Abstract
A magnetic enzyme nanosystem have been designed and constructed by a polydopamine (PDA)-modification strategy. The magnetic enzyme nanosystem has well defined core-shell structure and a relatively high saturation magnetization (Ms) value of 48.3 emu g−1. The magnetic enzyme system can realize rapid, efficient and reusable tryptic digestion of proteins by taking advantage of its magnetic core and biofunctional shell. Various standard proteins (e.g. cytochrome C (Cyt-C), myoglobin (MYO) and bovine serum albumin (BSA)) have been used to evaluate the effectiveness of the magnetic enzyme nanosystem. The results show that the magnetic enzyme nanosystem can digest the proteins in 30 minutes and the results are comparable to conventional 12 hours in-solution digestion. Furthermore, the magnetic enzyme nanosystem is also effective in the digestion of low-concentration proteins, even at as low as 5 ng μL−1 substrate concentration. Importantly, the system can be reused several times and has excellent stability for storage. Therefore, this work will be highly beneficial for the rapid digestion and identification of proteins in future proteomics.
Similar content being viewed by others
Introduction
Along with the rapid development of nanotechnology, functional nanomaterials with desirable components, designed structures and controlled morphologies have attracted much attention in material science1,2,3. As a type of important functional material, magnetic nanostructures have been widely applied in biomedicine, environmental remediation, energy generation and storage because of their unique physical and chemical properties (e.g. high surface to volume ratios, adjustable size, size-dependent magnetic properties, chemical stability and variable surface modifications, etc.)4,5,6. Recently, magnetic nanostructures with core-shell architecture have aroused intense interests in various biomedical applications including cancer therapeutics, disease detection and biofunctional carrier, due to their enhanced properties and the synergistic effect between multiple discrete components7,8,9,10. Specifically, by taking advantage of their unique magnetic properties, the high water dispersion and chemical durability, magnetic core-shell nanoparticles can be rapidly collected and dispersed in aqueous solution and are extremely favorable for collecting and carrying functional biomolecules (e.g. DNA, protein and peptide)11,12,13, which endow them with specific bioactivities and extend their biomedical applications.
With the advent of the post-genomic era, proteomics has drawn increasing attention in protein interaction studies, biomarker discovery and disease diagnosis and prevention14,15,16, because it focuses on the study of large scale proteins expression changes in structure and function via a global perspective. Tryptic proteolysis by sequence-specific proteases is a necessary and critical step in proteomics. Conventional in-solution or in-gel digestion by trypsin under certain restricted conditions is the commonly used proteolysis method17,18. However, these procedures often suffer from some disadvantages such as the low stability of trypsin due to the experimental condition changes, difficulty in the recovery of trypsin, prolonged digestion time and limitations for the digestion of low concentration proteins19,20,21. These problems can be partly addressed by the immobilization of enzymes on solid supports, which can enhance enzyme stability, increase enzyme to substrate ratio, improve enzymatic efficiency and facilitate separation and recovery for reuse22.
With the rapid development of nanotechnology, nanomaterials can provide an attractive solution due to their high surface area and designable functions23. Various mesoporous materials with nanostructures have been explored to carry enzymes by physical adsorption and they showed better performance than those using free enzymes alone24,25. Although promising, these enzyme-immobilized nanostructures are compromised by enzyme leaching because of the lack of specific interaction between the substrates and the target enzyme molecules and the centrifugal separation of the enzyme carriers after each step is inconvenient26,27. Therefore, the efficiency of the nanosystem would be negatively affected, while the recovery of these nanostructures with immobilized enzymes can hardly be achieved. To overcome these shortcomings, the proposed magnetic nanostructure is an ideal candidate by combing the merits of the magnetic property and the catalysis of trypsin. Normally, the chemical linkage based strategy was used to graft trypsin molecules and the magnetic particles were first modified with silane reagents, followed by the grafting of enzyme molecules under strict conditions28. However, the whole preparation process focuses on monolayer modification of the particles with small molecules, which would limit the immobilization amount and reduce the accessibility of protein substrates to the immobilized enzyme24,29. Although recent works attempted to increase the amount of the binding enzyme by coating a layer of functional polymer, complex, rigorous and longtime modification procedures were required30, which might change the conformation of the enzymes and cause a reduction of their catalytic activity. Thus, it is highly desirable to construct an enzyme-immobilized magnetic nanosystem for rapid protein digestion under mild conditions via a simple method.
