pH 4.5). Separation was done at a flow rate of 0.5 ml/min and chromatographic results were monitored at 214 nm. The active fraction was then applied on a Jupiter C5 (Phenomenex, USA) RP-HPLC column (4.6 × 250 mm) for separation in 0.1% v/v trifluoroacetic acid (TFA) buffer system in a linear gradient of acetonitrile concentration at a flow rate of 1 ml/min. The absorbance was monitored both at 210 nm and 280 nm. One active rough fraction was obtained as a result of the toxicity test on insects as described below. This active fraction was further fractionated on a Synergy Polar-RP400 (Phenomenex, USA) column (4.6 × 250 mm) in a linear gradient of acetonitrile (0.1% v/v TFA containing buffers) at a flow rate of 0.5 ml/min. Pure polypeptide elution time was monitored at the same wavelengths, 210 nm and 280 nm.
Characterization of polypeptide structure
Dry polypeptide was dissolved in 40 μL of solution which contained 6M guanidine hydrochloride, 3 mM EDTA and 0.5 M Tris-HCl (pH 8.5). After that, a hundred-fold molar excess of dithiothreitol (DTT) was added and reduction was performed at 40 °C in a nitrogen atmosphere for 4 hours. The polypeptide was alkylated by an isopropanol solution of 4-vinylpyridine (4 μL, 50% v/v; tenfold molar excess to DTT; 20 min in darkness) and purified from the reaction mixture on an RP-HPLC column. Proteolytic digestion of modified polypeptide was made by trypsin according to the procedure described27. Modified polypeptide molecules’ N-terminal sequence and tryptic peptide sequences were determined by Edman automated stepwise degradation using a Procise Model 492 protein sequencer (Applied Biosystems, USA) according to the manufacturer’s protocol.
Mass spectrometry
Fractions after separation, pure and modified polypeptide, were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI TOF-MS) on MALDI LR (Micromass UK) and Ultraflex TOF-TOF (Bruker Daltonik, Germany) instruments. ProteoMass peptide MALDI-MS calibration kit (with the range of 700–3500 Da) and protein MALDI-MS calibration kit (mass range of 700–66,000 Da; both by Sigma) were utilized for calibration of these spectrometers. Samples were prepared by the dried droplet method with α-cyano-4-hydroxycinnamic acid (in 50% acetonitrile +0.1% v/v TFA) matrix. Detection of molecular ions was performed in a linear positive mode.
NMR spectroscopy and spatial structure calculation
-
1
Two-dimensional 1H-1H double quantum filtered correlation spectroscopy (DQF-COSY), total correlation spectroscopy (TOCSY; 80-ms mixing time), nuclear Overhauser effect spectroscopy (NOESY; 150-ms mixing time) and 1H-13C heteronuclear single quantum coherence spectroscopy (HSQC; at natural abundance) NMR spectra28 (Supplementary Figs S2, S3, S4 and S5) of 1.5 mM recombinant ω-Tbo-IT1 toxin (solubilized in H2O or D2O with 20 mM phosphate buffer, pH 5.8) were acquired at 12 °C and 20 °C on NMR spectrometer Varian NMR System 700 MHz (Agilent, USA; provided by NBICS-Center at NRC “Kurchatov Institute”). The Watergate29 technique was used to suppress strong solvent resonance. 1H and 13C chemical shifts were referenced relative to the trimethylsilyl propionate. 1H and 13C resonance assignments for ω-Tbo-IT1 were obtained by a standard procedure30 using NMR spectra in the CARA software31. The 3JHNHα coupling constants were determined from a line shape analysis of the NOESY cross-peaks. The 3JHαHβ coupling constants were measured using ACME program32 in the DQF-COSY spectrum acquired in D2O. Temperature coefficients of amide protons (Δδ1HN/ΔT) were measured in a temperature range from 12 °C to 20 °C using 2D TOCSY spectra. The slowly exchanging amide protons were identified at 12 °C by reconstituting of lyophilized ω-Tbo-IT1 in D2O and immediately recording the one-dimensional proton and two-dimensional 1H-1H TOCSY spectra.
-
2
The spatial structure calculation of ω-Tbo-IT1 (two variants: recombinant with homoserine lactone or native with methionine on C-terminus) was performed using the simulated annealing/molecular dynamics protocol as implemented in the CYANA software package33. Upper inter-proton distance constraints were derived from nuclear Overhauser effect (NOE) cross-peaks via standard CYANA calibration procedure. Torsion angle restraints and stereospecific assignments were obtained by the analysis of local conformation in CYANA using sequential NOE data and J coupling constants. All Xxx-Pro peptide bonds were clearly identified as trans on the basis of characteristic NOE contacts30. The disulfide binding pattern (Cys residues 1–21, 8–25, 20–40 and 27–38) was uniquely determined from preliminary structure calculations and corresponding distance restraints were introduced in CYANA software. Other possible disulfide bond connectivity patterns were tested in a course of spatial structure calculations, but they did not correspond to the experimental NMR data. The 22 amide protons having temperature gradients less than 5 ppb/K and/or slow exchange with water protons were supposed to participate in hydrogen bonding and assigned as donors of backbone hydrogen bonds with related hydrogen-acceptor partners on the basis of preliminary structure calculations. Corresponding hydrogen bond restraints were employed in subsequent calculations for d(O,N) and d(O,HN) distances in accordance with hydrogen bond distance criteria in CYANA software. NOESY spectra back calculation was used to verify that the calculated structure did not have proton-proton contacts that were not seen in the spectra. At the final stages of structure calculations, inter-proton lower distance restraints of 4 Å were employed for such contacts. In the final calculation stage, the standard CYANA simulated annealing protocol was applied to 100 random structures using angle and distance restraints and resulting 20 NMR structures of ω-Tbo-IT1 with the lowest target functions were selected.
Visual analysis of calculated structures and figure drawings were performed using the MOLMOL software34.
Precursor determination
Total RNA was purified from the venom glands of T. oblongus using Trisol® Reagent (Ambion, Canada) according to manufacturer protocol. Total cDNA was synthesized from 5 μg of total RNA, using the MINT kit (Evrogen, Russia) following the manufacturer recommendations. Rapid amplification of cDNA ends was carried out using the universal primer T7cap (GTA ATA CGA CTC ACT ATA GGG CAA GCA GTG GTA ACA ACG CAG AGT) and degenerated primers To1 (TGT GCC AGC AAG AAY GAR MGN TGY GGN AAY) and To2 (GCG AGC AAG AAT GAR MGN TGY GGN AAY GCN) for 3′-terminus determination (3′-RACE) and To3 (ACG CGC AGT TTC TTR CTY KCN ACR CCN) for 5′-terminus determination (5′-RACE). DNA sequencing was carried out on ABI PRISM 3100-Avant.
