Abstract
Ulcerative colitis (UC) is a common inflammatory bowel disease (IBD) producingintestinal inflammation and tissue damage. The precise aetiology of UC remainsunknown. In this study, we applied a rank-based expression profile comparativealgorithm, gene set enrichment analysis (GSEA), to evaluate the expression profilesof UC patients and small interfering RNA (siRNA)-perturbed cells to predict proteinsthat might be essential in UC from publicly available expression profiles. We usedquantitative PCR (qPCR) to characterize the expression levels of those genespredicted to be the most important for UC in dextran sodium sulphate (DSS)-inducedcolitic mice. We found that bromo-adjacent homology domain (BAHD1), a novelheterochromatinization factor in vertebrates, was the most downregulated gene. Wefurther validated a potential role of BAHD1 as a regulatory factor for inflammationthrough the TNF signalling pathway in vitro. Our findings indicate thatcomputational approaches leveraging public gene expression data can be used to inferpotential genes or proteins for diseases and BAHD1 might act as an indispensablefactor in regulating the cellular inflammatory response in UC.
Similar content being viewed by others
Introduction
Ulcerative colitis (UC) and Crohn’s disease (CD), the two main subtypes ofinflammatory bowel disease (IBD), are immunologically mediated, idiopathic, chronic andrelapsing diseases1, the aetiology of which remains unclear. From thelatest Asian epidemiological investigation, the incidence of IBD, especially UC, isrising in parallel with the rapid socioeconomic development and westernization oflifestyle during the past two decades2,3.
Intestine epithelial cells (IECs), a layer of which forms a physical barrier separatingsubepithelial mucosal immune cells from a variety of antigenic substances present in theintestinal lumen, play a pivotal role in maintaining a balanced intestinalmicroenvironment4. Disruption or dysfunction of the intestinalbarrier promotes contact between immune cells and antigens and the excessive cytokinesproduced are involved in the pathogenesis of UC, causing mucosal tissue destruction andleading to bloody diarrhoea5. The inflammatory responses that mediateinflammatory signalling are driven by classical stimulus-regulated transcriptionfactors, including nuclear factor κ B (NF-κ B), activatorprotein-1(AP-1), IFN regulatory factors (IRFs)6 and mitogen-activatedprotein kinases (MAPKs)7.
Gene expression microarrays, which are frequently and widely applied in clinical studiesof human diseases, enable the measurement of genome-wide expression8.This method has been applied to investigate transcriptional signatures present ingastrointestinal tissue obtained from CD and UC patients for more than 10 years9. However, the majority of the genes identified by this method do notnecessarily play critical roles in the biological processes under investigation. Forthis reason, we adopted a systematic computational in silico approach to predict novelgenes or proteins on the basis of comprehensive testing of molecular signatures insiRNA-disease pairs: a pattern-matching strategy based on GSEA to more precisely selectgenes to study. GSEA is an analytical method that uses gene sets representing differentbiological processes to interpret gene expression data, producing a score that measuresthe similarity between two different processes by comparing their expressionprofiles10. This method has been used in drug discovery with genesets responsible for diseases to interpret the expression data from different drugs11,12.
In this study, we estimated the similarity between public expression data derived from UCpatients and that from siRNA perturbed cells by applying the parameter‘insensitive’ and the GSEA systematic algorithm13to estimate the distance between UC and different siRNA perturbations. We also verifiedthe predicted gene expression changes in a mouse model of acute DSS-induced colitis, acanonical IBD model. Our results revealed that the protein BAHD1 is downregulated themost in the colon tissue of the mouse model. As it has not been previously described tohave efficacy for UC or any related disorder of inflammation in the gastrointestinaltract, we evaluated the efficacy of BAHD1 in UC and explored its effect on the humanepithelial colorectal adenocarcinoma cell line Caco-2 exposed to inflammatory mediatorsand its related molecular mechanism.
Results
Computational Prediction and Assessment of a Novel Target in UC:BAHD1
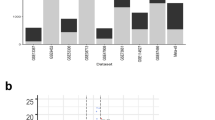
We calculated the distance values between UC and siRNA perturbation expressionprofiles based on GSEA. The siRNA perturbation expression profiles representedcell responses to the silencing of as many as 106 different genes (see Methods).By examining the relationships between UC and the 106 siRNAs based on predicteddistance scores, we identified major clusters of the entire data set [Fig. 1]. A low distance value indicates a similar generegulation tendency and the distance values between host responses to UC andthose to siRNA perturbations are shown in Supplementary Table S1. We picked out the top five genes whose singlesiRNA perturbations had the lowest distance values relative to UC, namely, EZH2,UPF1, FOXM1, NUDT6, BAHD1 (distance value = 0.868,0.878, 0.883, 0.885, 0.891, respectively) and we investigated the mRNA levelsof these most likely candidate genes in the DSS-induced colitis mouse model byqPCR. Among them, BAHD1 was found to be the most downregulated in the mousemodel compared with the control group [Fig. 2A],suggesting that this protein might be essential in this inflammatory boweldisease. An enrichment plot showing the enrichment score (ES) for the gene listrank for UC and BAHD1 is shown in Fig. 2B.