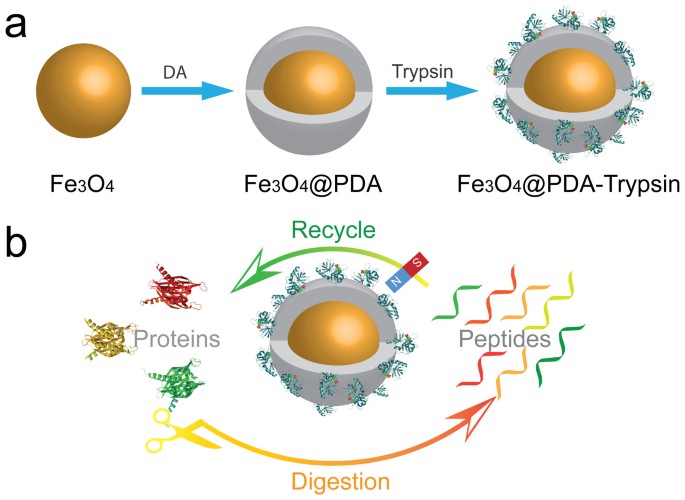
Inspired by the bio-adhesion properties of marine mussels, polydopamine, a novel coating material, has sparked research interest and been moved into the spotlight31,32. It can be easily coated on various inorganic and organic substrates with controllable film thickness and high stability33. More importantly, the chemical versatility, excellent dispersibility and extraordinary biocompatibility of polydopamine enable it to be a platform for diverse secondary reactions in biological applications (e.g. biomineralization, cell encapsulation, biomolecule immobilization)34,35,36. Herein, we propose to construct a magnetic enzyme nanosystem (denoted as MEN) via a PDA-modification strategy for the rapid digestion of proteins. As shown in Figure 1a, the Fe3O4 magnetic particles were coated with a layer of polydopamine after being dispersed in an alkaline solution of dopamine hydrochloride, after which the trypsin molecules can be easily immobilized on the surface of the magnetic particles by simply mixing under mild conditions. The composite nanosystem combines the merits of the easy separation of Fe3O4 particles and the specific activity of trypsin functionalized polymer shells. They show the following advantages: (1) a polymer shell with a high density of functional groups would ensure the large binding capacity of trypsin molecules; (2) the large exposed surface area allows rapid mass transfer and diffusion of reactants and products; (3) the stability of the magnetic enzyme system under typical proteolytic digestion conditions and fast magnetic separation enables the regeneration and reuse of the nanosystem.
Results
Figure 1a illustrates the construction strategy of the MEN. Well-dispersed Fe3O4 microspheres were synthesized by a previously reported solvothermal method. Then, the prepared magnetic particles were coated with a highly uniform polymeric shell by self-assembly of dopamine (DA) molecules in alkaline aqueous solution using a biomimetic adhesive method. In virtue of the excellent water-dispersibility and the presence of many catechol hydroxyl-groups on the PDA layer, the trypsin molecules can be easily grafted onto the polydopamine coated magnetic microspheres (denoted as Fe3O4@PDA) by taking advantage of the unique adhesive properties of PDA, thereby forming the multifunctional MEN31.
Figure 2a shows the scanning electron microscopy (SEM) image of the prepared Fe3O4 particles. Apparently, the well-dispersed Fe3O4 particles have a relatively coarse surface and regular spherical shape with an average size of 180 nm. Transmission electron microscopy (TEM) images (Figure 2b) further reveal that the Fe3O4 microspheres consist of many reunited nanoparticles. As shown in Figure 2c and d, after further coating of PDA shells, the Fe3O4@PDA microspheres are relatively dispersed, but are slightly larger in diameter and present different surfaces in comparison to the precursor Fe3O4 particles. The core-shell structure of the Fe3O4@PDA can be clearly observed in the TEM image (Figure 2e and f) and the thickness of the PDA shell is about 20 nm, confirming the formation of the uniform PDA shell. It should be noted that the coating time plays an important role in the thickness of PDA shell. As shown in Fig. S1, with the extension of the coating time, the thickness of polydopamine would increase; however, the microspheres also slightly aggregate. Therefore, magnetic microspheres with thin PDA layer are favorable due to well dispersibility and shorter reaction time without sacrificing the surface functionalization capability of the PDA layer. Thus thin PDA layer was used in the following application.