Gene construction
Production of a recombinant analogue of the toxin proceeded utilizing the pET-32-b(+) vector (Merck Biosciences, USA). Toxin gene was synthesized by PCR reaction in two stages on the basis of overlapping primers having an additional Met-codon for BrCN cleavage and a stop codon. The following primers were employed: TboIT1-D0 (AGC CAT GGC GAT GTG CGC TTC TAA GAA CGA ACG TTG), TboIT1-D1 (TCT AGG AAC GAA CGT TGC GGT AAC GCT CTG TAC GGT ACC AAA GGT CCG), TboIT1-D2 (GGT AAA TGC ATC TGC CGT ACC GTT CCG CGT AAA GGT GTT AAC TCT TGC), TboIT1-R4 (ACG GCA GAT GCA TTT ACC GTT GCA GCA ACC CGG ACC TTT GGT ACC GTA C) and TboIT1-R5 (CTC TCG AGT CAC ATG CAA CGG CAA GAG TTA ACA CC). PCR was performed using a (5:1) mixture of Taq DNA polymerase (Evrogene, Russia) and Pfu DNA polymerase (Promega, USA) under the following conditions: incubation at 94 °C for 5 min; 4 cycles (first stage PCR) or 24 cycles (second stage PCR) with each cycle including denaturation at 94 °C for 20 sec, annealing at 56 °C for 20 sec and extension at 72 °C for 20 sec; an additional extension step at 72 °C for 5 min and cooling at 10 °C for 1 min. Amplified products were separated in a 1% agarose gel and a target product was obtained by a GeneJet™ gel extraction kit (Thermo Scientific Fermentas, USA). The resulting gene product was ligated to the pET-32-b(+) vector on the EcoRV restriction site by blunt ends. Sequences of the genes were confirmed after cloning in XL21 E. coli cells.
Recombinant analog production
E. coli BL21(DE3) cells were transformed with expression vectors by electroporation. Transformed cells were cultured in LB medium with ampicillin (100 mg/ml) at 37 °C up to solution density 0.6–0.7 AU at 600 nm. T7-promoted expression was induced by addition of isopropyl-1-thio-β-D-galactopyranoside (IPTG) up to 0.2 mM concentration. Expression was performed for 12 h at 24 °C, after which cells were harvested and re-suspended in chromatogram buffer (300 mM NaCl, 20 mM Tris-HCl pH 7.5). The suspension was ultrasonicated and the supernatant, after centrifugation (15 min at 14,000g), was applied to TALON Metal Affinity resin (Clontech Laboratories, USA). Fusion protein purification was made in accordance with the manufacturer’s manual. Protein cleavage was performed overnight at room temperature in the dark in accordance with method35. HCl to a final concentration of 0.2 M and CNBr with a molar ratio to a fusion protein of 600:1 were added. Final purification of the recombinant polypeptide was made by RP-HPLC on Jupiter C5 (Phenomenex, USA) column and then its identity to natural polypeptide was confirmed by MALDI-MS.
Insect-toxicity tests
Toxicity was measured on M. domestica larvae of about 50 mg by weight. Lyophilized samples were dissolved in pure water and fixed aliquots were injected into larvae bodies using a 10 μL syringe (Hamilton, USA). To estimate concentration dependent effect, doses of 0.375, 0.75, 1.25, 3.0, 5.0, 7.0, 9.0, 12, 20, 25, 30, 40 and 60 μg/g were used. The paralyzing effect development was monitored during a period of 48 h. Each dose was measured on a group consisting of 16–20 larvae. The test was repeated five times and standard errors of the mean were calculated. Toxicity was also measured on G. portentosa cockroaches by the same protocol using 1.25, 2.5, 5.0, 10, 20, 40, 80 μg/g dose equivalents. In this case, the paralyzing effects development was monitored during a period of 24 h.
Electrophysiology
The effects on insect neuromuscular transmission were studied on late third stage larvae of Calliphora vicina. After dissection, the internal organs were removed so that the preparation consisted only of muscles attached to the cuticle. The ventral ganglion was excised and the segmental nerves were stimulated through a suction electrode. Recordings were made from ventral longitudinal fibers. To eliminate electrical contacts of the recorded fiber with its neighbors, the latter were dissected. The preparation was perfused with a saline solution containing (in mM): 172 NaCl, 2.5 KCl, 0.5 CaCl2, 8 MgCl2, 2.4 NaHCO3, 0.3 H2PO4 and 52 sucrose. pH was adjusted to 7.2 with NaOH or HCl. Experiments were performed at room temperature (20–24 °C). The excitatory postsynaptic currents (eEPSC) were evoked by nerve stimulation (0.5 Hz). Voltage-activated currents in muscle cells were evoked by 500 ms depolarizing voltage pulses from holding potential −90 mV to 0 mV. Potassium currents were inhibited both by 2 mM 4-aminopyridine (4-AP) and 10 mM tetraethylammonium. Sodium currents were eliminated by 10 mM lidocaine. To prevent muscle contraction, external Ca2+ was replaced by 10 mM Ba2+.
Adult male cockroaches of species Periplaneta americana were taken from stock colonies maintained at 29 °C with a photoperiod of 12 h light and 12 h darkness. Animals were cold anesthetized and immobilized on a dissection dish dorsal side up. Legs, dorsal cuticle gut and dorsolongitudinal muscles were removed to isolate the nerve cord. The preparation was placed in an experimental chamber with a solution containing (in mM): 160 N-methyl-D-aspartic acid (NMDG), 5 CaCl2, 10 HEPES, 20 TEA and 14 4-AP, with pH 7.3. Large (>50 μM) neurons were isolated by vibrodissociation from the terminal abdominal ganglion as described36 that use a local application of mechanical vibration directly on a ganglion instead of a tissue enzymatic treatment. The whole-cell voltage clamp recordings of Ca2+-currents were done with the pipette solution (in mM): 130 cesium methanesulfonate (CsMeSO4), 8 NaCl, 10 HEPES, 4 MgATP, 0.3 NaGTP, 0.5 ethylene glycol tetraacetic acid (EGTA) and 6 N-(2,6 dimethylphenylcarbamoylmethyl)-triethyl-ammonium bromide (QX314), with pH 7.3, using EPC8 amplifier (HEKA Elektronik). Currents were evoked by 500 ms depolarizing voltage pulses from holding potential −90 mV to 0 mV.
A cholinergic transmission was studied on a musculus sartorius neuromuscular preparation of the common frog Rana temporaria. The preparations were superfused with solution (in mM): 113 NaCl, 2.5 KCl, 0.6 CaCl2, 3 NaHCO3 and 4 MgCl2 (pH 7.3). Currents were recorded by a conventional two-electrode voltage clamp using an Axoclamp 2B (Axon Instr., USA) amplifier. The data were filtered at 2 kHz and stored on a computer.