(A) qPCR analysis of the five most likely causative genes in the mouseUC model, namely, EZH2, UPF1, FOXM1, NUDT6 and BAHD1. Bars indicate themean ± SEM;n = 3 per group;*P < 0.05;**P < 0.01. (B) Enrichment plot for UCand BAHD1. The enrichment plot presents the hit position information ofsignature genes in a reference PRL. The size of signature genes is n, andPRL length is N. The function value goes up by 1/n at the hit positions andgoes down by 1/(N-n) at the miss hit positions. The line coloured cyanrepresents the running enrichment score plot of BAHD1 upregulated signaturegenes in UC’s PRL and the final enrichment score (i.e., maximumdeviation of running enrichment score from zero) is 0.0557( ). The line coloured yellow represents the runningenrichment score plot of BAHD1 downregulated signature genes inUC’s PRL and the final enrichment score is−0.0706(
). The line coloured yellow represents the runningenrichment score plot of BAHD1 downregulated signature genes inUC’s PRL and the final enrichment score is−0.0706( ). The enrichmentscore of BAHD1’s signature in UC’s PRL is
). The enrichmentscore of BAHD1’s signature in UC’s PRL is . The line coloured blue represents therunning enrichment score plot of UC upregulated signature genes inBAHD1’s PRL and the final enrichment score is0.1738(
. The line coloured blue represents therunning enrichment score plot of UC upregulated signature genes inBAHD1’s PRL and the final enrichment score is0.1738( ). The line coloured greenrepresents the running enrichment score plot of UC downregulated signaturegenes in BAHD1’s PRL and the final enrichment score is–0.1331(
). The line coloured greenrepresents the running enrichment score plot of UC downregulated signaturegenes in BAHD1’s PRL and the final enrichment score is–0.1331( ). The enrichmentscore of UC’s signature in BAHD1’s PRL is
). The enrichmentscore of UC’s signature in BAHD1’s PRL is . We can infer that the top- (orbottom-) ranked genes in UC are strongly positively (or negatively)regulated in BAHD1, resulting in a high enrichment score forUC’s signature in BAHD1’s PRL.
. We can infer that the top- (orbottom-) ranked genes in UC are strongly positively (or negatively)regulated in BAHD1, resulting in a high enrichment score forUC’s signature in BAHD1’s PRL.
Decreased BAHD1 Expression in in vivo and in vitro Modelsand UC Patients
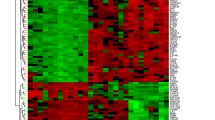
To characterize BAHD1 expression in colon tissue, intestinal samples from healthyhumans were evaluated by using immunohistochemistry (IHC). As shown in Fig. 3A, BAHD1 protein was expressed normally in the healthyhuman large intestine, including crypt and epithelial cells, mainly in thenucleus. As for the protein’s expression in colitis, we found thatBAHD1 was significantly decreased in IECs and crypt cells in the large intestineof UC patients compared with control patients who had no history of intestinalinflammation [Fig. 3B], indicating that dysregulatedexpression of BAHD1 in the intestine may be associated with regions of activedisease in UC. Western blotting using in vivo and vitro models supportedthis inference: Caco-2 cells showed a significantly reduced level of BAHD1protein in a cell model, in which Caco-2 cells were exposed to inductive factorsfor 24 h (see Methods). A similar observation was made in the mouse model ofacute colitis (see Methods) in comparison with the control group [Fig. 3C]. Therefore, we next explored BAHD1’s functionsand unveiled its possible molecular mechanisms in the cell model, which mightgive some hints regarding the development of UC.
Reduced BAHD1 expression in in vivo and in vitro models and inUC patients.
(A) Nuclear localization of BAHD1 in normal human large intestine.Histological sections of colon samples taken from healthy adjacent ordistant colon from subjects of certain human cancers were stained for BAHD1.Magnification of 10*10 (upper) and 10*40 (lower) show that BAHD1 (brownstaining) is universally present in IECs and crypt cells in the largeintestine. (B) IHC for BAHD1 in UC patient colonic tissue (lower)compared with the control group (upper), 10*20 magnification. (C)Western blot analysis of BAHD1 expression in vitro and invivo. The blots shown represent at least three independent experiments;GAPDH was used as a loading control.
Associated Inflammatory Mediators were Enhanced in BAHD1-deficient Caco-2Cell Model
To explore the relationship between our predicted protein BAHD1 and the responsesof IECs in an inflammatory microenvironment, we used the Caco-2 cell lineexposed to several inflammatory mediators to establish a cell model to mimic gutinflammation for IECs (see Methods). Caco-2 cells were treated with siBAHD1 for48 h before 24 h of exposure. The cells were collectedfor mRNA extraction and the cell culture supernatant was reserved to measurecytokine secretion. Notably, the cell model pre-treated with siBAHD1 displayedincreased mRNA and protein expression of associated cytokines, includingpro-inflammatory cytokines such as TNF-α, IL-6, IL-1β,IFN-β and IFN-γ [Fig. 4A,B] andchemokines such as IL-8, CCL3, CCL4, CCL5, CX3CL1 and CXCL10 [Fig. 4B,C]. In addition, certain cell adhesion molecules, includingimmunoglobulin superfamily intercellular adhesion molecule 1 (ICAM-1) andvascular cell adhesion molecule 1 (VCAM-1), displayed similar increases in thesiBAHD1 group [Fig. 4D] and they are important factorsmediating leukocyte migration and local inflammation in IBD14.However, although most cytokines showed a marked increase in the Caco-2 cellmodel and even in the siBAHD1 group without any stimulus (like IFN-βand CX3CL1), with decreased expression of BAHD1, several chemokines such asCXCL3 and CXCL5 did not display a similar trend [Fig. 4E].As for other types of inflammatory mediators, cyclooxygenase-2 (COX-2), anenzyme that catalyses the inflammatory response factor prostaglandin, had higherexpression in the cell model pre-treated with siBAHD1, as did two isoforms ofnitric oxide (NO) synthases, inducible NOS (iNOS) and endothelial NOS (eNOS)[Fig. 4F].
Associated inflammatory mediators were enhanced by BAHD1 deficiency invitro.