The crystal structure of the magnetic composite microspheres was investigated by powder X-ray diffraction (XRD), as shown in Figure 3a. The XRD pattern of the Fe3O4 microspheres indicates that all diffraction peaks in the XRD pattern of Fe3O4 microspheres are consistent with the magnetite (JCPDS card No. 19-0629). Notably, the XRD pattern of the Fe3O4@PDA microspheres has similar diffraction peaks as those of precursor Fe3O4 microspheres, indicating that the polymer shell is amorphous in nature and the main magnetite phase of magnetic cores was not destroyed during the fabrication procedure. To evidently confirm the formation of PDA shell, Raman spectroscopy was used in investigating the changes of the interface of magnetic composite microspheres. As shown in Figure 3b, three strong and broad bands around 370, 489 and 681 cm−1 can be observed in the Raman spectrum of bare magnetic cores, which correspond to the T2g and A1g modes of symmetry37,38. However, after coating with the PDA polymer shell, the intensity of these characteristic bands decreased, while two new broad bands around 1355 cm−1 and 1577 cm−1 are present in the Raman spectrum. They are characteristic bands of polydopamine, which can be attributed to the deformation of the catechol group39. It should be noted that catechol groups serve as active sites for the grafting of targets and therefore the high abundance of residual catechol groups would contribute to the highly efficient immobilization of trypsin molecules.
The magnetic properties of MEN microspheres are beneficial for their convenient and fast separation, removing the need for repeated centrifugation in practical applications. As shown in Figure 4, the hysteresis loops of the MEN microspheres and the precursor Fe3O4 microspheres were recorded by a superconducting quantum interface device (SQUID) magnetometer. Apparently, both the Fe3O4 and Fe3O4@PDA microspheres have strong magnetism at room temperature. The saturation magnetization (Ms) values of the Fe3O4 and Fe3O4@PDA microspheres are 66.6 emu g−1 and 45.0 emu g−1, respectively. Because of the introduction of nonmagnetic species, the saturation magnetization (Ms) values of Fe3O4@PDA microspheres are smaller than that of Fe3O4 microspheres; however, they can still be rapidly separated by a magnet. As shown in the Figure 5 inset, the MEN microspheres can be easily dispersed in aqueous solution without an external magnetic field and they can then be rapidly collected from the mixture within 30 s by a magnet. These results demonstrate that the MEN microspheres simplify the separation process in practical applications, because of their excellent magnetic response.
The construction of a magnetic enzymatic nanosystem was easily conducted at room temperature and the amount of immobilized trypsin was monitored by an UV/Vis spectrophotometer. Figure 5 shows the time-dependent curve of trypsin immobilization. It is apparent that the maximum immobilization of trypsin can be realized in less than 30 minutes. This could be ascribed to the large exposed surface area of Fe3O4@PDA composite microspheres, which may provide more chances to interact with large guest molecules. The immobilization capacity of trypsin was calculated to be 85.9 μg mg−1 (enzyme/carrier).
The successful grafting of trypsin molecules was confirmed by the zeta-potential analysis of the resulting composite microspheres. As shown in Figure 6a, the naked magnetic cores show positive values (8.74 ± 0.87 mV), while the Fe3O4@PDA composite microspheres present a negative zeta-potential of −28.23 ± 0.47 mV because of the deprotonation of the phenolic group on the polydopamine shells40. However, after immobilization of trypsin, the zeta potential values of the microspheres increased to −5.73 ± 0.20 mV because of the introduction of positive trypsin molecules41. In addition, FT-IR spectra were also recorded to study the transformation of surface properties and the evidence of the successful immobilization of trypsin. As shown in Figure 6b, the strong absorption peak at around 590 cm−1 in FT-IR of Fe3O4 microspheres can be assigned to the stretching vibration of Fe-O from the Fe3O4. For the Fe3O4@PDA composite microspheres, new strong and broad absorption bands can be observed in the range of 1800–1000 cm−1, which can be attributed to the stretching vibration of the aromatic rings and the C-O stretching of phenol compounds and further confirms the formation of the PDA shell. After the grafting of the trypsin, besides the characteristic adsorption peaks of the Fe3O4 core and the PDA shell, several characteristic absorption peaks of trypsin with weak intensities were also observed in the range of 1000–1800 cm−1 and 2750–3000 cm−1 in the FTIR of MEN microspheres (Figure S2), compared to the Fe3O4@PDA microspheres. The above results demonstrate the successful construction of the MEN system.
