Abstract
Thaumarchaeota and the gene encoding for a subunit of ammonia monooxygenase (amoA) are ubiquitous in Polar Seas and some Thaumarchaeota also have a gene coding for ureC, diagnostic for urease. Using quantitative PCR we investigated the occurrence of genes and transcripts of ureC and amoA in Arctic samples from winter, spring and summer. AmoA genes, ureC genes and amoA transcripts were always present, but ureC transcripts were rarely detected. Over a 48 h light manipulation experiment amoA transcripts persisted under light and dark conditions, but not ureC transcripts. In addition, maxima for amoA transcript were nearer the surface compared to amoA genes. Clone libraries using DNA template recovered shallow and deep amoA clades but only the shallow clade was recovered from cDNA (from RNA). These results imply environmental control of amoA expression with direct or indirect light effects and rare ureC expression despite its widespread occurrence in the Arctic Ocean.
Similar content being viewed by others
Introduction
The global nitrogen cycle depends on the remineralization of organic nitrogen to ammonium and nitrate, with microbially mediated ammonia oxidation by ammonia monooxygenase being a key intermediate step. This process is commonly inferred by the presence of the ammonia monooxygenase α-subunit (amoA). The amoA gene, found in some Bacteria and some Archaea, is part of a larger gene family of copper-containing membrane-bound monooxygenases1. Evidence to date indicates that oceanic archaeal amoA is involved in converting ammonia to nitrite, a rate limiting step in aerobic nitrification2. The discovery of non-extremophile marine group I (MGI) Archaea that could grow chemolithoautotrophically by aerobically oxidizing ammonia to nitrite3 and the occurrence of the amoA gene in other uncultivated, non-extremophile MGI4,5, provided an added perspective on the nitrogen cycle. The widespread detection of both MGI and archaeal amoA in the ocean combined with the rarity of bacterial nitrifiers has led to the new paradigm that Archaea are largely responsible for marine nitrification6,7,8 and in particular may contribute substantially to nitrification in the upper few hundred meters of the ocean9. Interestingly, nitrification by bacteria is inhibited by light, but ammonia oxidizing Archaea (AOA) are often retrieved from the euphotic zone and the effect of light on amoA is not resolved10,11.
MGI are sufficiently distinct that they are considered a phylum of Archaea, Thaumarchaeota12, but the diversity and metabolic capacity of these largely uncultivated microorganisms remains to be fully elucidated. In addition to potentially oxidizing ammonia, genomic surveys indicate that some MGI may use urea, broken down to ammonium (NH4+) and inorganic carbon (C), as an energy and carbon source. The genes that code for urease have been reported in Cenarchaeum symbiosum4 and “Candidatus Nitrosopumilus sediminis” AR2 from Svalbard13. Recently ureC, the gene that codes for a subunit of urease has been amplified from the deep Mediterranean Sea14 and detected in metagenomes from a deep hydrothermal plume15 and the deep Pacific Ocean16. The ureC gene has also been reported from surface waters of the Gulf of Maine17 and in the winter Arctic Ocean18. Alonso-Saez and colleagues18 amplified the ureC gene from deep Antarctic waters, but they failed to detect it in summer Antarctic surface waters. In that same study the Archaeal amoA gene was amplified from the same surface waters, suggesting that the potential for MGI Archaea to use urea is not universal. In fact, ureC and the urease are not found in cultivated N. maritimus19.
Over much of the Arctic, nitrogen availability limits photosynthetic productivity, with nitrate rapidly depleted in surface waters during the short summer growing season. However, nitrate concentrations are substantial in and below the halocline20. Interestingly, Northern Baffin Bay stands out from much of the Arctic, with high photosynthetic rates continuing throughout the Arctic summer, despite low standing stocks of inorganic nitrogen in the euphotic zone21,22. Galand and collegues23 found that MGI and archaeal amoA genes were abundant in Northern Baffin Bay, particularly in the Arctic halocline, leading to the hypothesis that the archaeal community could influence photosynthetic community structure by rapidly recycling inorganic nitrogen in the euphotic zone. Nitrate concentrations in the surface waters of Baffin Bay are often below the detection limits (ca. 0.03 μmol N L−1)24,25, but ammonium concentrations can be substantial26, suggesting bacterial degradation of fixed organic material and possible release of ammonium. The high affinity of MGI Archaea for ammonium27 could enable MGI to successfully compete with phytoplankton for ammonium and result in amoA expression nearer the surface. In contrast to NH4, there have been few measures of urea in the Arctic, but in general, concentrations are variable and sporadic28,29, which leads to the hypothesis that expression of genes coding for urease would also be variable. The presence of archaeal ureC genes in Arctic MGI Archaea is intriguing18 but to date there is no evidence of ureC being expressed in polar waters.