Four groups are involved here (NC = negative controlsiRNA-transfected group; NC+Mix = Caco-2 cell modelgroup; siBAHD1 = siBAHD1-transfected group;siBAHD1 + Mix = Caco-2 cellmodel pre-treated with siBAHD1). (A) Effects of BAHD1 repression onthe expression of proinflammatory factors (TNF-α, IL-6,IL-1β, IFN-β and IFN-γ) in the Caco-2cell model. (B) Detection of IL-6, IL-8 and MCP-1 secretion in cellculture supernatant by ELISA, the siBAHD1 group is reported as the foldincrease compared with the simple cell model. (C) BAHD1 inhibition inthe cell model influenced the mRNA levels of chemokines such as IL-8, CCL3,CCL4, CCL5, CX3CL1 and CXCL10. (D) Expression of the cell adhesionmolecules ICAM-1 and VCAM-1 displayed similar increasing trends in the cellmodel with siBAHD1 interference. (E) The expression of CXCL3 andCXCL5 did not show the predicted trend at the mRNA level. (F) COX-2and NO synthases, including iNOS and eNOS, showed higher expression in thecell model pre-treated with siBAHD1. All data above are shown as themean ± SEM from three independentmeasurements. Statistical significance was determined byStudent’s t-test(*P < 0.05;**P < 0.01). The western blots shown arerepresentative of at least three independent experiments.
Activation of Ikappa B (IκB) Kinase (IKK)/NF-κ Band JNK/AP-1 Pathways in the siBAHD1-treated Caco-2 Cell model
In response to stimulation by the various factors described above, activitythrough two important inflammation pathways, the IKK/NF-κ B andJNK/AP-1 pathways, was measured by using relevant key phospho-specificantibodies to investigate the underlying molecular mechanism of BAHD1 deficiencyin the cell model. Activation was comparable in the stimulation only group andthe siBAHD1 group. In vitro, siBAHD1 in Caco-2 cells resulted in strongerphosphorylation of IKK α/β, IκBαand NF-κ B subunit p65 in the IKK/NF-κ B pathway.Moreover, the AP-1 protein c-JUN and upstream regulator JNK showed higherphosphorylation levels in the siRNA-treated group [Fig.5A]. Regarding other MAPK pathways active during the acute phase in cellinjury, we did not observe activation of either the ERK1/2 pathway or the p38pathway in the siBAHD1 group [Fig. 5B]. By contrast, STAT3phosphorylation was clearly increased, indicating that there might someconnection between BAHD1 and the Janus kinase/signal transducer and activator oftranscription (JAK/STAT) pathway [Fig. 5B]. These resultswere in accordance with the enhanced levels of cytokines in the siRNA-treatedgroup, indicating that BAHD1 might be associated with cytokine expression inintestinal cells through a certain inflammatory pathway.
Activation of the IKK/NF-κ B and JNK/AP-1 pathways, but not theERK1/2 and p38 pathways, in siBAHD1-treated Caco-2 cells.
Caco-2 cells were treated with negative control siRNA or BAHD1 siRNA. At 48h, they were exposed to the stimulating mixture (inflammatory mediatorsincluding TNF-α, IFN-γ, IL-1β and LPSdescribed before) for 5 minutes (NC+Mix group, siBAHD1+Mixgroup) or not (NC group, siBAHD1 group). (A) The siBAHD1 group showedstronger activation of key proteins in the NF-κ B (the level ofp-IKK α/β, p-IκBα, p-p65increased) and JNK/AP-1 (phosphorylation level of JNK and c-JUN enhanced)pathways. (B) The phosphorylation level of STAT3 showed the sametrend as in (A). Other MAPK pathways such as P38 and ERK1/2 did notshow a significant difference between the NC and siBAHD1 groups in theinflammatory environment. Representative western blots of cell lysates areshown from at least three independent experiments.
An NF-κ B inhibitor Blocked the Effects caused by BAHD1Repression in Caco-2 Cell model
We had found that IKK and IκBα were activated in the cellmodel system; therefore, we wondered whether its downstream target,NF-κ B, participated in the regulation of cytokine gene expression.To confirm that the NF-κ B pathway was involved in the regulation byBAHD1 of inflammatory gene expression in the cell model, we used anNF-κ B inhibitor, parthenolide (PTN, 30 μm),which is a sesquiterpene lactone that inhibits activation of theNF-κ B pathway15. It significantly inhibited thestimulus-induced activation of the NF-κ B pathway and repressed theexpression of associated inflammatory genes [Fig. 6A,C].After treatment with siRNA for 48 h, Caco-2 cells were incubatedwith PTN for 1 hour before the addition of the mixture of variousstimulators described above. The phosphorylation levels ofIKKα/β, IκBα andNF-κ B p65 were reduced significantly in both the NC and siBAHD1groups [Fig. 6B], which was consistent with a decrease incytokines such as TNF-α, IL-6, IL-8, CCL3 [Fig.6C]. Therefore, the results suggested that cytokines were highlyactivated mainly through the NF-κ B pathway.
An NF-κ B inhibitor blocked the effects caused by BAHD1repression in Caco-2 cells.
Caco-2 cells were pre-treated with PTN before exposure to the inflammatorymediators mixture (Mix). (A) PTN inhibited the activation of theNF-kB pathway: the phosphorylation of IKK α/β andIκBα decreased significantly at a concentration of30 μm with little effect on cell viability[Supplementary Fig. S6]. (B) The NF-κ B inhibitor PTNeliminated the phosphorylation difference of IKK,IκBα and NF-κ B p65 in theNC/siBAHD1-treated cell model. (C) PTN repressed the expression ofassociated inflammatory genes such as TNF-α, IL-6, IL-8 and CCL3in vitro. The values represent themean ± SEM of three independentexperiments. Representative blots of cell lysates are shown from at leastthree independent experiments.