As shown in Figure 1b, the MEN can be used as a digestion platform for rapid protein digestion as a result of its unique enzymatic activity; and the MEN microspheres can be easily separated and collected from the digestion solution for recycling by taking advantage of their magnetic properties. The MEN microspheres were mixed with the protein solution. Then, with the temperature at 37°C for 30 min, the proteins were digested. Notably, all the trypsin molecules were presented on the large exposed surface of the magnetic composite microspheres and therefore the target proteins could easily interact with them. After digestion, the resulting peptides could diffuse rapidly out of the surface so as to expose the active sites for interaction with undigested proteins, which would promote catalytic efficiency. More importantly, the MEN microspheres can be conveniently isolated from the mixture by a magnet, while leaving the protein digest ready for detection. Being washed with buffer several times can regenerate the MEN, at which point it is ready for reuse.
The effectiveness of the constructed MEN system was evaluated by digesting the model protein cytochrome c under heating at 37°C. As the digestion time and the amount of trypsin are critical for protein digestion, herein, the digestion time and the amount number of MEN microspheres were optimized. Figure 7a presents the identification results of Cyt-c digested by the MEN system under different heating times. The proteins can be effectively digested and identified with relatively high sequence coverage (>70%) even though the proteins were only treated with the MEN system for 5 min. Further increasing the digestion time would lead to an increased number of matched peptide and sequence coverage and excellent performance of protein digestion and identification can be realized in half an hour.
The amount of MEN microspheres can also affect the performance of protein digestion. As shown in Figure 7b, the MEN digestion system was highly effective, as the Cyt-c could be digested by 25 μg MEN microspheres and the protein could be identified with sequence coverage of 79% and 14 matched peptides. Better performance could be further realized via increasing the amount of MEN microspheres and the highest number of matched peptides and amount of sequence coverage was observed when 100 μg MEN microspheres were used. However, the number of matched peptides decreased slightly while sequence coverage was still high when the amount of MEN microspheres was increased to 200 μg. This could be ascribed to some peptide fragments being absorbed by the redundant microspheres. According to the above results, the optimal time and enzymatic microsphere quantity are 30 min and 100 μg respectively and should be used for the MEN digestion system.
Figure 8a shows the MALDI-TOF mass spectrum of Cyt-c digested by the MEN digestion system. Apparently, the target proteins were well digested and detected with a high intensity and signal to noise (S/N) ratio and 17 peptides can be matched and identified with a sequence coverage of 92%. Table S1 lists the detailed information for the identified peptides. For comparison, conventional in-solution digestions of 30 minutes and 12 hours were also conducted. As shown in Figure 8b, for the short time (30 minutes) in-solution digestion, only a few peptides from Cyt-c could be detected and the S/N ratio and intensity were very low, leading to the poor quality of the MS. As expected, mass spectra with enhanced S/N ratios and intensity can be obtained (Figure 8c) when the proteins were digested for a long-time (12 hours) in-solution. 15 target peptides with sequence coverage of 83% can be identified, which were comparable to but slightly lower than those obtained using the MEN system. Reduction of protein digestion time from 12 hours to 30 minutes by the MEN system can significantly improve the throughput and turnaround time of proteomic analysis. However, the digestion time was extended to 12 hours when using the in-solution technique and it was difficult to separate or recover the free trypsin molecules in the mixture solution afterwards.
Compared with the high-abundance proteins, the concentrations of most protein biomarkers are extremely low and rapid digestion and detection of low concentration protein samples is critical and highly desirable in practical proteomics research and biomedical diagnosis. However, quite notoriously, in-solution digestion of small quantity proteins is often compromised by the low concentration, small sample volume and low enzyme/protein ratio. Figure S3 shows the MS of low concentration (10 ng μL−1 and 5 ng μL−1, respectively) Cyt-c digested using conventional in-solution digestion. It is apparent that only a few peaks with low S/N ratio and intensity were detected, while it is hard to identify the target protein. However, as shown in Figure 9a, when it was digested using the MEN system, peptides (marked with ‘*’) with sequence coverage of 60% could be detected and identified for the target protein by the MS. Furthermore, even when the protein concentration was lowered to 5 ng μg−1, 6 peptides (marked with ‘*’) from Cyt-c could still be detected after digestion using the MEN system (Figure 9b). This could can be attributed to high local concentrations of substrate proteins near immobilized trypsin on the surface of MEN microspheres, which was absent from the in-solution digestion system.