The main objective of our study was to quantify the amoA and ureC transcripts in three regions of the Arctic Ocean (Amundsen Gulf, Lancaster Sound and Baffin Bay) and to gain an understanding of conditions associated with gene expression. To this end, we investigated amoA and ureC transcripts by qPCR during polar darkness and around the vernal equinox. We then experimentally investigated amoA and ureC expression in natural communities exposed to typical irradiances of the Arctic Ocean euphotic zone. Finally we investigated the depth distribution of genes and transcripts from MGI Thaumarchaeota, archaeal amoA and ureC, along a north to south gradient of the hydrographically complex center of the Northern Baffin Bay30, targeting water masses with different bio-physical properties. Phylogenetic differences between the more likely active and more likely dormant Thaumarchaeota with amoA were also examined through cloning and sequencing the amoA gene from both DNA and cDNA (from total RNA).
Results
Effect of light on ureC and amoA transcripts
In the Amundsen Gulf region (Western Canadian Arctic), amoA, transcripts, ureC transcripts and MGI 16S rRNA were quantified on 3 dates as ambient light conditions increased over time (January, March and the beginning of April). In surface waters, amoA and MGI copies mL−1 were greatest in January. At the halocline, more transcripts were recorded in April. There were always more transcripts at the halocline than at surface. UreC transcripts were only detected in January (Figure 1).
In the initial samples from Lancaster Sound (Eastern Canadian Arctic) used for the light-manipulation experiment, amoA transcripts were detected at T0 from both 1% (80 m deep) and 25% (25 m shallow) surface irradiance (Figure 2), with an order of magnitude (ca. 105 versus 104) fewer transcripts near the surface. UreC transcripts were successfully quantified from the freshly collected deeper sample (T0), with 10 copies per mL of seawater, but not in the sample from 25% irradiance. There were no significant changes in MGI 16S rRNA copies mL−1 over the 48 hours and no effect of light (3 way ANOVA) on MGI 16S rRNA (Figure 2). There were, however, significantly more copies of MG1 16S rRNA in the deeper sample compared to the 25 m water over the 48 h incubation (P < 0.001).
The amoA transcripts were significantly greater in the dark than in the light overall, but the effect of collection depth was not significant (see below). The multiple pairwise comparisons revealed several interesting trends. After 24 hours, amoA transcripts were always greater in the dark (ANOVA- pairwise multiple comparison, P = 0.008) compared to bottles exposed to light. After 48 h, the amoA transcripts from the two depths diverged, with a significant increase under both light and dark conditions in the 25 m origin treatments (P < 0.001), conversely in the 80 m origin treatments, amoA transcripts decreased significantly (P < 0.001) (Figure 2). Transcripts of ureC were not detected in the experimental treatments.
Environmental and biological conditions in Baffin Bay
The Baffin Bay water columns (Stns. 129, 123 and 109) were density stratified with a subsurface chlorophyll maximum (SCM) just above a pycnocline (from the maximum rate of change in sigma theta) at 25–30 m (Figure 3). Secondary chlorophyll fluorescence peaks were also detected at ca. 70–100 m. The highest extracted Chl a values were 1.4 μg L−1 at Stn 129–22 m and 1.3 μg L−1 at Stn 109–25 m. Most other values were below 1 μg L−1 (Supplementary Table S1). Dissolved oxygen concentrations ranged from 279 to 356 μmol kg−1 (Figure 3). At Stn123, there was a low oxygen intrusion between 200 and 300 m (Figure 3) coinciding with slight increases in salinity and temperature (not shown). Bacteria concentrations were ca. 105 cells mL−1 and tended to decrease with depth, with slightly higher concentrations above the pycnocline (Supplementary Table S1). Nitrite concentrations ranged from 0.05 to 0.17 μmol kg−1, nitrate concentrations ranged from below the detection to 15.34 μmol kg−1 (Supplemenary Table S1). The nitrite maxima were above nitrate maxima and between 100–200 m at the three stations (Fig. 4). Ammonium concentrations were variable with higher values in the upper waters (Supplementary Table 1).
Depth profiles of Northern Baffin Bay Stations 129, 123 and 109.
Density (sigma theta) is indicated by the solid black line, circles embedded in these lines show sample depths for DNA and RNA (open circles) and for DNA only (filled circles). In situ chlorophyll fluorescence (relative units) is shown by the green line and oxygen concentrations (μmol kg −1) by the dotted line.
In September 2006, the sun was above the horizon for ca. 15 h on Sept. 11 to ca. 12 h on Sept. 20. Surface values of photosynthetically active radiation (PAR) were lowest at station 129 (1.74 μmol photons m−2 s−1), which was sampled at 05:17 UTC, during twilight prior to apparent sunrise at the latitude of sampling. The other three stations were sampled with the sun higher above the horizon and surface PAR values were much greater, especially at Station 109, sampled on a clear day ca. 4 h after apparent sunrise (Table 1). Subsurface PAR values for samples collected at depth were consequently greater at Stn 109 compared to the other stations.
Transcript and gene quantification in the water column
MGI 16S rRNA (from cDNA) qPCR values ranged from ca. 175 to 4.5 × 105 copies mL−1 of seawater collected and amoA transcripts from 18 to 1829 copies mL−1 (Supplementary Fig. S1). In contrast to amoA, where transcripts were detected in all samples tested, ureC transcripts were never detected from the Baffin Bay samples.