BAHD1 Differentially Modulated the TNF Signalling Pathway by AlteringTNFR1 Expression
Downregulation of BAHD1 correlated positively with cytokine secretion; therefore,we turned our attention to the starting point of this secretory process. TheNF-κ B and AP-1 pathways, which are critical for expression of theproinflammatory cytokine cascade, might be mediated by tumour necrosis factorreceptor 1 (TNFR1)16. Consistent with the high activation of keyproteins in intracellular cell signalling pathways, we detected that theexpression of TNFR1 increased dramatically in the siBAHD1 group, at both theprotein and mRNA levels, compared with the negative control group [Fig. 7A,B]. High levels of cytokines secreted by colonicepithelium that had lost BAHD1 expression might in turn result in persistentactivation of the NF-κ B and JNK/AP-1 pathways. Taken together,these results suggest an essential role of BAHD1 in the negative regulation ofthe starting point of the pathway through the TNF signalling pathway.
BAHD1 modulated the TNF signalling pathway by altering TNFR1expression.
Caco-2 cells were incubated with BAHD1 siRNA (siBAHD1) and a negative controlsiRNA (NC) before incubation with the mixture (Mix) for 24 h tomimic gut inflammation. (A) mRNA and (B) protein levels showedsignificantly increased contents of TNFR1 in the siBAHD1 group compared withthe NC group. (C) A hypothesis regarding BAHD1 negative regulation ofthe TNF signalling pathway. The data are expressed as themean ± SEM;n = 3, *p < 0.05;**p < 0.001 versus NC group. For the blots,independent experiments were repeated at least three times.
Discussion
Here, we used a systematic computational approach based on publicly available geneexpression signatures to predict multiple previously undescribed molecules forUC10. Distance values derived from comparing public UC microarraydata against a compendium of gene expression signatures comprising 106 siRNAs wereevaluated in the datasets based on GSEA. All siRNAs were manually filtered accordingto their quality from a whole range of data in the GEO platform. A small distanceindicates a similar gene regulation tendency and among the smallest-scoring siRNAspredicted from our approach were EZH2, UPF1, FOXM1, NUDT6 and BAHD1.
The exact functions of these five factors in IBD have not been previously reported.EZH2 (enhancer of zeste homolog 2) is a histone methyltransferase associated withtranscriptional repression and its overexpression promotes tumour development17. UPF1 (up-frameshift mutant 1) is the regulator of nonsensetranscripts 1 in humans and participates in both nuclear mRNA export and mRNAsurveillance18. The third one, FOXM1, is a transcription factorinvolved in cell cycle progression that regulates the expression of a large numberof G2/M-specific genes19, while NUDT6 (nucleoside diphosphate-linkedmoiety X motif 6) is thought to be a fibroblast growth factor antisense geneassociated with cell cycle progression and tumour proliferation20.Further study into the potential genes driving this clustering could reveal newinformation regarding UC’s pathogenesis and new molecular therapeuticdirections.
Among them, BAHD1 is involved in gene silencing21 and was the mostdownregulated in the UC mouse model, which was inferred to indicate the mostpotential as a regulatory protein for the disease. The experimental validation weperformed in vitro confirmed that the loss of BAHD1 activated variouscytokines during a cellular immune response through associated signalling pathways.Intestinal inflammation and tissue damage is a direct result of increasedcirculating inflammatory cytokines, which are secreted at sites of inflammation andimpact during the onset, progression and resolution of UC22. Thosecytokines and also COX-2, iNOS and eNOS, are mediated by several signallingpathways23,24.
Transcription factors, including NF-κ B and AP-1, play critical roles inthe expression of genes involved in inflammation and carcinoma development in thegastrointestinal tract25,26. In the present study, we showed thatkey proteins in the NF-κ B pathway, including IKKα/β, IκBα and NF-κ Bsubunit p6527,28, were activated to a higher level in stimulatedCaco-2 cells with BAHD1 knocked down compared with a purely stimulated group. AP-1is a member of a family of transcription factors mainly belonging to the JUN and Fosfamilies whose activation is involved in inflammatory gene expression29. A similar phenomenon was observed in the JNK/AP-1 pathway, in which thephosphorylation levels of JNK and c-JUN increased. Taken together, the data gave thestrongest hint that a link might exist between activation of the transcriptionfactors NF-κ B and AP-1 and the reduction in BAHD1 expression inIECs.
Pathogen-associated molecular patterns are sensed by specific receptors, which inturn activate signalling cascades to induce the synthesis of inflammatory mediatorssuch as TNF, IL-1 and IFN30. TNFR1, which is ubiquitously expressed,has pleiotropic functions related to cell immunity, survival, apoptosis and necrosisand can be activated via both membrane-bound and soluble TNF31,32.The TNF receptor is primarily responsible for initiating inflammatory responses bymediating TNF-α- induced NF-κ B activation33,34. In this study, TNFR1 transcription increased significantly inCaco-2 cells after downregulation of BAHD1, causing TNF signalling pathwayactivation in IECs during inflammatory mediator exposure. As a result, morecytotoxic inflammatory factors were produced in response to the activation of thepathway, which in turn resulted in inflammatory status aggravation with moresecreted cytokines, especially TNF combined with TNFR1. The continuous excessivecytokine secretion caused injury and dysfunction of the IECs. A hypothesis of BAHD1negatively regulating the TNF signalling pathway by altering TNFR1 expression isshown in Fig. 7C, in which the inflammatory microenvironmentinduces downregulation of BAHD1 in IECs, which in turn can increase the productionof various cytokines through the IKK/NF-κ B and JNK/AP-1 pathway.
As a novel heterochromatinization factor in vertebrates, BAHD1 participates in genesilencing by promoting the formation of heterochromatin through interaction withHP1, MBD1, HDAC5 and several transcription factors to control cell differentiationand maintenance of homeostasis21. We speculate that in some way,BAHD1 connects with other repressive core complex factors to mediate TNFR1 genesilencing.
As for the MAPK pathways, the JNK pathway and the p38 MAPK pathway regulate apoptoticcell death, whereas ERK1/2 acts as a prosurvival factor that contributes to theregulation of cell proliferation and differentiation35. Therefore,JNK activation in IECs with downregulated BAHD1 levels during the acute phase ofinjury may result directly in apoptosis, not survival.