Magnetic microspheres are characterized by easy separation and collection by an external magnetic field and therefore the MEN microspheres can be conveniently collected and regenerated. Mass spectra and identification results of digested proteins were used to evaluate the reusability of the MEN. Figure 10a shows the results of protein digestion using the recycled MEN microspheres. The target proteins could be effectively identified even when the MEN microspheres were reused 5 times and 15 peptides with a sequence coverage of 81% were matched on the fifth trial. Figure 10b and c represent the typical MS obtained from the third and fifth recycling digestion. Apparently, high quality mass spectra were observed and most of the target peptides were detected with strong intensities. These results demonstrate that the MEN microspheres have excellent recyclability.
Normally, the trypsin solution is very stable in the refrigerator. The digestion result using trypsin solution stored for 25 days is very similar to that obtained using the freshly prepared trypsin solution (Figure S4). In order to investigate their stability, MEN microspheres were stored in a refrigerator for different time intervals and then used for protein digestion. The digests were submitted for MS detection and the number of matched peptides and sequence coverages for protein identification were summarized in Figure 11. Interestingly, even with the prolongation of storage time, the digestion and detection results were not greatly deteriorated. Even for the MEN microspheres stored for 50 days, 16 peptides with sequence coverage of 87% can still be observed, indicating the high proteolytic activity of the MEN microspheres after long storage time. Therefore, it can be safely concluded that the MEN microspheres have excellent stability.
To confirm the universality of the MEN system for digestion of various proteins, bovine album serum (BSA) and myoglobin (MYO) were also digested using the MEN microspheres. As shown in Figure 12a, many peptides with S/N ratios and high intensities can be observed in the MS of BSA. After data search by the mascot, 36 peptides with sequence coverage 55% could be matched for the identification of the BSA protein. High quality MS could also be observed for the MYO proteins after MEN digestion and detection (Figure 12b) and 14 peptides are matched while the sequence coverage is high at 83%. Table S2 lists the detailed information regarding the identified peptides. For comparison, in-solution digestion was also conducted (Figure S5). 35 matched peptides with sequence coverage of 42% can be identified for BSA and 15 matched peptides with sequence coverage of 77% can be identified for MYO. Apparently, the results obtained using the 30 min MEN digestion approach is quite comparable to and in general better than the results obtained using the 12-hour in-solution method. The above results further demonstrate that the MEN system can be used for rapid digestion of proteins.
Discussion
A novel magnetic enzymatic nanosystem was constructed via a simple mussel-inspired polydopamine linking method. The MEN system can realize the rapid digestion of proteins, convenient magnetic separation and MEN recycling, thanks to the exposed tryptic surface, the Fe3O4 core and the stable PDA linker. Various proteins were used to evaluate the performance of the MEN system. The proteins can be digested in 30 minutes and the system is also effective for the digestion of extremely low-concentration proteins. Furthermore, the MEN system can be recycled several times with high digestion efficiency being maintained. In addition, the MEN microspheres are very stable in refrigerator storage, as the digestion efficiency is almost unchanged even after storage for 50 days. Therefore, the MEN system has high potential for rapid and high throughput protein digestion in proteomics.
Methods
Chemicals
Ferric chloride (FeCl3·6H2O), ethylene glycol (EG), trisodium citrate (H3Cit), tris (hydroxymethyl) minomethane hydro-chloride (Tris-HCl), ethanol (EtOH), sodium acetate (NaAc), ammonium bicarbonate (NH4HCO3), acetonitrile (ACN), trifluoroacetic acid (TFA) and ammonium hydroxide (NH3·H2O) were obtained from Alfa Aesar. Dopamine (DA), 2, 5-dihydroxybenzoic acid (2, 5-DHB), myoglobin from equine heart (MYO), cytochrome C (Cty-c), bovine serum albumin (BSA) and trypsin (from bovine pancreas, TPCK treated) were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Preparation of Fe3O4 and Fe3O4@PDA microspheres
The magnetic Fe3O4 particles were prepared according to a reported solvothermal approach with minor modification42. Typically, FeCl3·6H2O (0.81 g) and trisodium citrate (0.20 g) were first dissolved in ethylene glycol (20 mL). Then, NaAc (1.20 g) was added and the mixture was vigorously stirred to form a transparent solution. Afterward, the solution was transferred to a 50 mL Teflon-lined stainless-steel autoclave. The autoclave was sealed and heated to 200°C and maintained for 8 hours and then allowed to cool down to room temperature. The products were washed with ethanol and deionized water several times and dried at 60°C for 12 hours.