Effects on (gamma-aminobutyric acid-ergic) GABAergic and glutamatergic transmission were studied on brain slices of male Wistar rats (20–22 postnatal days). Animals were killed by cervical dislocation and then were decapitated. Slices containing medial frontal (prelimbic) cortex were cut with a vibratome (Vibroslice 752M, Campden Instruments, UK) and incubated at room temperature for at least 1 h and then were transferred to a recording chamber perfused with artificial cerebrospinal fluid (ACSF) at 24–25 °C. Whole-cell recordings were made from layer 3 pyramidal neurons which were identified visually using infrared transmitted illumination from Axioscop microscope (Karl–Zeiss, Germany) equipped with differential interference contrast optics and Sanyo video camera (model VCB-3512P, Japan) for contrast enhancement. Pyramidal neurons were clamped at −80 mV during recording and synaptic responses were evoked with a monopolar glass electrode. For recording of eEPSC in rat brain neurons, the pipette solution contained (in mM): 114 K-gluconate, 6 KCl, 10 HEPES, 4 ATP-Mg and 0.3 GTP (pH was adjusted to 7.25 with KOH). To reverse and increase the amplitude of GABAA receptor-mediated evoked inhibitory post synaptic currents (eIPSCs) in postsynaptic cells at negative membrane potential, we used an internal solution of high chloride concentration, which contained (in mM): 120 KCl, 10 HEPES, 4 ATP-Mg and 0.3 GTP (pH was adjusted to 7.25 with KOH). The external solution contained (in mM) 143 NaCl, 5 KCl, 2.5 CaCl2, 2 MgCl2, 10 D-glucose, 10 HEPES (pH was adjusted to 7.4 with HCl). All recordings were made on an EPC8 amplifier (HEKA Elektronik, Germany) operated in a bridge-balance mode with capacitance neutralization. Signals were filtered with 5 kHz and digitized with a sampling frequency of 10 kHz for analysis.
Computation
Homological polypeptides search in Uniprot Protein Knowledgebase was done using BLASTp tool (BLOSUM-80 matrix). Offline computation, sequence alignments and E. coli expression codon optimization were performed with Lasergene® (DNAStar, USA) programs.
Results
Toxin isolation
Crude T. oblongus venom was selected as a source of insect-selective components because it had shown high toxicity to insects in a preliminary test (data not shown). For isolation of active compounds, we employed the proven tactic based on step-by-step separation by different chromatographic methods following biological activity trials of fractions21,37. Acute toxicity to insects was used as the test.
Measured activity was concentrated in polypeptide compounds that were separated from low and high molecular weight components by SEC (Fig. 1A). Rough separation by RP-HPLC (Fig. 1B) resulted in a selection of a moderately hydrophobic fraction that finally was separated into individual polypeptides (Fig. 1C). The purity of the most active polypeptide (ω-Tbo-IT1) obtained after third separation was confirmed by MALDI-MS that detected one component with molecular weight 4332.8 Da.
Multistage isolation of ω-Tbo-IT1 from crude venom by chromatoraphy.
(A) On a size-exclusion column TSK 2000SW column (Beckman, 7.5 × 600 mm) in 20 mM sodium phosphate (pH 4.5) with 150 mM NaCl buffer, at flow rate 0.5 ml/min; (B) on RP-HPLC column Jupiter C5 (Phenomenex, 4.6 × 250 mm) in 0.1% TFA containing buffers with a linear gradient of acetonitrile concentration at the flow rate 1 ml/min; (C) on Synergy Polar-RP400 column (Phenomenex, 4.6 × 250 mm) in 0.1% TFA containing buffers with a linear gradient of acetonitrile concentration at the flow rate 0.5 ml/min. Elution time of fractions toxic to insects are shadowed.
Primary structure determination
N-terminal Edman sequencing of the isolated polypeptide revealed first eleven residues to be: CASKNERCGNA. The mass difference between native and alkylated polypeptides prove an eight-cysteine content that is very common among spider toxins having ICK folding38,39. To complete structure elucidation, rapid cDNA ends amplification (RACE) with following sequencing approach was utilized.
As a result, it was deduced that ω-Tbo-IT1 precursor protein consisted of the signal peptide, the propeptide fragment and 41 residue-length mature sequence followed by a stop codon (Fig. 2A). The prepropeptide structure was in good accordance with spider toxin maturation principles40,41 and no other post-translation modifications were identified. Molecular weight difference for native toxin compared with deduced sequence was 0.3 Da and tryptic peptide masses were also in close agreement.
Primary structure features of the novel polypeptide toxin: (A) cDNA coded precursor protein sequence and derived protein structure of ω-Tbo-IT1, where residues equal to those defined by Edman degradation are underlined. (B) Alignment of mature ω-Tbo-IT1 sequence with three best BLASTp hits retrieved from UniProt databank. Residues similar to ω-Tbo-IT1 are shown by bold letters.
The BLASTp search for primary structure homologs for mature polypeptide sequence revealed some structural identity with three spider toxins (Fig. 2B). There was one insect sodium channel inhibitor μ-agatoxin II from funnel-web spider Agelenopsis aperta venom42 and there were two inhibitors of insect sodium channels inactivation, δ-palutoxin IT1 and δ-palutoxin IT4 from Pireneitega luctuosa spider venom43.
Recombinant analogues production
To produce a sufficient amount of ω-Tbo-IT1 for further analysis, a recombinant analog was synthesized in E. coli BL21(DE3) cells. Thioredoxin was chosen as a fusion partner for expression, as it had shown a good potency for enhancing polypeptide toxin expression earlier44,45,46. Thioredoxin–ω-Tbo-IT1 fusion protein production and purification were followed by CNBr cleavage to release target polypeptide finally purified by RP-HPLC. The yield of the purified recombinant polypeptide was very poor –0.46 mg/L of cell culture. The molecular weight of the recombinant product was equal to the natural one but with a difference referred to homoserine lactone on the C-terminus that is a common modification for CNBr treated polypeptides. The identity of recombinant analog by biological function to the natural ω-Tbo-IT1 was confirmed by biological tests both in acute toxicity on M. domestica larvae and in electrophysiological assays on C. vicina preparations.
Toxicity assay
Evaluative toxicity tests were performed for identification of the most toxic fractions during whole process of T. oblongus spider venom fractionation when small lyophilized part of each fractions was injected in order to give a rough estimate of its activity.
Crude venom toxicity to M. domestica larvae was estimated as LD50 10 μg/g for polypeptide fractions (separation steps one and two). Doses were a little bit higher for pure toxin ω-Tbo-IT1, which possessed toxic activity of about 15 μg/g (data not shown). More precise measurements were made for recombinant analogs (Fig. 3) that have demonstrated activity equal to those measured earlier for the native toxin. The LD50 dose calculated from this dose-dependent curve was approximately 950 ng/larvae, that is, equal to 19 μg/g or 4.4 nmol/g of body weight.