The JAK/STAT pathway is a central mediator of the responses of various extracellularcytokines and has been implicated in the pathogenesis of many human immunitydisorders, including IBD. JAK inhibitors have the potential to treat theinflammation associated with colitis36,37. The detailed mechanismwhereby repression of BAHD1 in enteric cells leads to higher levels of STAT3phosphorylation requires further exploration.
Although we found some interesting results, it should be noted that follow-upinvestigative work will be necessary. First, in vivo evidence regarding theprecise role of BAHD1 in UC is lacking. In addition, although exposure of Caco-2cells to inflammatory stimuli is a common approach to investigating inflammatorysignalling, it is not the disease per se. Finally, the detailed mechanism of howBAHD1 represses TNF receptor expression is unclear and will require deepexploration.
In conclusion, the results raise the intriguing possibility that a computationallypredicted protein, BAHD1, might act as an important regulator of the classicalinflammation pathways in UC and there might be a functional association betweenintestinal cell inflammatory responses and BAHD1. Additionally, the computationalapproaches we used based on public gene expression databases show potential for thediscovery of genes and proteins that might be vital factors in the pathogenesis ofcertain diseases.
Methods
Human Tissue
Human paraffin-embedded colonic mucosa pinches were obtained from surgicalpatients with active UC or healthy adjacent or distant colonic tissue fromsubjects with certain cancers. Pathological analysis verified the diagnoses ofUC. This study was approved by the Ethics Committee of the First AffiliatedHospital, College of Medicine, Zhejiang University, Hangzhou, China. Informedconsent was obtained from each subject before the study. The study protocols andthe consent form were administered in accordance with the approved guidelines ofthe Ethics Committee.
Animal Treatment
Six- to eight-week-old littermate female C57BL/6 mice were purchased fromZhejiang Experimental Animal Centre, Hangzhou, China. Mice were housed in ananimal room with air-conditioned specific pathogen-free (SPF) conditions at23 ± 2 °C with a12 h light/dark cycle and they were acclimated for 7 days beforeexperimentation. The Animal Care and Use Committee of Zhejiang Universityapproved all the mouse studies, which were performed in accordance with theChinese guidelines for the care and use of laboratory animals.
Computation of Distances between Different Expression Data
UC patient tissue expression profiles and siRNA-perturbed cell expressionprofiles were collected from the National Center for Biotechnology Information(NCBI) Gene Expression Omnibus (GEO)38. To make full use ofcollected expression data, the data platforms were restricted to the AffymetrixHuman Genome U133A Array (GPL96) and the Affymetrix Human Genome U133 Plus 2.0Array (GPL570), the most widely used Homo sapiens platform. Any probesnot numbered in all the datasets were excluded in our next analysis, resultingin 22,215 validated probes. Expression profiles representing different siRNApermutations were collected by searching in the GEO with the key words siRNA,shRNA and by manual checking. Due to the dearth of siRNA perturbation expressionprofiles, the cell types used were not restricted to particular cell types [Supplementary Table S3].
To properly compare expression profiles from different datasets, the validatedprobes were ranked by their expression change compared to the control asdescribed below.
First, we paired each experiment’s (UC patient samples or siRNAprocessed cells) expression profile to a control (healthy controls or untreatedcells).
Second, for each pair of samples, we ranked the probes considering both theirfold-changes and their absolute values relative to a probe rank list (PRL)11. The detailed steps are list below.
(1) Expression values less than the primary threshold value (the lower quartile of expressionvalues of these two samples) were set to that value. (2) The probeswere then ranked in descending order of corresponding experiment-to-controlratio values. (3) For probes where the ratio value equals one, thesecondary threshold value (one-tenth of the primary threshold value) was used toreset the value of these probes.(4) These probes were sub-sorted in descendingorder of the new ratios to produce the final probe rank list for each pair ofsamples.
This hierarchical sort strategy avoids the inappropriately high or low ranks thatcan be caused by large fold changes resulting from dividing by small values.
Third, PRLs representing the same permutation (same disease or same genesilencing) were combined with the R package GeneExpressionSignature39 into final PRLs to represent the cell’s responses tothem according to a hierarchical majority-voting scheme13,40.
Then, the distance between UC and the different siRNA perturbations was estimatedby GSEA10 in the following steps:
(1) The distanceswere calculated by comparing two PRLs through investigating whether the top- (orbottom-) ranked probes in one PRL was also the top- (or bottom-) ranked in theother PRL. Thus, for each PRL, a gene set containing its top- (or bottom-)ranked probes was generated as its signature. The top and bottom 250 probes wereselected as the signature of each PRL. The sizes of the signatures werechangeable and their effect on the final prediction was limited; therefore, thesize was set to an empirical value from a reference paper41. (2) When comparing two PRLs A and B, the enrichment score ofA’s (or B’s) signature in reference PRL B (or A) wascalculated by equal weighted GSEA10,11 and the enrichment scorecould be presented as ESAB(or ESBA) ranging from−1 to 1. A high enrichment score indicates that the signature genesalso tend to appear at the top or bottom of the reference PRL, indicatingsimilarity between them. (3) The distance between each two PRLs wasdefined as 1-(ESAB+ESBA)/2 ranging from 0 to 2.
Immunohistochemistry (IHC)
Colon sections (4 μm) cut from paraffin-embeddedintestine tissues were deparaffinized with xylene and rehydrated with ethanol.For IHC, tissue sections were preincubated with 10% normal goat serum (ZSGB-BIO,Beijing, China) in PBS (pH 7.5) and then incubated with primary antibodiesagainst BAHD1 (dilution: 1:200, Abcam, Cambridge, UK) overnight at4 °C. Tissue sections were stained with HPR secondaryantibody (dilution: 1:1000, ZSGB-BIO, Beijing, China) for 1 h at37 °C in an incubator. Immunoreactivity was detectedusing a DAB kit (ZSGB-BIO, Beijing, China) and visualized as brown staining.