For the preparation of Fe3O4@PDA core-shell microspheres, the as-synthesized Fe3O4 particles (25 mg) were fully dispersed in 25 mL of 20 mM tris-HCl (pH = 8.0) by ultrasonication for 30 min. Dopamine (50 mg) was dissolved into 25 mL deionized water. The dopamine solution was quickly injected into the Fe3O4 dispersion under continuous magnetic stirring at 30°C for 2 hours. After that, the products were collected and separated with the help of a magnet and then washed several times with deionized water.
Construction of magnetic enzymatic nanosystem (MEN)
Fe3O4@PDA microspheres (10 mg) were added into 2.5 mL of 25 mM NH4HCO3 buffer to form a homogeneous suspension by ultrasonication for 2 hours, while trypsin (5 mg) was dissolved in another 2.5 mL of 25 mM NH4HCO3 buffer. Then, the particle suspension was added into the trypsin solution and the mixture was shaken for varying times. Subsequently, the microspheres loaded with trypsin were collected and isolated from the mixture by magnetic sorting and then the microspheres were washed three times with the NH4HCO3 buffer to remove any weakly bound trypsin. The amount of trypsin immobilized was measured using an UV/visible absorption spectrophotometer at 280 nm for comparing the protein concentration before and after immobilization. The obtained microspheres were redispersed into NH4HCO3 buffer (10 mg mL−1) via vibration to form a homogeneous suspension.
Rapid digestion of proteins using MEN
Cytochrome C (Cyt-c), myoglobin (MYO) and bovine serum albumin (BSA) were used to evaluate the efficiency of protein digestion by MEN. The proteins were first dissolved into NH4HCO3 buffer (25 mM) to form protein solutions of different concentrations. 100 μg of the MEN microspheres suspension was added into 50 μL of the protein solution and the mixture was then shaken at 37°C for 30 minutes. Subsequently, the supernatant was isolated and collected by magnetic separation for the following detection process. For regeneration, the collected MEN microspheres were washed with the NH4HCO3 buffer three times. The in-solution digestion with free trypsin was also conducted for comparison. Trypsin was then added into the protein solution with an optimized enzyme/substrate ratio of 1:50 (w/w) and the solution was incubated at 37°C for 12 hours.
MALDI-TOF MS analysis
1 μL of proteolytic digest was mixed with 1 μL of matrix solution containing 20 mg mL−1 DHB (in 50% acetonitrile aqueous solution, v/v) and 1% (v/v) H3PO4 aqueous solution by pipetting and 0.5 μL of mixture was deposited onto the MALDI target. MALDI-TOF mass spectrometry analysis was performed on an AB SCIEX MALDI-TOF/TOF 5800 mass spectrometer (Foster City, CA, USA) in positive ion mode with a 355 nm Nd:YAG laser, 200 Hz repetition rate and 20 kV acceleration voltage. Search parameters of fragment ion spectra were submitted to MASCOT (http://www.matrixscience.com/) for a database search and identification of corresponding peptides.
Characterization
Scanning electron microscopy (SEM) images were obtained on a field emission scanning electron microscope (FESEM; NanoSEM 630, NOVA). Transmission electron microscopy (TEM) images were taken with a JEOL-2010 microscope at the accelerating voltage of 200 kV. Powder X-ray diffraction (XRD) patterns were collected on a PANalytical Empyrean X-ray powder diffractometer (Cu Kα radiation, 45 kV, 40 mA) with the detective range from 5 to 80 degrees. Raman spectra were recorded on a WITec Confocal Raman instrument with a 514 nm laser wavelength. The zeta potential of the particles was recorded using a Malvern Zetasizer ZS. Fourier transform infrared spectra were determined on a Bruker Vertex V70 FTIR spectrometer over a potassium bromide pellet and then scanned from 400 to 4000 cm−1 at a resolution of 6 cm−1. Magnetization measurement was carried out with a superconducting quantum interface device (SQUID) magnetometer at 300 K. The UV-Vis adsorption spectral values were performed with a Perkin-Elmer Lambda 950 Spectrophotometer.
References
Biju, V. Chemical modifications and bioconjugate reactions of nanomaterials for sensing, imaging, drug delivery and therapy. Chem Soc Rev 43, 744–764 (2014).
Liu, J., Qiao, S. Z., Hu, Q. H. & Lu, G. Q. Magnetic nanocomposites with mesoporous structures: synthesis and applications. Small 7, 425–443 (2011).
Wang, W., Dahl, M. & Yin, Y. Hollow Nanocrystals through the Nanoscale Kirkendall Effect. Chem Mater 25, 1179–1189 (2012).