Crude T. oblongus venom injection displayed immediate paralysis approximately one minute after injection. After this time elapsed, larvae revealed brief physical activity of about thirty seconds in duration, followed by paralysis. This paralysis lasted for about 2 h and then if they had been subjected to a low dose of venom, larvae started responding weakly to stimuli; but they died at higher doses. The same effect was observed for active fraction and the pure toxin as well as for recombinant analogue. For low doses of recombinant toxin (less than LD50), time for paralysis to occur increased from 30 s to 2 min.
Paralyzing action towards cockroaches was demonstrated both on adult and juvenile specimens of G. portentosa. Dose 20 μg/g resulted in fast partial paralysis of adult cockroaches, which died 24 h after injection. Lower doses did not induce any paralysis or lethal indications while doses 20, 40 and 80 μg/g led to 50, 75 and 100% mortality during 24 h. So the activity of the toxin to both analyzed insects was similar.
Electrophysiology
Activity of native toxin was manifested as reversible inhibition of eEPSC in C. vicina neuromuscular junction (Fig. 4A). The inhibition was not accompanied by significant change of rise time (rt) or decay (τ) of the eEPSC (Fig. 4B). Parameters of spontaneous EPSC (sEPSC) were not affected by the toxin, suggesting that the effect was due to the quantum content decrease. Interestingly, frequency of sEPSC was also unaffected. Thus, the toxin strictly discriminated evoked and spontaneous release. Recombinant analog effect on neuro-muscular junction was identical to the natural toxin (data not shown). Stock solution of recombinant polypeptide was used for the concentration dependence analysis that resulted in the calculation of IC50 value 40 ± 10 nM and Hill coefficient 3.4 ± 0.3 (Fig. 4C,D).
Action of ω-Tbo IT1 on C. vicina neuro-muscular junction.
(A) Reversible time dependent inhibition of eEPSC (30 min washout presented) by 0.1 μM natural ω-Tbo-IT1. (B) Parameters chart for measured amplitude (A), rise time (rt), decay time constant (dt) and frequency of sEPSC (F) after 0.1 μM natural toxin application. Standard deviation are shown (n = 6). (C) Concentration dependent eEPSC amplitude decrease and (D) inhibition curve fitted by Hill equation evoked by the toxin application. Standard deviation are shown (n = 5). (E) Ca2+ concentration in external solution controls the eEPSC amplitude but not the sEPSC frequency (n = 6). (F) Co2+, the non-selective Ca2+ channel blocker, inhibits the eEPSC like ω-Tbo-IT1 (see panel A).
The revealed difference between the toxin effects on evoked and spontaneous neuromediator release suggests that presynaptic Ca2+ channels are the most obvious molecular target of the action. Indeed, for the classical blocker of these insect presynaptic calcium channels palutoxin-II (PLTX-II), similar effects were measured in an electrophysiological study on Drosophila melanogaster neuromuscular junctions47. To check the regulatory role of Ca2+ ions on the eEPSC amplitude and sEPSC frequency, we varied external Ca2+ concentration in our preparation. Decrease of Ca2+ concentration caused steep decrease of eEPSC amplitude with the apparent EC50 of 0.7 ± 0.2 mM and Hill coefficient 3.8 ± 0.2 (Fig. 4E). The Hill coefficient was similar to the one calculated for ω-Tbo-IT1 inhibition. On the contrary, the frequency of sEPSC was found insensitive to the Ca2+ concentration (Fig. 4E). To further confirm that toxin action mode may be due to inhibition presynaptic calcium channels, we studied the action of Co2+ ions, which is a non-selective Ca2+ channel blocker (Fig. 4F). 1 mM Co2+ caused strong inhibition of eEPSC amplitude with only minor action on the frequency and other characteristics of sEPSC. Thus, the effect of ω-Tbo-IT1, the effect of Ca2+ reduction and the effect of Co2+ are similar.
To directly demonstrate action of ω-Tbo-IT1 on presynaptic Ca2+ channels, we used neurons isolated from the abdominal ganglion of the cockroach P. americana. The Ca2+ currents were evoked in these neurons by depolarization from −90 to 0 mV. In all 28 neurons tested 50 μM Cd2+ caused strong and reversible current inhibition. In 16 cells the currents were completely insensitive to ω-Tbo-IT1 in 1 μM concentration (Fig. 5A). In the other cells Ca2+ currents demonstrated significant inhibition 48 ± 17%, (n = 12) (Fig. 5B). The inhibition was partially reversible during a 20 min period. Thus, among the different subtypes of Ca2+ channels expressed in cockroach neurons, there is a significant fraction, which is sensitive to ω-Tbo-IT1.
ω-Tbo IT1 action on Ca2+ channels in cockroach neurons.
Representative recordings of Ca2+ currents evoked by 500 ms membrane depolarization from –90 mV to 0 mV in control (gray) and 1 μM concentration of ω-Tbo-IT1. (A) Example of the toxin-sensitive current. (B) Example of the toxin-insensitive current, which however is inhibited by Cd2+.
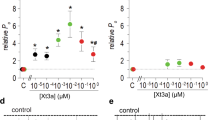
To analyze selectivity of ω-Tbo-IT1 and to reveal other possible targets, several additional preparations were used. In all preparations, a 1 μM concentration, which caused fast and complete inhibition of eEPSC on the fly neuromuscular synapse, was used. First, we studied ω-Tbo-IT1’s effect on Ca2+ channels in insect muscle. Fig. 6A demonstrates the lack of toxin effect on barium-mediated currents (n = 6), which were readily blocked by 50 μM Cd2+. Thus, a subtype of Ca2+ channels sensitive to ω-Tbo-IT1 has neuronal specificity.
Selectivity of ω-Tbo-IT1 action.
Two-electrode voltage clamp records in the fly muscle cell for the Ba2+ mediated currents evoked by voltage jumps from −90 mV to −20 mV (A) as well as synaptic currents in frog neuro-muscular junction (B) and both GABA-ergic (C) and glutamate-ergic transmission (D) in rat brain slices were not affected after application of ω-Tbo IT1 in 1 mM concentration. Currents inhibited by 50 mM Cd2+ and control currents (grey) are presented.
To determine the toxin’s selectivity for insects, we tested ω-Tbo-IT1 action on non-insect preparations such as frog neuromuscular junction (Fig. 6B) and rat brain glutamate-mediated (Fig. 6C) and GABA-mediated (Fig. 6D) neuronal synaptic transmission in the prefrontal cortex. To isolate (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) AMPA receptor-mediated component of EPSCs, all recordings were performed in the presence of GABAA receptor antagonist bicuculline methyl-bromide (10 μM) and (isopropyl-1-thio-β-D-galactopyranoside-) NMDA-receptor antagonist D-(-)-2-amino-5-phosphono-pentanoic acid (d-AP5, 50 μM). To isolate GABAA, receptor-mediated inhibitory post-synaptic current recordings were performed in the presence of AMPA-receptor antagonist 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX, 10 μM) and d-AP5 (50 μM). No significant action of the toxin was revealed in all these preparations (n = 6, for each preparation). In all cases 50 μM Cd2+ strongly inhibited the post-synaptic currents. Thus, we identified ω-Tbo-IT1 to be a specific inhibitor of insect presynaptic Ca2+ channels.