Baffin Bay amoA transcripts varied down the water column and between stations. The station (Stn) 129 maximum for amoA transcripts was at 35 m, which was above the nitrite maximum (Figure 4; note the log scale for depth). For Stn 123, the maximum amoA transcripts were just below the nitrite maximum. Finally for Stn 109, the amoA transcript maximum was at 90 m, which was above the nitrite maximum (Fig. 4).
In Baffin Bay, gene abundance (from DNA) also varied with depth and station. Stn 129 maxima for amoA, ureC and MGI 16S rRNA genes were detected in the 120 m sample (Figure 5a). At Stn. 123, the maxima for MGI, amoA and ureC genes were at 280 m and coincided with the nitrate maximum at the base of a low oxygen intrusion (Figs. 3 and 5a). At station 109, gene copies of MGI, amoA and ureC were also all greatest nearer the nitrate maximum at 195 m (Fig. 5a).
Panel (a) Northern Baffin Bay, qPCR copies mL−1 of MGI, amoA and ureC genes (DNA template). Station number and depth of sampling are indicated along the X axis. Panel (b) Log-log plots of gene copies of ureC (open circles) and amoA (closed circles) compared the 16S rRNA genes of MGI. Lines indicate significant linear regressions of the log transformed data; the dashed line is ureC versus MG1 (r2 = 0.76, p < 0.001), the solid line is amoA versus MGI (r2 = 0.78, p < 0.001). Standard errors of triplicates and standards were smaller than the size of the symbols.
Baffin Bay qPCR values from DNA template ranged from 16 to 3756 copies of MGI (16S rRNA) genes mL−1, 24 to 5270 amoA gene copies mL−1 and 45 to 2780 ureC gene copies mL−1. Overall amoA gene copy numbers were significantly correlated with MGI rRNA genes (Tau, T = 0.74, p < 0.001) as were ureC gene copies to MGI rRNA genes (T = 0.66, p < 0.001) (Fig. 5b). UreC gene copy numbers also correlated with amoA gene copies (T = 0.79, p < 0.001) and genes followed depth related variables, such as silica, phosphate and nitrate (Supplementary Fig. S2). The ratio of amoA genes to MGI 16S rRNA genes was on average 1.8 ranging from 0.2 to 5.5. There were fewer ureC gene copies than amoA copies with an average ratio of 0.4 ureC genes to MGI 16S rRNA genes (Figure 5b).
The maximum abundance of amoA transcripts did not correspond to the same depths as the maximum gene copies in any of the water column profiles. Furthermore there was no significant correlation between archaeal amoA transcripts and gene copy numbers estimated from the DNA template, nor between MG1 16S rRNA genes and 16S rRNA copies from cDNA.
Across stations and depths from both sides of the Canadian Arctic we found a significant positive relationship between MG1 16S rRNA and amoA transcripts (analysis on log transformed data with r2 = 0.80 and p < 0.001) (Supplementary Fig. S1). The PCA indicated that nutrients, including silica and phosphate, which were associated with the deepest samples, best explained gene concentrations along the X axis. Chl a and ammonium were more associated with amoA transcripts and MGI 16S rRNA (Supplementary Fig. S2).
Diversity and phylogeny of amoA in Baffin Bay
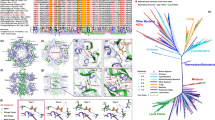
A total of 423 cloned amoA gene sequences with the correct target size (635 nt) from 8 libraries passed chimera and other quality tests. The two libraries constructed using cDNA as a template were asymptotic. There were five OTUs at the 97% similarilarity level out of 32 clones at Stn 123–200 m and a single OTU from 46 clones at station 109–110 m (Table 2). The DNA libraries were also asymptotic. At 97% similarity, there were 4 OTUs out of 19 clones from Station 109–110 m. Only 6 clones were recovered from the shallowest library (station 129–35 m) and each clone was unique. At the two stations and depths where both DNA and cDNA were used as templates, the DNA derived communities were more diverse (Table 2). All closest matches were to uncultured environmental archaeal amoA genes (Fig. 6).
Distance tree of the amoA gene from the Northern Baffin Bay.
Major clades A and B from (Francis et al. 2005) are indicated; A.a coincides with A in Kalanetra et al. (2009). All reference sequences except N. maritimus were from open water (Francis et al. 2005). Sequence names in this study consist of station number and sample depth followed by d for DNA or r for RNA (from cDNA).
Maximum likelihood (not shown) and Neighbor joining phylogenies (Figure 6) placed all of the amoA sequences into previously defined marine groups, corresponding to clades A and B defined by Francis et al.31, with no sequences within their group C. All amoA sequences from RNA source libraries grouped within clade A, which is thought to represent shallow ecotypes32. The most common cDNA sourced amoA sequences grouped together in a sub-clade that we annotated A.A, which corresponds to the group A of Kalanetra et al.33 (Figure 6). Our sub-clade A.A sequences were > 99% similar to other polar sequences (not shown). At Stn 123–200 m, amoA clones from two clusters (indicated as A.B. and A.A.a in Figure 6) were most often from cDNA (representing > 50% of cDNA clones). At Stn. 109–110 m, only A.A.a was retrieved from cDNA. A third Arctic sub-cluster in the A clade (A.C. in Figure 6) had no cDNA representatives. DNA sequences fell into both A and B clades32. Overall 40% of the sequences from the DNA libraries fell into the B clade.