Wu, Z., Li, W., Webley, P. A. & Zhao, D. General and Controllable Synthesis of Novel Mesoporous Magnetic Iron Oxide@Carbon Encapsulates for Efficient Arsenic Removal. Adv Mater 24, 485–491 (2011).
Pan, Y., Du, X., Zhao, F. & Xu, B. Magnetic nanoparticles for the manipulation of proteins and cells. Chem Soc Rev 41, 2912–2942 (2012).
Behrens, S. Preparation of functional magnetic nanocomposites and hybrid materials: recent progress and future directions. Nanoscale 3, 877–892 (2011).
Hao, R. et al. Synthesis, Functionalization and Biomedical Applications of Multifunctional Magnetic Nanoparticles. Adv Mater 22, 2729–2742 (2010).
Yang, C., Wu, J. & Hou, Y. Fe3O4 nanostructures: synthesis, growth mechanism, properties and applications. Chem Commun 47, 5130–5141 (2011).
Wang, C. et al. Iron Oxide@Polypyrrole Nanoparticles as a Multifunctional Drug Carrier for Remotely Controlled Cancer Therapy with Synergistic Antitumor Effect. ACS Nano 7, 6782–6795 (2013).
Li, L. et al. Multifunctional magnetic-fluorescent eccentric-(concentric-Fe3O4@SiO2)@polyacrylic acid core-shell nanocomposites for cell imaging and pH-responsive drug delivery. Nanoscale 5, 2249–2253 (2013).
Chung, H. J., Castro, C. M., Im, H., Lee, H. & Weissleder, R. A magneto-DNA nanoparticle system for rapid detection and phenotyping of bacteria. Nat Nanotechnol 8, 369–375 (2013).
Chen, J. et al. Guidance of Stem Cells to a Target Destination in Vivo by Magnetic Nanoparticles in a Magnetic Field. ACS Appl Mater Interfaces 5, 5976–5985 (2013).
Li, Z. et al. A Smart Nanoassembly for Multistage Targeted Drug Delivery and Magnetic Resonance Imaging. Adv Funct Mater 24, 3612–3620 (2014).
Doerr, A. Clinical proteomics on target. Nat Meth 6, 560 (2009).
Ray, S. et al. Proteomic technologies for the identification of disease biomarkers in serum: Advances and challenges ahead. Proteomics 11, 2139–2161 (2011).
Beretta, L. Proteomics from the clinical perspective: many hopes and much debate. Nat Methods 4, 785–786 (2007).
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J. V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1, 2856–2860 (2006).
Wisniewski, J. R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat Methods 6, 359–362 (2009).
Hartmann, M. & Jung, D. Biocatalysis with enzymes immobilized on mesoporous hosts: the status quo and future trends. J Mater Chem 20, 844–857 (2010).
Yao, G., Deng, C., Zhang, X. & Yang, P. Efficient Tryptic Proteolysis Accelerated by Laser Radiation for Peptide Mapping in Proteome Analysis. Angew Chem 122, 8361–8365 (2010).
Slysz, G. W. & Schriemer, D. C. Blending Protein Separation and Peptide Analysis through Real-Time Proteolytic Digestion. Anal Chem 77, 1572–1579 (2005).
Xue, T. et al. Graphene-Supported Hemin as a Highly Active Biomimetic Oxidation Catalyst. Angew Chem Int Ed 51, 3822–3825 (2012).
Torres-Salas, P. et al. Immobilized Biocatalysts: Novel Approaches and Tools for Binding Enzymes to Supports. Adv Mater 23, 5275–5282 (2011).
Hudson, S., Cooney, J. & Magner, E. Proteins in Mesoporous Silicates. Angew Chem Int Ed 47, 8582–8594 (2008).
Liu, W.-L. et al. Novel trypsin-FITC@MOF bioreactor efficiently catalyzes protein digestion. J Mater Chem B 1, 928–932 (2013).
Ma, J. et al. Efficient proteolysis using a regenerable metal-ion chelate immobilized enzyme reactor supported on organic–inorganic hybrid silica monolith. Proteomics 11, 991–995 (2011).
Lee, J. et al. Magnetically-separable and highly-stable enzyme system based on crosslinked enzyme aggregates shipped in magnetite-coated mesoporous silica. J Mater Chem 19, 7864–7870 (2009).
Li, D., Teoh, W. Y., Gooding, J. J., Selomulya, C. & Amal, R. Functionalization Strategies for Protease Immobilization on Magnetic Nanoparticles. Adv Funct Mater 20, 1767–1777 (2010).