Spatial structure of ω-Tbo-IT1
Spatial structure of recombinant ω-Tbo-IT1 was studied by NMR spectroscopy in aqueous solution at pH 5.8. The set of 20 structures (Fig. 7A) was calculated using the following NMR experimental data: upper and lower NOE-based distance restraints, J coupling-based torsion angle restraints and hydrogen bond restraints (see a survey of the structural statistics in Table 1). The obtained structural ensemble (Fig. 7A) demonstrates no large distance violations, good Ramachandran statistics and quite low values of the root mean square deviation (RMSD) for backbone and heavy atoms (Table 1).
Spatial structure of ω-Tbo-IT1.
(A) Ensemble of 20 NMR-derived structures of ω-Tbo-IT1 after superposition of the backbone atoms of the “cystine knot” residues (regions 1–8, 20–27 and 38–40). The heavy side chain and backbone bonds are shown in black and gray, respectively. (B) Ribbon representation of the ω-Tbo-IT1 structure. Extended β-sheet and 310-helical turn of the major and small β-hairpin loops are highlighted in green and pink, respectively. The backbone and side-chain heavy bonds are shown in violet and cyan, respectively. The carbonyl, carboxyl, amide and disulfide bonds are additionally depicted in red, green and yellow, respectively. (C) Spatial structure alignment of the major (in green) and minor (in cyan) conformation of ω-Tbo-IT1 with ω-ACTX-Hv1a (in magenta). The arrows indicate the postulated twist and scissors-like mutual motions of the β-hairpin loops of ω-Tbo-IT1. The side-chains of the residues alternatively charged the tips of ω-Tbo-IT1 (positive R32/R32* and K33/K33* for major/minor conformations) and ω-ACTX-Hv1a (negative E26 and E28) are shown. In the bottom, sequence alignment for region of β-hairpin tip of two insect calcium channel inhibitors (ω-Tbo-IT1 and ω-ACTX-Hv1a) is presented. Positions of positively charged residues marked in the consensus as “+”, polar uncharged residues as “0” and hydrophobic–residues as “&”. Completely homologous residues are bold and shaded. Similar residues are only shaded.
The ω-Tbo-IT1 structure (Fig. 7B) involves two loops protruding from a “globular core” cross-linked by three disulfide bonds (Cys1-Cys21, Cys8-Cys25 and Cys20-Cys40) presenting the “cystine knot” arrangement (I–IV, II–V, III–VIII) typical for spider venom toxins with eight cysteines. The toxin is therefore concluded to assume the conventional ICK fold. The short β-hairpin loop (Gly9-Lys16) has on its tip a reverse turn (Ala11-Gly14) in the 310-helix conformation with the capping side chain to backbone (Asn10-Leu12) hydrogen bond. The major β-hairpin is formed by two extended β-strands (Lys24-Val30 and Gly34-Met41) having bulge regions (Thr29-Val30 and Gly34-Asn36) which are connected by an inverse γ-turn (Pro31-Lys33) on the hairpin tip and “stapled” by the additional disulfide bond (Cys27-Cys38) in the center. In addition to 4 disulfide bonds, the toxin structure is stabilized by 21 backbone-to-backbone hydrogen bonds.
Nevertheless, some heterogeneity exists in the toxin spatial structure, which is revealed by the disproportional peak doubling and resonance broadening observed in the NMR spectra (Supplementary Figs S1, S2 and S3) for some residues situated near the “cystine knot” disulfide bridges, the short β-hairpin loop and the bulge regions of the extended β-strands. Enhanced intramolecular mobility with slow (in the NMR time scale) conformational exchange between the major and minor similar forms can be caused by disulfide bridge isomerization or hydrogen bond switching. These would result in moderate mutual movements of the β-hairpin loops and alternative twisting of the β-sheet structure in the major β-hairpin loop, which are observed in the structural ensemble of ω-Tbo-IT1 (Fig. 7A,B). At the same time, the obtained structure set reveals also that the β-hairpin loops may be connected by the hydrogen bonding of the side chain of Asn36 with the backbone carbonyl group of Tyr13 and the side-chain hydroxyl group of Thr15 that would restrict the scissors-like mutual motions of the β-hairpin loops, in favor of the major conformation.
Discussion
Initially, the present work was aimed at finding new insect-selective toxins with acute toxicity toward pests. T. oblongus venom was chosen as a source based on the hunting features of this spider. The 41-residue toxin called ω-Tbo-IT1 was isolated and it demonstrated significant toxicity to insects and neutrality to vertebrates.
Primary structure and toxicity
The toxin ω-Tbo-IT1 has a classical ICK motif and demonstrates moderate sequence homology with the insect sodium channel inhibitors from the venom of funnel-web spider Agelenopsis aperta42 and Australian tangled nest spider Pireneitega luctuosa43. Following structural classification for spider venom peptide48, this new insect toxin belongs to structural class 2.1.1.0, for members of which classical distribution of cysteines is characteristic. Using another proposed classification49, a novel toxin may be named ω-philotoxin-To1a, which reflects biological action on Ca2+ channels and appurtenance of source spider to Philodromidae family.
Toxin primary structure is substantially different compared to other spider calcium toxin structures. As shown in Fig. 2B, three polypeptides share homology of less than 50% and the maximal disagreement is situated in the loop sequences located between Cys II-Cys III and Cys VI-Cys VII. Both regions are shorter in other ICK toxins under consideration; these regions were detected by NMR as regions of ω-Tbo-IT1 with intramolecular mobility (310-helix and tip of β-hairpin loop on Fig. 7B,C). This allows making the assumption about the unique evolutionary advantage of ω-Tbo-IT1 structure over other spider ICK family members. It may mean that twist- and scissors-like motions help ω-Tbo-IT1 adjust its binding site to the environment, so the toxin likely has dynamic rather than static epitope.
As noted above, the primary structure homology is not related to the biological properties of toxins. However, it was interesting to compare the acute toxicity measured in insects to the nearest structural homologs and for some of the most active insect sodium channel inhibitors from spider venom (Table 2). Presented data let us distinguish three groups. Extremely active toxins with LD50 in the picomolar range, active toxins−LD50 in few nanomoles (this group includes ω-Tbo-IT1) and one moderately active toxin, which has LD50 more than 10 nmol/g. Cellular target and mode of action as seen from the table do not correlate with insect toxicity.
The toxin’s dosage in insects is in good agreement with measured activity on neuro-muscular junction in C. vicina (IC50 value of 40 ± 10 nM). Low IC50 value could have set a new toxin in line with candidate base bioinsecticides; however, its measured dose is inferior to one measured for “extremely active” toxins. Further point mutagenesis should remove this limitation because spatial structure data produce a good opportunity for modeling.