There was some amoA clone clustering according to depth. The shallow A.A.a group was, for example, was absent from the 400 m sample and one deep B sub-clade was overrepresented in the deepest samples (Figure 6).
Discussion
Taken together, the lines of evidence suggest that amoA expression was influenced by light but amoA containing MGI persisted in the upper waters of the Arctic Ocean and were possibly more active when irradiance (PAR) was lower. Our qPCR results indicated that amoA transcripts were more abundant in surface waters during January, the darkest winter month and later more abundant in the halocline where light levels were lower. In the light manipulation experiment amoA transcript levels were significantly higher in the dark. We found that actual irradiance levels at the time of sample collection likely influenced transcript abundance, with more transcripts detected in samples from the euphotic zone (defined as >1% of surface PAR), when surface irradiance was lower. Light may inhibit amoA activity directly, promote more rapid degradation of transcripts, or have secondary influence on substrate availability, for example phytoplankton may be better able to compete with ammonium oxidizing Archaea for ammonium when light levels are greater. Similarly, in the Pacific, Church et al.34 reported peak amoA transcript abundance at the bottom of the euphotic zone.
Our results indicated that amoA transcriptional activity probably occurs throughout winter. Compared to other oceanic regions, Arctic and Antarctic MGI 16S rRNA genes are more abundant nearer the surface, especially in winter and early spring35, with values lower in summer36. Collins et al.37 also reported MGI 16S rRNA genes in newly formed sea ice in the Beaufort Sea in October when light levels are rapidly decreasing. The lower expression that we observed nearer the surface compared to the halocline in March and April could be due to direct light inhibition or lack of substrate. The net effect was a decline in amoA near surface activity in spring when phytoplankton activity begins38,39.
In Baffin Bay, distribution profiles of the amoA transcripts down the water column were closely correlated with the MGI 16S rRNA indicating that the MGI were in a state consistent with being metabolically active and with ammonium oxidation having occurred around the time of sampling. This was notable since the transcript distribution differed from the gene distribution (from DNA) (Figure 3); suggesting that environmental gene distribution may not accurately reflect activity. In the Pacific, Church et al.34 reported similar results with relatively more amoA transcripts nearer the surface, despite higher gene numbers at depth. The relationship between the euphotic zone and nitrification activity was reported in a North-South transect of the Atlantic Ocean, ammonia oxidizing activity was greatest at the base of the euphotic zone and decreased with depth40. But this depth dependent distribution, did not occur in the Black Sea and the highest expression of amoA is within a nitrification zone coinciding with low oxygen levels41. At Stn 123, where we sampled a lower oxygen layer, amoA transcripts were high compared to other stations sampled at the same depth, but higher still at the base of the euphotic zone. While we did not directly measure nitrification, we speculate that in the Arctic amoA gene maximum activity differed from DNA gene distribution due to substrate availability over relevant time spans, with amoA transcripts associated with Chl a and ammonium concentrations in the PCA analysis (Supplementary Fig. S2). In Baffin Bay, dissolved organic matter (DOM) is released by phytoplankton leakage21 and zooplankton and protist grazing42. This DOM is quickly broken down by bacterial activity43. The higher probable microbial activity at the base of the euphotic zone stands out as the overiding factor associated amoA transcript abundance. In contrast, the surface MGI populations, are most likely maintained by diurnal activity during periods of low incoming solar radiation each day.
In early September, the greatest concentrations of MGI 16S rRNA and amoA genes in Baffin Bay were at the nitrate maximum, similarly in the Pacific, the same two genes tracked nitrate concentrations down the water column11. However, our PCA showed that phosphorus and silica, with higher concentrations in deeper waters, had a stronger relationship with gene copy numbers in Northern Baffin Bay, consistent with the oceanography of the region21. The abundance of genes, but not transcripts, below the subsurface chlorophyll maximum layer may be linked to loss rates. For example, protists that graze on prokaryotes are also more abundant above the nitracline44. Alternatively, the lower numbers of amoA transcripts per MGI 16S RNA gene deeper in the water column could also be due to inherent biological properties of dominant clades in deeper waters45.
For ureC, we detected the gene at all depths and stations and in a parallel study, the gene was reported in Amundsen Gulf in March and February18, indicating that MGI with the ureC gene are widely distributed in the Arctic, irrespective of season or depth. In contrast to amoA transcripts, which were detected in all samples, we only recorded ureC transcripts in samples from January, when photosynthetic primary production was not possible because there is no light and from a sample, collected from the bottom of the euphotic zone in Lancaster Sound, which is a region of high biological activity25.