Cell Culture
The heterogeneous human epithelial colorectal adenocarcinoma cell line, Caco-2(Institute of Biochemistry and Cell Biology, China Academy of Sciences,Shanghai, China) was maintained in Minimum Essential Medium (MEM, Invitrogen,Carlsbad, CA, USA), supplemented with 10% fetal calf serum (FCS, Invitrogen,Carlsbad, CA, USA) and 100 U/ml penicillin-streptomycin(Sigma-Aldrich, St. Louis, MO, USA) at 37 °C in ahumidified 5% CO2 atmosphere.
RNA Mediated Interference
Small-interfering RNA (siRNA)-mediated knockdown in human Caco-2 cells wasperformed using negative control (NC) siRNA (Invitrogen, Carlsbad, CA, USA), anda specifically designed siRNA with the sequence 5′-AUA GCA CUU CUCCUC AAU GCA GGC C-3′ to target human BAHD1. Lipofectamine 2000(Invitrogen, Carlsbad, CA, USA) was used as the siRNA transfection reagent, asper the manufacturer’s instructions. siRNAs were used at a finalconcentration of 15 nM. The interfering effect was verified by qPCRand western blotting for the mRNA or protein, respectively [Supplementary Fig. S4].
Cytokines Measurement
To evaluate proinflammatory factors in the DSS-induced colitis in mice, distalcolons cut longitudinally were washed in phosphate buffered saline (PBS). Stripsof 100 mg of colon tissue were placed in clean EP tubes containing0.5 ml cold PBS and ground with a pestle. The samples werecentrifuged at 14,000 g for 10 minutes at4 °C. The supernatant was collected to quantify theproduction of TNF-α, IL-6 and IFN-γ by enzyme-linkedimmunosorbent assay (ELISA) kits (eBioscience, San Diego, CA, USA), according tothe manufacturer’s protocol. Similarly, for cell culture, cell-freesupernatant was harvested after treatment and analyzed for IL-6, IL-8 and MCP-1contents.
Gene Expression Measure
RNAiso Plus (Takara, Otsu, Japan) was used to isolate mRNA from cells or tissue.The PrimeScript RT Master Mix (Takara, Otsu, Japan) was used to generate cDNA.qPCR was performed using a SYBR Green Premix DimerEraser (Takara, Otsu, Japan)on the 7900HT Fast Real-Time PCR system (Applied Biosystems). All reactions hada melting curve with a single peak. The cycle threshold (CT) values for thetriplicate samples were averaged and the data were analyzed using theΔΔCT method, where foldchange = 2−ΔΔCT.All data analyzed were normalized to beta-Actin or GAPDH expression. For thespecific primer sequences (Sangon Biotech, Shanghai, China), see Supplementary Table S4.
Western Blot Analysis
To detect the expression of associated proteins, Caco-2 cells and distal colontissue of mice were washed with cold PBS, collected and lysed in RIPA buffer(Pulilai BioTech, Beijing, China) containing protease inhibitor cocktail(Sigma-Aldrich, St. Louis, MO, USA). Proteins(30 μg/sample) were separated by SDS/PAGE andtransferred to a polyvinylidene difluoride membrane (0.45 mm pore;Millipore, Bedford, MA, USA). After being blocked with 5% skim milk powderdiluted in TBS containing 5% Tween-20 for 1 h, the membrane wasincubated with a primary antibody, anti-p-NF κB (S536), p-IKK-alpha(S176)/IKK-beta (S177), anti-p-IκB-alpha, anti-p-c-JUN (S73),anti-p-STAT3 (Y705), anti-TNF-R1, anti-GAPDH (Cell Signaling Technology,Danvers, MA, USA); anti-p-p38 (Y182+T180) anti-BAHD1 (Abcam, Cambridge, UK); oranti-JNK1 (pY185)/JNK2 (pY185)/JNK3 (pY223) (Epitomics, Burlingame, CA, USA), at4 °C overnight. Immunoreactive proteins were detectedusing an enhanced chemiluminescence light (ECL) detecting kit (Lianke MultiSciences, Hangzhou, China). GAPDH acted as a loading control. The experimentswere replicated at least three times and representative results are shown.
Establishment of DSS-induced Colitis in Mice
Chemically induced murine models of intestinal inflammation are the most commonlyused and best-described models for investigating the pathophysiologicalmechanisms and immunological processes underlying chronic mucosalinflammation42. The protocol to develop acute colitis inC57BL/6 mice was as described previously43. The drinking supplyof the mouse cages was filled with DSS (MW: 36,000–50,000; MPBiochemicals, Solon, OH, USA) at 3% weight/volume, while control mice receivedautoclaved water for 7 days. Mice receiving DSS orally developed acute UC-likeclinical and pathological manifestations. The mice developed colonic mucosalinflammation limited to the mucosa and contained large numbers ofimmunoglobulin-secreting plasma cells, accompanied by body weight loss, bloodydiarrhoea during the acute phase and shortening of the colon. In accord withthese manifestations, the inflamed colon tissue secreted much moreTNF-α, IL-6 and IFN-γ. [Supplementary Fig. S5] Disease activity index(see Supplementary Materials) andhistological analysis of haematoxylin & eosin (H&E)-stainedcolon sections [Supplementary TableS5] were the two main standards that identified the successfulestablishment of the UC-like mouse model.