Molecular target of action
Using different electrophysiology approaches, we demonstrated that ω-Tbo-IT1 acts as an inhibitor of insect Cav channels. Therefore, we compared ω-Tbo-IT1 with other calcium-channel inhibitors from spider venoms. More than 100 spider toxins acting on different calcium channels have already been described in scientific papers and deposited in the UniProt Database, (http://www.uniprot.org/). A few of them, however, were experimentally characterized in research papers as insect-selective calcium channel inhibitors. These are: ω-plectoxin-Pt1a (PLTX-II) from Plectreurys tristis47,50,51; ω-atracotoxin-Hv1a (ω-ACTX-Hv1a)10,11,13,52,53 and ω-atracotoxin-Hv2a (ω-ACTX-Hv2a) from Hadronyche versuta54; ω-atracotoxin-Ar1a (ω-ACTX-Ar1a) from Athrax robustus10; and huwentoxin-V (ω-TRTX-Hs2a)55,56.
Effect of ω-Tbo-IT1 demonstrates remarkable similarity with action of well-known toxin PLTX-II on Drosophila neuro-muscular junction47. Indeed, PLTX-II affected evoked, but not spontaneous, synaptic release. It was directly demonstrated that PLTX-II blocks Ca2+ currents in cultured embryonic Drosophila neurons50. More recent study suggests that PLTX-II selectively blocks the Drosophila Cav2 channel (Dmca1A)51. Like ω-Tbo-IT1 in our experiments, PLTX-II has no effect on Drosophila muscle and it is inactive on synaptic transmission at frog neuromuscular junctions50. Summarizing foregoing effects affinity we assume that ω-Tbo-IT1 shares the same biological target as PLTX-II – insect Cav2 channels.
In preparations of cockroach P. americana neurons we found that at least one type of Ca2+-currents in these neurons is strongly inhibited by ω-Tbo-IT1. It is known that four different kinds of Ca2+ currents were distinguished on the basis of their kinetics, voltage range of activation and pharmacological profile and can be evoked in dorsal unpaired median neurons isolated from abdominal ganglia of the American cockroach P. americana57. These currents are likely mediated by different types (or isoforms) of Cav channels (Cav1, Cav2 and Cav3). Presumably the Cav2 subtype of insect Ca2+ channels is sensitive to the toxin application.
The closest vertebrate analog of Cav2 channels are N-type channels responsible for synaptic release of neurotransmitters and so inhibitors of Cav2 insect channel can potentially affect presynaptic N-type vertebrate channels. Therefore, action of ω-Tbo-IT1 was tested in several vertebrate synaptic preparations. Our electrophysiological experiments have demonstrated that the action was highly selective. This is not very surprising since sequence similarity between vertebrate and invertebrate channels is limited and the channels often differ by their sensitivity to pharmacological agents58. For instance, ω-ACTX-Hv1a strongly affects insect Cav1 currents but has no effect on calcium currents in rat trigeminal neurons and rat Cav1.2, Cav2.1 and Cav2.2 HVA channels59.
Certainly, we cannot exclude that ω-Tbo-IT1 can influence other channels. For instance, its action on sodium channels was not studied systematically.
Structural comparison to ICK toxins
Among insect-selective calcium channel inhibitors from spider venoms, only for ω-atracotoxin-Hv1a and ω-atracotoxin-Hv2a spatial structures were identified52,54. In the most general terms, all three toxins resemble each other. They contain a globular hydrophobic core and a С-terminal tail that protrudes from it. ω-ACTX-Hv1a and ω-ACTX-Hv2a both contain three N-terminal residues extending beyond their central core that are very dynamic in solution52,54. In contrast, ω-Tbo-IT1 starts straight from the hydrophobic core. We think that short flexible N-terminal regions in ω-ACTX-Hv1a and ω-ACTX-Hv2a hardly influence activity of toxins.
The C-terminal region of ω-ACTX-Hv1a is a finger-like β-hairpin that is formed by residues 22–3752. It doesn’t share primary structure homology to the same region of ω-Tbo-IT1 but is identical to it by backbone fold (Fig. 7C). Two anti-parallel β-strands in the hairpin are stabilized not only by hydrogen bonds but also one disulfide bond, in the case of ω-Tbo-IT1.
Despite a good alignment of the structures of ω-Tbo-IT1 and ω-ACTX-Hv1a, the major conformation of ω-Tbo-IT1 has an inverse direction of the polypeptide chain in the tip of the major β-hairpin loop due to different local structure of the bulge regions of the β-strands. Nevertheless, the ω-Tbo-IT1 minor conformation’s twisted major β-hairpin loop and β-hairpin loop of ω-ACTX-Hv1a are comparable. The β-hairpin in ω-Tbo-IT1 core has an unusual link between two strands, which is not a short β-turn (as usual in ICK-toxins), but γ-turn instead. Notably, their tips are alternatively charged, ω-Tbo-IT1 has two positive residues Arg32 and Lys33, whereas in contrast ω-ACTX-Hv1a has negative residues Glu26 and Glu28.
For ω-ACTX-Hv1a it was shown experimentally that β-hairpin is indispensable for insecticidal activity. Thus, a β-hairpinless mutant of this toxin is devoid of insecticidal activity60 and two residues Asn27 and Arg35 together with Pro10 are essential for activity59. For ω-ACTX-Hv2a it was experimentally revealed by truncation that C-terminal β-hairpin is also indispensable for an activity like the one in ω-ACTX-Hv1a54. By analogy we assume that the following residues in ω-Tbo-IT1 may be involved in binding: Asn10, Arg32 and/or Lys33.
As it was mentioned above, the small β-hairpin loop with 310-helical turn is absent in ω-ACTX-Hv1a since its Cys II-Cys III interval is shorter by 6 residues. Our hypothesis is that the short β-hairpin helps major C-terminal β-hairpin during the process of interaction of ω-Tbo-IT1 with the channel. This speculation might be fortified by some NMR signal doubling and broadening (Supplementary Figs S1 and S2) we observed (e.g. for Thr15 and Thr29 from short β-hairpin and major β-hairpin, correspondingly). Therefore, we observed minor cross peak signals, which may imply simultaneous movements of two hairpins (these movements are shown by arrows in Fig. 7C). Moreover, we hypothesize here that twist- and scissors-like motions might be coordinated, because we observed formation of additional hydrogen bonds, namely those between residues Asn36-Tyr13 and Asn36-Thr15 correspondingly. Such formation would be impossible if motions under consideration could have been non-simultaneous. Postulated β-hairpin loop movements in ω-Tbo-IT1 may serve for searching the proper binding site in the target calcium channel.