The phylogeny of the amoA gene implies the presence of at least two distinct MGI ecotypes. All amoA transcripts grouped within the clade A31, which is classed as a shallow group32. More specifically, in our clone libraries the most abundant cDNA amoA clones were closely related to each other and within a single sub-clade (A.A). This sub-clade was originally identified by Kalanetra et al33 from the central Arctic Ocean, with representatives from the Southern Ocean and may be typical for cold waters. In contrast, amoA sequences from cluster B, which is thought to be more representative of the deep ocean32, were never recovered from cDNA clone libraries suggesting this clade was dormant in Baffin Bay at the time of sampling. DNA based and cDNA based clone libraries from the central California Current recovered both A and B clusters and the A cluster dominated in all but the deepest samples9. Recently Sintes et al.45 explored amoA gene diversity using amoA primers designed to detect two distinct clades of the AOA; one associated mostly with less deep waters and high ammonium concentrations (HAC) and the second clade, occurring in deeper waters and with low ammonium concentrations (LAC). Both primer pairs detected high concentrations of amoA HAC and LAC in an Arctic coastal site with co-occurrence down the water column.
The co-occurrence of the two AOA clusters in the Arctic Ocean, even near the surface, is intriguing. Inspecting gene ratios from our qPCR results we note the potential of 2 copies of the amoA gene within ca. 60% of the MGI (with a 1.8 to 1 ratio of amoA genes to 16S rRNA genes), but fewer MGI containing the ureC gene (with a ratio of 0.4). In effect, the persistence of a dormant B clade at similar occurrence rates (40% of our DNA template clones) and the observation of a ureC gene to MGI 16S rRNA gene ratio of 0.4 may be linked.
Apart from its initial discovery in the coral reef endosymbiont46, the archaeal ureC gene has been most often reported from deeper waters or winter surface waters where concentrations of fresh organic material would seem to be low47. The sporadic amplification of ureC transcripts over a range of samples from the Arctic, is telling. As mentioned earlier, there appeared to be a light effect, but the rarity and weak concentrations of ureC transcripts may also be a result of the ephemeral occurrence of urea itself in the Arctic environment29. Zooplankton are the main source of urea in the Arctic, with some copepod species persisting and feeding near the surface and at mid-depths throughout the Arctic winter48. Interestingly, one copepod species, Acartia tonsa, releases different forms of nitrogen depending on diet; with a mixed diet of heterotrophic protists and diatoms resulting in greater release of urea compared to either dietary source alone49. Copepod species in the Arctic are mostly opportunists and able to switch to the dominant food source50 and thus the taxonomy of the protist plankton prey could also result in the variable urea concentrations. The episodic but poorly understood input of urea into the surrounding waters could be sufficient for maintaining Thaumarchaeota with the capacity to scavenge and use urea as a carbon and energy source. While the archaeal ureC gene has been reported in the Arctic and elsewhere18, transcriptional activity to date has only been reported in very deep waters14 and a coral symbiont4. The similarity between the two types of environments is not obvious, except possibly the availability of animal generated urea.
Overall we found evidence of two MGI populations, one active nearer the surface and a second, deeper more dormant population, which may be reliant on sporadic inputs of urea as a source of energy and carbon. Finally, while amoA transcript abundance was greater in the dark, light inhibition was not complete and transcripts persisted under low irradiance. The distribution of AOA in the Arctic suggests multiple links between biological and physical properties in this region.
Methods
Sampling
Samples were collected from Amundsen Gulf during the Circumpolar Flaw Lead System Study (CFL)51 in winter 2008–2009, the North Water region of Northern Baffin Bay between Ellesmere Island and Greenland from 11–17 September 2006 and from near Lancaster Sound on 20 Sept 2006 (Table 1). Samples for both missions were collected from the research icebreaker CCGS Amundsen. Water from the Amundsen Gulf was sampled from 2 depths: surface (ca. 10 m) and at the halocline that is indicative of Pacific origin waters39 (ca. 50 m) in January, March and April. The three Northern Baffin Bay stations, 109, 123 and 129, were sampled along a south-north transect. Since the cruise was multidisciplinary, with constrained logistics, samples were taken opportunistically over the 24 h period (Table 1). All seawater samples were collected with 12-L Niskin type bottles (Ocean Test Equipment) mounted on carousel rosette system equipped with a SBE-911 plus conductivity-temperature-depth (CTD) profiler with a dissolved oxygen sensor. Other instruments on the rosette system recorded fluorescence (Seapoint), transmissivity (WetLabs C-Star Transmissometer), photosynthetically active radiation (PAR; Biospherical Instruments) and relative nitrate concentrations from an in situ ultraviolet spectrometer (ISUS) probe (Satlantic). Water sample depths were selected on the downward cast52. In particular, we followed the nitrate profile and targeted peak and minimum concentrations indicated from the ISUS trace. Eight depths at each station were selected for DNA and four to six for RNA analysis. DNA and RNA samples were filtered separately, both using a peristaltic pump system. Both filtrations included an in-line 3 μm pre-filtration through a 47 mm polycarbonate filter. The entire system used to filter RNA was treated with RNaseZap® prior to filtration. One to seven L were filtered for RNA at 4°C in the dark, with a maximum filtration time of 3 hours. Samples for RNA were collected on 47 mm diameter 0.2 μm pore size polycarbonate (PC) filters, which were placed in 1.8 mL cryovials with RLT buffer (QIAGEN RNeasy) mixed with 1% beta-mercaptoethanol. The vials were immediately flash frozen in liquid nitrogen and stored at −80°C. For DNA, six L of water were filtered through 0.2 μm Sterivex filter units (Millipore) and stored in buffer at −80°C23.