Establishment of a Cell Model to Simulate Inflammatory Environment forIECs
To develop an vitro model to investigate inflammatory signalling in IECs, Caco-2cells were exposed to the inflammatory mediators LPS(1 μg/ml) (Sigma-Aldrich, St. Louis, MO, USA), tumournecrosis factor-α (rh TNF-α, 50 ng/ml),recombinant human interferon-γ (rh IFN-γ,50 ng/ml) and interleukin-1 beta (rh IL-1β,25 ng/ml) (Peprotech, Rocky Hill, NJ, USA) for 24 h44. Using qPCR and ELISA, we found that exposure to a mixture ofthese factors together (MIX) in Caco-2 cells for 24 h mimickedclosely the environment of gut inflammation as the cells expressed the highestamounts of proinflammatory factors (TNF-α, IL-6) and chemokines(IL-8, MCP-1) [Fig. 8].
Establishment of a cell model to simulate the inflammatory environment ofIECs.
An in vitro model to estimate gut inflammation using variousproinflammatory factors including TNF-α, IFN-γ andIL-1β and the gram-negative bacterial product LPS. Caco-2 cellswere incubated with each factor separately and with their mixture (MIX) for24 hours. RNA was extracted and qPCR was performed. (A)qPCR analysis of TNF-α, IL-6 and IL-8 expression. The data werenormalized to the expression of GAPDH. (B)(C)(D)Stimulated production of IL-6, MCP-1 and IL-8 in culture supernatant ofCaco-2 cells in different exposure groups measured using ELISA kits. Thevalues are expressed as the mean ± SEMof three separate experiments performed in triplicate.*P < 0.05,**P < 0.01. The increase in the MIX group wasmore significant than in any of the other stimulated groups after24 h of exposure.
Statistical Analysis
All data are shown as the mean ± SEM or SDvalue from at least three independent experiments. Significant differences wereevaluated by the unpaired Student’s t-test with two-taileddistributions. P-values below 0.05 were considered significant. The results wereconsidered significant at *P < 0.05;**P < 0.01. Prism version 5.0 (Graph PadSoftware) was used to perform the statistical analyses.
Additional Information
How to cite this article: Zhu, H. et al. Computational Prediction andValidation of BAHD1 as a Novel Molecule for Ulcerative Colitis. Sci. Rep.5, 12227; doi: 10.1038/srep12227 (2015).
References
Ordas, I., Eckmann, L., Talamini, M., Baumgart, D. C. & Sandborn, W. J. Ulcerative colitis. Lancet. 380, 1606–1619 (2012).
Ng, S. C. Epidemiology of inflammatory bowel disease: Focus on Asia. Best. Pract. Res. Cl. Ga. 28, 363–372 (2014).
Molodecky, N. A. et al. Increasing Incidence and Prevalence of the Inflammatory Bowel Diseases With Time, Based on Systematic Review. Gastroenterology. 142, 46–54 (2012).
Wirtz, S. & Neurath, M. F. Mouse models of inflammatory bowel disease. Adv. Drug. Deliver. Rev. 59, 1073–1083 (2007).
Sartor, R. B. Mechanisms of disease: pathogenesis of Crohn’s disease and ulcerative colitis. Nat. Clin. Pract. Gastr. 3, 390–407 (2006).
Smale, S. T. Selective transcription in response to an inflammatory stimulus. Cell. 140, 833–844 (2010).
Feng, Y. J. & Li, Y. Y. The role of p38 mitogen-activated protein kinase in the pathogenesis of inflammatory bowel disease. J. Digest. Dis. 12, 327–332 (2011).
Schena, M., Shalon, D., Davis, R. W. & Brown, P. O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 270, 467–470 (1995).
Warner, E. E. & Dieckgraefe, B. K. Application of genome-wide gene expression profiling by high-density DNA arrays to the treatment and study of inflammatory bowel disease. Inflamm. Bowel. Dis. 8, 140–157 (2002).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. P. Natl. Acad. Sci. USA. 102, 15545–15550 (2005).
Lamb, J. et al. The connectivity map: Using gene-expression signatures to connect small molecules, genes and disease. Science. 313, 1929–1935 (2006).
Dudley, J. T. et al. Computational Repositioning of the Anticonvulsant Topiramate for Inflammatory Bowel Disease. Sci. Transl. Med. 3 10.1126/scitranslmed.3002648 (2011).
Iorio, F. et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. P. Natl. Acad. Sci. USA. 107, 14621–14626 (2010).
Panes, J. Adhesion molecules: their role in physiopathology and treatment of inflammatory bowel disease. Gastroenterol. Hepatol. 22, 514–524 (1999).
Sheehan, M. et al. Parthenolide, an inhibitor of the nuclear factor-kappa B pathway, ameliorates cardiovascular derangement and outcome in endotoxic shock in rodents. Mol. Pharmacol. 61, 953–963 (2002).
Chu, W. M. Tumor necrosis factor. Cancer letters. 328, 222–225 (2013).
Vire, E. et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 439, 871–874 (2006).
Cheng, Z. H., Muhlrad, D., Lim, M. K., Parker, R. & Song, H. W. Structural and functional insights into the human Upf1 helicase core. Embo. J. 26, 253–264 (2007).
Wierstra, I. & Alves, J. FOXM1, a typical proliferation-associated transcription factor. Biol. Chem. 388, 1257–1274 (2007).
Baguma-Nibasheka, M., Macfarlane, L. A. & Murphy, P. R. Regulation of fibroblast growth factor-2 expression and cell cycle progression by an endogenous antisense RNA. Genes. 3, 505–520 (2012).
Bierne, H. et al. Human BAHD1 promotes heterochromatic gene silencing. P. Natl. Acad. Sci. USA. 106, 13826–13831 (2009).
Christophi, G. P., Rong, R., Holtzapple, P. G., Massa, P. T. & Landas, S. K. Immune markers and differential signaling networks in ulcerative colitis and Crohn’s disease. Inflamm. Bowel. Dis. 18, 2342–2356 (2012).