Qin, W. et al. Trypsin Immobilization on Hairy Polymer Chains Hybrid Magnetic Nanoparticles for Ultra Fast, Highly Efficient Proteome Digestion, Facile 18O Labeling and Absolute Protein Quantification. Anal Chem 84, 3138–3144 (2012).
Shen, Y. et al. Immobilization of trypsin via reactive polymer grafting from magnetic nanoparticles for microwave-assisted digestion. J Mater Chem B 1, 2260–2267 (2013).
Lee, H., Dellatore, S. M., Miller, W. M. & Messersmith, P. B. Mussel-Inspired Surface Chemistry for Multifunctional Coatings. Science 318, 426–430 (2007).
Lynge, M. E., van der Westen, R., Postma, A. & Stadler, B. Polydopamine-a nature-inspired polymer coating for biomedical science. Nanoscale 3, 4916–4928 (2011).
Liu, Y., Ai, K. & Lu, L. Polydopamine and Its Derivative Materials: Synthesis and Promising Applications in Energy, Environmental and Biomedical Fields. Chem Rev 114, 5057–5115 (2014).
Yang, S. H. et al. Mussel-Inspired Encapsulation and Functionalization of Individual Yeast Cells. J Am Chem Soc 133, 2795–2797 (2011).
Martin, M. et al. Preparation of core-shell Fe3O4@poly(dopamine) magnetic nanoparticles for biosensor construction. J Mater Chem B 2, 739–746 (2013).
Liu, Y. et al. Dopamine-Melanin Colloidal Nanospheres: An Efficient Near-Infrared Photothermal Therapeutic Agent for In Vivo Cancer Therapy. Adv Mater 25, 1353–1359 (2013).
Mandal, M. et al. Magnetite nanoparticles with tunable gold or silver shell. J Colloid Interface Sci 286, 187–194 (2005).
Kyeongse, S., Youngmin, L., Mi Ru, J., Ki Min, N. & Yong-Mook, K. Comprehensive design of carbon-encapsulated Fe3O4 nanocrystals and their lithium storage properties. Nanotechnology 23, 505401 (2012).
Thakur, V. K., Lin, M.-F., Tan, E. J. & Lee, P. S. Green aqueous modification of fluoropolymers for energy storage applications. J Mater Chem 22, 5951–5959 (2012).
Liu, Q., Yu, B., Ye, W. & Zhou, F. Highly Selective Uptake and Release of Charged Molecules by pH-Responsive Polydopamine Microcapsules. Macromol Biosci 11, 1227–1234 (2011).
Peng, G., Zhao, C., Liu, B., Ye, F. & Jiang, H. Immobilized trypsin onto chitosan modified monodisperse microspheres: A different way for improving carrier's surface biocompatibility. Applied Surface Science 258, 5543–5552 (2012).
Liu, J. et al. Highly Water-Dispersible Biocompatible Magnetite Particles with Low Cytotoxicity Stabilized by Citrate Groups. Angew Chem Int Ed 48, 5875–5879 (2009).
Acknowledgements
Research reported in this publication was partially supported by the National Cancer Institute of the National Institutes of Health under Award Number DP2CA174508. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
G.C. designed and performed the experiments. G.C. and S.Y.Z. conceived the idea, discussed and wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Cheng, G., Zheng, SY. Construction of a high-performance magnetic enzyme nanosystem for rapid tryptic digestion. Sci Rep 4, 6947 (2014). https://doi.org/10.1038/srep06947
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep06947
This article is cited by
-
Heterojunction catalysts g-C3N4/-3ZnO-c-Zn2Ti3O8 with highly enhanced visible-light-driven photocatalytic activity
Journal of Sol-Gel Science and Technology (2020)
-
BRAF protein immunoprecipitation, elution, and digestion from cell extract using a microfluidic mixer for mutant BRAF protein quantification by mass spectrometry
Analytical and Bioanalytical Chemistry (2019)
-
A facile hydrothermal synthesis of CeO2 nanocubes decorated ZnO nanostructures: optical and enhanced photocatalytic properties
Journal of Materials Science: Materials in Electronics (2019)
-
Application of polydopamine in biomedical microfluidic devices
Microfluidics and Nanofluidics (2018)
-
Hyaluronic acid-modified [19F]FDG-conjugated magnetite nanoparticles: in vitro bioaffinities and HPLC analyses in organs
Journal of Radioanalytical and Nuclear Chemistry (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.