Nutrient concentrations: nitrite (NO2−), nitrate (NO3−), silicic acid (Si(OH)4) and soluble reactive phosphate (SRP), were determined on board with a Bran and Luebbe Autoanalyzer III using standard protocols53 and detection limit of 0.03 μmol L−1 for the three nutrients. Ammonium was determined manually with the sensitive fluorometric method of Holmes et al.54 and a detection limit of 0.01 μmol L−1. Oxygen values from the CTD were calibrated at the Institut National Researche Scientifique in Québec, Canada, using samples taken at discrete depths. Samples for total chlorophyll a (Chl a) were collected onto Whatman GF/F filters. These were stored at −80°C until extraction in 95% ethanol55 and concentrations determined by spectrofluorometry. Flow cytometry (FCM) samples were fixed with paraformaldehyde, flash frozen in liquid nitrogen and stored at −80C56,57. This method does not distinguish Bacteria and Archaea and we use the term bacteria for convenience.
Light manipulation experiment
Samples from two photic depths, 25% and 1% of surface PAR, were collected from near Lancaster Sound, where Arctic water enters Northern Baffin Bay (Stn 301, Table 1). The water from 25 m (25% PAR) and 80 m (1% PAR) was collected into 18 1−L PC bottles and incubated at 4°C, at either in the dark or at 0.60 μmol photons m−2 s−1 in bottles shaded with blue filters, which is typical for September at the 25% photic depth (25% of surface PAR) in the study region. One bottle from each depth was immediately filtered for RNA, as above. After incubations of 24 and 48 h, two replicate bottles for each treatment were filtered as above.
Molecular biological techniques
DNA was extracted using a salt extraction method58 as modified by Harding et al.59 and RNA extracted as in Church et al.60. RNA extracts were reverse transcribed and PCR amplified (RT-PCR) by using a high capacity cDNA archive kit (Applied Biosystems) as specified by the manufacturer.
Quantitative PCR was performed on a Dyad Disciple thermal cycler with Chromo 4 Real-Time Detector (Bio-Rad) in 96 well opaque qPCR plates with adhesive seals (Bio-Rad). Primers and qPCR conditions, as well as amoA, ureC and MGI positive controls and standard curves were the same as in Alonso-Saez et al.18. Primer specificity was verified by running PCR products on gels and sequencing the resulting single bands from the individual primer sets. The triplicate standard curves and negative controls (without DNA added) were subjected to qPCR along with the samples. Copy number per mL of seawater was calculated as described in Zhu et al.61.
Eight clone libraries targeting the archaeal amoA gene and transcripts were constructed using Arch-amoAF and Arch-amoAR primers31. Three separate PCR reactions containing 2, 3 or 4 μL of template were amplified to counter primer competition and increase the probability of recovering common gene variants62,63. The three PCR products were pooled and then purified with Qiaquick PCR Purification Kit (Qiagen) and cloned using the TA cloning kit (Invitrogen) according to the manufacturer's instructions. Inserts were amplified with the vectors' M13 primers and resulting amplicons were analyzed by restriction fragment length polymorphism (RFLP) following digestion with RsaI (New England Biolab). Up to 10 clones per pattern were sequenced to cover potential RFLP variants. Sequencing (single read) used the vector's T7p universal primer and was performed at Centre Hospitalier de l'Université Laval (CHUL), Québec, with an ABI 3730xl system (Applied Biosystems).
The sequences were compared to the NCBI GenBank nr/nt database using NCBI BLAST64. Non-redundant nucleotide and translated amino acid sequences are deposited under accession numbers HQ713534-HQ713542 for the cDNA clones and JF272615-JF272702 for the DNA clones.
Phylogenetic and statistical analysis
In addition to the 202 archaeal amoA gene sequences from this study, 148 reference sequences were selected from Francis et al.31 and the resulting 350 sequences were aligned using Clustal X v.1.83, then manually checked and trimmed using GeneDoc, v. 2.6.0.2. After trimming, additional sequences including nearest matches to our sequences, were added for phylogenetic tree construction using the DNAML program from PHYLIP (available at http://evolution.genetics.washington.edu/phylip.html). The topology of dendrograms using maximum likelihood (ML) and neighbor-joining (NJ) were similar. This larger tree placed all of our sequences fell into Marine Group I (MGI), clades A and B defined in Francis et al.31. Non-marine and redundant sequences from our study were then removed from the tree leaving only representative A and B sequences in the final tree where evolutionary history was inferred by NJ, with evolutionary distances computed using the Maximum Composite Likelihood method. Phylogenetic analyses were conducted in MEGA465.