Hobbs, S. S. et al. TNF transactivation of EGFR stimulates cytoprotective COX-2 expression in gastrointestinal epithelial cells. Am. J. Physiol-Gastr. L. 301, G220–229 (2011).
Pelletier, J. P. & Martel-Pelletier, J. The Novartis-ILAR rheumatology prize 2001 osteoarthritis: from molecule to man. Arthritis. Res. 4, 13–19 (2002).
Schmid, R. M., Adler, G. & Liptay, S. Activation of NF kappaB in inflammatory bowel disease. Gut. 43, 587–588 (1998).
Ben-Neriah, Y. & Karin, M. Inflammation meets cancer, with NF-kappa B as the matchmaker. Nat. Immunol. 12, 715–723 (2011).
Hayden, M. S. & Ghosh, S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Gene. Dev. 26, 203–234 (2012).
Smale, S. T. Hierarchies of NF-kappaB target-gene regulation. Nat. Immunol. 12, 689–694 (2011).
Schonthaler, H. B., Guinea-Viniegra, J. & Wagner, E. F. Targeting inflammation by modulating the Jun/AP-1 pathway. Ann. Rheum. Dis. 70, I109–I112 (2011).
Gaestel, M., Kotlyarov, A. & Kracht, M. Targeting innate immunity protein kinase signalling in inflammation. Nat. Rev. Drug. Discov. 8, 480–499 (2009).
Varfolomeev, E. et al. Cellular inhibitors of apoptosis are global regulators of NF-kappaB and MAPK activation by members of the TNF family of receptors. Sci Signal. 5, 10.1126/scisignal.2001878 (2012).
Naude, P. J. W., den Boer, J. A., Luiten, P. G. M. & Eisel, U. L. M. Tumor necrosis factor receptor cross-talk. Febs. J. 278, 888–898 (2011).
Loetscher, H., Stueber, D., Banner, D., Mackay, F. & Lesslauer, W. Human Tumor-Necrosis-Factor-Alpha (Tnf-Alpha) Mutants with Exclusive Specificity for the 55-Kda or 75-Kda Tnf Receptors. J. Biol. Chem. 268, 26350–26357 (1993).
Chen, G. & Goeddel, D. V. TNF-R1 signaling: a beautiful pathway. Science. 296, 1634–1635 (2002).
Xia, Z. G., Dickens, M., Raingeaud, J., Davis, R. J. & Greenberg, M. E. Opposing Effects of Erk and Jnk-P38 Map Kinases on Apoptosis. Science. 270, 1326–1331 (1995).
Coskun, M., Salem, M., Pedersen, J. & Nielsen, O. H. Involvement of JAK/STAT signaling in the pathogenesis of inflammatory bowel disease. Pharmacol. Res. 76, 1–8 (2013).
Sandborn, W. J. et al. Phase 2 Randomized Study of CP-690,550, an Oral Janus Kinase Inhibitor, in Active Crohn’s Disease. Gastroenterology. 140, S124–S124 (2011).
Iorio, F., Tagliaferri, R. & di Bernardo, D. Identifying network of drug mode of action by gene expression profiling. J. Comput. Biol. 16, 241–251 (2009).
Li, F. et al. GeneExpressionSignature: an R package for discovering functional connections using gene expression signatures. Omics. 17, 116–118 (2013).
Ni, M. et al. ExpTreeDB: Web-based query and visualization of manually annotated gene expression profiling experiments of human and mouse from GEO. Bioinformatics. 30, 3379–3386 (2014).
Burczynski, M. E. et al. Molecular classification of Crohn’s disease and ulcerative colitis patients using transcriptional profiles in peripheral blood mononuclear cells. J. Mol. Diagn. 8, 51–61 (2006).
Alex, P. et al. Distinct Cytokine Patterns Identified from Multiplex Profiles of Murine DSS and TNBS-induced Colitis. Inflamm. Bowel. Dis. 15, 341–352 (2009).
Wirtz, S., Neufert, C., Weigmann, B. & Neurath, M. F. Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2, 541–546 (2007).
Van De Walle, J., Hendrickx, A., Romier, B., Larondelle, Y. & Schneider, Y. J. Inflammatory parameters in Caco-2 cells: Effect of stimuli nature, concentration, combination and cell differentiation. Toxicol. In. Vitro. 24, 1441–1449 (2010).
Acknowledgements
We thank Dr. Min Yue for her critical inputs on the project. We also thank Han Zhang,Linjie Xu and Shengwen Song for their outstanding technical assistance. This studywas supported by The Ministry of Education of Talents Scheme (NCET-13-0525).
Author information
Authors and Affiliations
Contributions
The co-first authors H.T.Z, X.Y.W. and J.L. were responsible for performing the mainexperiment, the analysis and the writing of the manuscript. Co-authors X.C.B. andL.H. were involved in computational prediction part. W.G.C. and C.L. performed themouse model colitis assessments. Z.S. and C.F.X. provided advice in designing theexperiments. G.Q.X., C.H.Y. and L.H.C. are the principal investigators andcorresponding authors for these studies.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0International License. The images or other third party material in this article areincluded in the article’s Creative Commons license, unless indicatedotherwise in the credit line; if the material is not included under the CreativeCommons license, users will need to obtain permission from the license holder toreproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhu, H., Wan, X., Li, J. et al. Computational Prediction and Validation of BAHD1 as a Novel Molecule for UlcerativeColitis. Sci Rep 5, 12227 (2015). https://doi.org/10.1038/srep12227
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep12227
This article is cited by
-
EZH2 Regulates Intestinal Inflammation and Necroptosis Through the JNK Signaling Pathway in Intestinal Epithelial Cells
Digestive Diseases and Sciences (2019)
-
Dysregulated Up-Frameshift Protein 1 Promotes Ulcerative Colitis Pathogenesis Through the TNFR1-NF-κB/MAPKs Pathway
Digestive Diseases and Sciences (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.