Conclusion
In summary, work discussed describes structural and electrophysiological characterization of novel inhibitor of insect calcium channels (presumably, insect Cav2 type). This inhibitor is a toxin, by primary structure related to some sodium channel inhibitors, but its target is neuronal insect voltage-gated Ca2+ channels. We suspect that the interaction of ω-Tbo-IT1 with channels is mediated by twist and scissors-like motions of two β-hairpin loops. Loop movement results in distance variation between the ‘hot spots’ of the toxin-receptor binding and facilitate interaction. Such ‘dynamic epitope’ may be regarded as an evolutionary step forward from the static epitope.
Additional Information
How to cite this article: Mikov, A. N. et al. ω-Tbo-IT1 - New Inhibitor of Insect Calcium Channels Isolated from Spider Venom. Sci. Rep. 5, 17232; doi: 10.1038/srep17232 (2015).
References
Escoubas, P. & King, G. F. Venomics as a drug discovery platform. Expert Rev Proteomics 6, 221–224 (2009).
Escoubas, P., Quinton, L. & Nicholson, G. M. Venomics: unravelling the complexity of animal venoms with mass spectrometry. J Mass Spectrom 43, 279–295 (2008).
Escoubas, P., Sollod, B. & King, G. F. Venom landscapes: mining the complexity of spider venoms via a combined cDNA and mass spectrometric approach. Toxicon 47, 650–663 (2006).
Favreau, P. et al. Mass spectrometry strategies for venom mapping and peptide sequencing from crude venoms: case applications with single arthropod specimen. Toxicon 47, 676–687 (2006).
Jiang, L. et al. Transcriptome analysis of venom glands from a single fishing spider Dolomedes mizhoanus. Toxicon 73, 23–32 (2013).
Liang, S. Proteome and peptidome profiling of spider venoms. Expert Rev Proteomics 5, 731–746 (2008).
Baconguis, I. & Gouaux, E. Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature 489, 400–405 (2012).
Bohlen, C. J. et al. A bivalent tarantula toxin activates the capsaicin receptor, TRPV1, by targeting the outer pore domain. Cell 141, 834–845 (2010).
Bosmans, F. & Swartz, K. J. Targeting voltage sensors in sodium channels with spider toxins. Trends Pharmacol Sci 31, 175–182 (2010).
Chong, Y. et al. The omega-atracotoxins: selective blockers of insect M-LVA and HVA calcium channels. Biochem Pharmacol 74, 623–638 (2007).
Klint, J. K. et al. Spider-venom peptides that target voltage-gated sodium channels: pharmacological tools and potential therapeutic leads. Toxicon 60, 478–491 (2012).
Lee, S. et al. Solution structure of GxTX-1E, a high-affinity tarantula toxin interacting with voltage sensors in Kv2.1 potassium channels. Biochemistry 49, 5134–5142 (2010).
Nicholson, G. M. Insect-selective spider toxins targeting voltage-gated sodium channels. Toxicon 49, 490–512 (2007).
Sousa, S. R., Vetter, I. & Lewis, R. J. Venom peptides as a rich source of cav2.2 channel blockers. Toxins (Basel) 5, 286–314 (2013).
Vassilevski, A. A., Kozlov, S. A. & Grishin, E. V. Molecular diversity of spider venom. Biochem. 74, 1505–1534 (2009).
King, G. F. & Hardy, M. C. Spider-venom peptides: structure, pharmacology and potential for control of insect pests. Annu Rev Entomol 58, 475–496 (2012).
Herzig, V. et al. ArachnoServer 2.0, an updated online resource for spider toxin sequences and structures. Nucleic Acids Res. 39, D653–7 (2011).
Kozlov, S. A. et al. Comprehensive analysis of the venom gland transcriptome of the spider Dolomedes fimbriatus. Sci. Data 1, 140023 (2014).
Hardy, M. C., Daly, N. L., Mobli, M., Morales, R. A. & King, G. F. Isolation of an orally active insecticidal toxin from the venom of an Australian tarantula. PLoS One 8, e73136 (2013).
Vassilevski, A. A. et al. Cyto-insectotoxins, a novel class of cytolytic and insecticidal peptides from spider venom. Biochem J 411, 687–696 (2008).
Lipkin, A. et al. Novel insecticidal toxins from the venom of the spider Segestria florentina. Toxicon 40, 125–30 (2002).
Windley, M. J. et al. Spider-venom peptides as bioinsecticides. Toxins (Basel) 4, 191–227 (2012).
Fang, W. et al. Development of transgenic fungi that kill human malaria parasites in mosquitoes. Science 331, 1074–1077 (2011).
Ch’ng, J. H. et al. A whole cell pathway screen reveals seven novel chemosensitizers to combat chloroquine resistant malaria. Sci Rep 3, 1734 (2013).
Van den Berg, A. & Dippenaar-Schoeman, A. S. A revision of the Afrotropical species of the genus Tibellus Simon (Araneae: Philodromidae). Koedoe 37, 67–114 (1994).
Rowan, D. H. & Barret, P. D. N. H. Identifying spiders through DNA barcodes. Can J. Zool. 83, 481–491 (2005).
Vassilevski, A. A., Kozlov, S. A., Egorov, T. A. & Grishin, E. V. Purification and characterization of biologically active peptides from spider venoms. Methods Mol Biol 615, 87–100 (2010).
Cavanagh, J., Fairbrother, W. J., Palmer, A. G., Rance, M. & Skelton, N. J. Protein NMR Spectroscopy. Protein NMR Spectroscopy (Elsevier, 2007). 10.1016/B978-012164491-8/50008-7
Piotto, M., Saudek, V. & Sklenár, V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2, 661–5 (1992).
Wüthrich, K. NMR of Proteins and Nucleic Acids. 14–105 (Wiley-Interscience, 1986).
Keller, R. L. J. The Computer Aided Resonance Assignment Tutorial. 9–58 (CANTINA Verlag, 2004).
Delaglio, F., Wu, Z. & Bax, A. Measurement of homonuclear proton couplings from regular 2D COSY spectra. J. Magn. Reson. 149, 276–81 (2001).
Güntert, P. Automated NMR protein structure calculation. Prog. Nucl. Magn. Reson. Spectrosc. 43, 105–125 (2003).
Koradi, R., Billeter, M. & Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55 (1996).
Andreev, Y. A., Kozlov, S. A., Vassilevski, A. A. & Grishin, E. V. Cyanogen bromide cleavage of proteins in salt and buffer solutions. Anal. Biochem. 407, 144–146 (2010).
Vorobjev, V. S. Vibrodissociation of sliced mammalian nervous tissue. J. Neurosci. Methods 38, 145–50 (1991).
Dubovskii, P. V. et al. Novel lynx spider toxin shares common molecular architecture with defense peptides from frog skin. FEBS J 278, 4382–4393 (2011).
Norton, R. S. & Pallaghy, P. K. The cystine knot structure of ion channel toxins and related polypeptides. Toxicon 36, 1573–1583 (1998).