To estimate library coverage using Chao1, sequences were clustered at a similarity level ≥97% using DOTUR66. For the light experiment, significance of the 3 way (depth of collection, light level and time of incubation) ANOVA was tested on log transformed qPCR data following a general linear model after testing for main effects. Pairwise multiple comparisons using the Holm-Sidak method were used to test the significance of interactions.
Kendall's correlation analysis was used to test correlations among all quantitative variables from Northern Baffin Bay. A principal components analysis (PCA) was conducted using environmental and qPCR data and plotted in a coordinate system, given by the two main components, to explore possible relationships between variables. Missing data were handled by pairwise deletion. PCA was done with PAST67. ANOVA, Kendall's correlation and regression analysis on log transformed qPCR data were carried using SigmaPlot version 11 (Systat Software Inc).
Additional information
Accession codes: Non-redundant nucleotide and translated amino acid sequences are deposited under accession numbers HQ713534-HQ713542 for the cDNA clones and JF272615-JF272702 for the DNA clones.
Change history
07 July 2015
A correction has been published and is appended to both the HTML and PDF versions of this paper. The error has not been fixed in the paper.
References
Pester, M., Schleper, C. & Wagner, M. The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr. Opin. Microbiol. 14, 300–306, 10.1016/j.mib.2011.04.007 (2011).
Bouskill, N. J., Eveillard, D., Chien, D., Jayakumar, A. & Ward, B. B. Environmental factors determining ammonia-oxidizing organism distribution and diversity in marine environments. Environ. Microbiol. 14, 714–729, 10.1111/j.1462-2920.2011.02623.x (2012).
Konneke, M. et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546, 10.1038/nature03911 (2005).
Hallam, S. J. et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proceed. Natl. Acad. Sci. USA 103, 18296–18301, 10.1073/pnas.0608549103 (2006).
Wuchter, C. et al. Archaeal nitrification in the ocean. Proceed. Natl. Acad. Sci. USA 103, 12317–12322, 10.1073/pnas.0600756103 (2006).
Beman, J. M., Sachdeva, R. & Fuhrman, J. A. Population ecology of nitrifying Archaea and Bacteria in the Southern California Bight. Environ. Microbiol. 12, 1282–1292, 10.1111/j.1462-2920.2010.02172.x (2010).
Francis, C. A., Beman, J. M. & Kuypers, M. M. M. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 1, 19–27, 10.1038/ismej.2007.8 (2007).
Santoro, A. E. & Casciotti, K. L. Enrichment and characterization of ammonia-oxidizing archaea from the open ocean: phylogeny, physiology and stable isotope fractionation. ISME J. 5, 1796–1808, 10.1038/ismej.2011.58 (2011).
Santoro, A. E., Casciotti, K. L. & Francis, C. A. Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ. Microbiol. 12, 1989–2006, 10.1111/j.1462-2920.2010.02205.x (2010).
Hu, A. Y., Jiao, N. Z., Zhang, R. & Yang, Z. Niche Partitioning of Marine Group I Crenarchaeota in the Euphotic and Upper Mesopelagic Zones of the East China Sea. Appl. Environ. Microbiol. 77, 7469–7478, 10.1128/aem.00294-11 (2011).
Mincer, T. J. et al. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ. Microbiol. 9, 1162–1175, 10.1111/j.1462-2920.2007.01239.x (2007).
Brochier-Armanet, C., Boussau, B., Gribaldo, S. & Forterre, P. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6, 245–252, 10.1038/nrmicro1852 (2008).
Park, S.-J. et al. Draft Genome Sequence of an Ammonia-Oxidizing Archaeon, “Candidatus Nitrosopumilus sediminis” AR2, from Svalbard in the Arctic Circle. J. Bacteriol. 194, 6948–6949, 10.1128/JB.01869-12 (2012).
Yakimov, M. M. et al. Contribution of crenarchaeal autotrophic ammonia oxidizers to the dark primary production in Tyrrhenian deep waters (Central Mediterranean Sea). ISME J. 5, 945–961, 10.1038/ismej.2010.197 (2011).
Baker, B. J., Lesniewski, R. A. & Dick, G. J. Genome-enabled transcriptomics reveals archaeal populations that drive nitrification in a deep-sea hydrothermal plume. ISME J. 6, 2269–2279, 10.1038/ismej.2012.64 (2012).
Konstantinidis, K. T., Braff, J., Karl, D. M. & DeLong, E. F. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at Station ALOHA in the North Pacific Subtropical Gyre. Appl. Environ. Microbiol. 75, 5345–5355, 10.1128/aem.00473-09 (2009).
Tully, B. J., Nelson, W. C. & Heidelberg, J. F. Metagenomic analysis of a complex marine planktonic thaumarchaeal community from the Gulf of Maine. Environ. Microbiol. 14, 254–267, 10.1111/j.1462-2920.2011.02628.x (2012).
Alonso-Saez, L. et al. Role for urea in nitrification by polar marine Archaea. Proceed. Natl. Acad. Sci. USA 109, 17989–17994, 10.1073/pnas.1201914109 (2012).