Craik, D. J., Daly, N. L. & Waine, C. The cystine knot motif in toxins and implications for drug design. Toxicon 39, 43–60 (2001).
Kozlov, S. A., Vassilevski, A. A. & Grishin, E. V. In Protein biosynthesis (eds. Esterhouse, T. E. & Petrinos, L. B. ) 225–248 (Nova Biomedical Books, 2009).
Kozlov, S. A. & Grishin, E. V. The universal algorithm of maturation for secretory and excretory protein precursors. Toxicon 49, 721–726 (2007).
Adams, M. E., Bindokas, V. P., Hasegawa, L. & Venema, V. J. Omega-agatoxins: novel calcium channel antagonists of two subtypes from funnel web spider (Agelenopsis aperta) venom. J Biol Chem 265, 861–867 (1990).
Corzo, G. et al. Isolation, synthesis and pharmacological characterization of delta-palutoxins IT, novel insecticidal toxins from the spider Paracoelotes luctuosus (Amaurobiidae). Eur J Biochem 267, 5783–5795 (2000).
Osmakov, D. I. et al. Sea anemone peptide with uncommon β-hairpin structure inhibits acid-sensing ion channel 3 (ASIC3) and reveals analgesic activity. J. Biol. Chem. 288, 23116–27 (2013).
Andreev, Y. A. et al. Analgesic compound from sea anemone Heteractis crispa is the first polypeptide inhibitor of vanilloid receptor 1 (TRPV1). J. Biol. Chem. 283, 23914–21 (2008).
Shlyapnikov, Y. M., Andreev, Y. A., Kozlov, S. A., Vassilevski, A. A. & Grishin, E. V. Bacterial production of latarcin 2a, a potent antimicrobial peptide from spider venom. Protein Expr. Purif. 60, 89–95 (2008).
Branton, W. D., Kolton, L., Jan, Y. N. & Jan, L. Y. Neurotoxins from Plectreurys spider venom are potent presynaptic blockers in Drosophila. J. Neurosci. 7, 4195–200 (1987).
Kozlov, S. & Grishin, E. Classification of spider neurotoxins using structural motifs by primary structure features. Single residue distribution analysis and pattern analysis techniques. Toxicon 46, 672–86 (2005).
King, G. F., Gentz, M. C., Escoubas, P. & Nicholson, G. M. A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon 52, 264–76 (2008).
Leung, H. T., Branton, W. D., Phillips, H. S., Jan, L. & Byerly, L. Spider toxins selectively block calcium currents in Drosophila. Neuron 3, 767–72 (1989).
Kuromi, H., Honda, A. & Kidokoro, Y. Ca2+ influx through distinct routes controls exocytosis and endocytosis at drosophila presynaptic terminals. Neuron 41, 101–11 (2004).
Fletcher, J. I. et al. The structure of a novel insecticidal neurotoxin, omega-atracotoxin-HV1, from the venom of an Australian funnel web spider. Nat Struct Biol 4, 559–566 (1997).
Bloomquist, J. R. Mode of action of atracotoxin at central and peripheral synapses of insects. Invert. Neurosci. 5, 45–50 (2003).
Wang, X. H. et al. Discovery and structure of a potent and highly specific blocker of insect calcium channels. J Biol Chem 276, 40306–40312 (2001).
Zhang, P.-F., Chen, P., Hu, W.-J. & Liang, S.-P. Huwentoxin-V, a novel insecticidal peptide toxin from the spider Selenocosmia huwena and a natural mutant of the toxin: indicates the key amino acid residues related to the biological activity. Toxicon 42, 15–20 (2003).
Deng, M., Luo, X., Meng, E., Xiao, Y. & Liang, S. Inhibition of insect calcium channels by huwentoxin-V, a neurotoxin from Chinese tarantula Ornithoctonus huwena venom. Eur. J. Pharmacol. 582, 12–16 (2008).
Grolleau, F. & Lapied, B. Dorsal unpaired median neurones in the insect central nervous system: towards a better understanding of the ionic mechanisms underlying spontaneous electrical activity. J. Exp. Biol. 203, 1633–48 (2000).
King, G. F. Modulation of insect Ca(v) channels by peptidic spider toxins. Toxicon 49, 513–30 (2007).
Tedford, H. W. et al. Scanning mutagenesis of omega-atracotoxin-Hv1a reveals a spatially restricted epitope that confers selective activity against insect calcium channels. J. Biol. Chem. 279, 44133–40 (2004).
Tedford, H. W., Fletcher, J. I. & King, G. F. Functional significance of the beta hairpin in the insecticidal neurotoxin omega-atracotoxin-Hv1a. J. Biol. Chem. 276(28), 26568–76 (2001).
Skinner, W. S. et al. Purification and characterization of two classes of neurotoxins from the funnel web spider, Agelenopsis aperta. J. Biol. Chem. 264, 2150–5 (1989).
Figueiredo, S. G. et al. Purification and amino acid sequence of the insecticidal neurotoxin Tx4(6-1) from the venom of the ‘armed’ spider Phoneutria nigriventer (Keys). Toxicon 33, 83–93 (1995).
Acknowledgements
Authors show gratitude to Prof. Magazanik L.G. from I.M. Sechenov Institute of Evolutionary Physiology and Biochemistry for consulting in experiment organization and to Ivanov I.A. and Kost V.Yu. from Shemiakin-Ovchinnikov Institute for stimulating discussions. SK is grateful to Russian scientific foundation (grant No 14-24-00118) for financial support. Other authors want to thank the Program of Presidium of RAS «Molecular and Cell Biology» and the Grant of the President of the Russian Federation for (NSh-1924.2014.4).
Author information
Authors and Affiliations
Contributions
A.N.M., I.M.F., N.N.P., E.E.M., A.V.Z., K.K.K. and E.V.B. performed experiments; A.N.M., T.N.B., D.A.A., A.V.L., E.V.B., D.B.T. and Y.A.A. analyzed data; Y.A.A. gave critical evaluation of experimental results; A.N.M., E.V.B., D.B.T. and S.A.K. wrote the manuscript; A.V.L., S.A.K. and E.V.G. supervised research. All of the authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Mikov, A., Fedorova, I., Potapieva, N. et al. ω-Tbo-IT1–New Inhibitor of Insect Calcium Channels Isolated from Spider Venom. Sci Rep 5, 17232 (2015). https://doi.org/10.1038/srep17232
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep17232
This article is cited by
-
Venom-gland transcriptomics and venom proteomics of the Tibellus oblongus spider
Scientific Data (2023)
-
Lidocaine and carbamazepine inhibit while phenytoin and lamotrigine paradoxically enhance the insect neuromuscular transmission
Invertebrate Neuroscience (2019)
-
Identification of unusual peptides with new Cys frameworks in the venom of the cold-water sea anemone Cnidopus japonicus
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.