Walker, C. B. et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proceed. Natl. Acad. Sci. USA 107, 8818–8823, 10.1073/pnas.0913533107 (2010).
Carmack, E. C. The Alpha/Beta ocean distinction: A perspective on freshwater fluxes, convection, nutrients and productivity in high-latitude seas. Deep-Sea Res. Pt. II 54, 2578–2598, 10.1016/j.dsr2.2007.08.018 (2007).
Mei, Z. P. et al. Physical control of spring-summer phytoplankton dynamics in the North Water, April-July 1998. Deep-Sea Res. Pt. II 49, 4959–4982, doi.org/10.1016/S0967-0645(02)00173-X (2002).
Lewis, E. L., Ponton, D., Legendre, L. & LeBlanc, B. Springtime sensible heat, nutrients and phytoplankton in the Northwater Polynya, Canadian Arctic. Cont. Shelf Res. 16, 1775–+, 10.1029/JC088iC04p02565 (1996).
Galand, P. E. et al. Archaeal diversity and a gene for ammonia oxidation are coupled to oceanic circulation. Environ. Microbiol. 11, 971–980, 10.1111/j.1462-2920.2008.01822.x (2009).
Tremblay, J. E., Gratton, Y., Fauchot, J. & Price, N. M. Climatic and oceanic forcing of new, net and diatom production in the North Water. Deep-Sea Res. Pt. II 49, 4927–4946, 10.1016/s0967-0645(02)00171-6 (2002).
Martin, J., Tremblay, J. E. & Price, N. M. Nutritive and photosynthetic ecology of subsurface chlorophyll maxima in Canadian Arctic waters. Biogeosci. 9, 5353–5371, 10.5194/bg-9-5353-2012 (2012).
Tremblay, J. E. et al. Trophic structure and pathways of biogenic carbon flow in the eastern North Water Polynya. Prog. Oceanogr. 71, 402–425, 10.1016/j.pocean.2006.10.006 (2006).
Martens-Habbena, W., Berube, P. M., Urakawa, H., de la Torre, J. R. & Stahl, D. A. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461, 976–U234, 10.1038/nature08465 (2009).
Codispoti, L. A. An oceanic fixed nitrogen sink exceeding 400 Tg Na-1 vs the concept of homeostasis in the fixed-nitrogen inventory. Biogeosci. 4, 233–253, 10.5194/bg-4-233-2007 (2007).
Simpson, K. G., Tremblay, J. E., Gratton, Y. & Price, N. M. An annual study of inorganic and organic nitrogen and phosphorus and silicic acid in the southeastern Beaufort Sea. J. Geophy. Res.-Oceans 113, C07016 10.1029/2007jc004462 (2008).
Bacle, J., Carmack, E. C. & Ingram, R. G. Water column structure and circulation under the North Water during spring transition: April-July 1998. Deep-Sea Res. Pt. II 49, 4907–4925, 10.1016/s0967-0645(02)00170-4 (2002).
Francis, C. A., Roberts, K. J., Beman, J. M., Santoro, A. E. & Oakley, B. B. Ubiquity and diversity of ammonia-oxidizing Archaea in water columns and sediments of the ocean. Proceed. Natl. Acad. Sci. USA 102, 14683–14688, 10.1073/pnas.0506625102 (2005).
Beman, J. M., Popp, B. N. & Francis, C. A. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2, 429–441, 10.1038/ismej.2007.118 (2008).
Kalanetra, K. M., Bano, N. & Hollibaugh, J. T. Ammonia-oxidizing Archaea in the Arctic Ocean and Antarctic coastal waters. Environ. Microbiol. 11, 2434–2445, 10.1111/j.1462-2920.2009.01974.x (2009).
Church, M. J., Wai, B., Karl, D. M. & DeLong, E. F. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ. Microbiol. 12, 679–688, 10.1111/j.1462-2920.2009.02108.x (2010).
Hollibaugh, J. T., Lovejoy, C. & Murray, A. E. Microbiology in Polar Oceans. Oceanogr. 20, 140–145 (2007).
Alonso-Saez, L., Sanchez, O., Gasol, J. M., Balague, V. & Pedros-Alio, C. Winter-to-summer changes in the composition and single-cell activity of near-surface Arctic prokaryotes. Environ. Microbiol. 10, 2444–2454, 10.1111/j.1462-2920.2008.01674.x (2008).
Collins, R. E., Rocap, G. & Deming, J. W. Persistence of bacterial and archaeal communities in sea ice through an Arctic winter. Environ. Microbiol. 12, 1828–1841, 10.1111/j.1462-2920.2010.02179.x (2010).
Forest, A. et al. Biogenic carbon flows through the planktonic food web of the Amundsen Gulf (Arctic Ocean): A synthesis of field measurements and inverse modeling analyses. Prog. Oceanogr. 91, 410–436, 10.1016/j.pocean.2011.05.002 (2011).
Terrado, R. et al. Protist community composition during spring in an Arctic flaw lead polynya. Polar Biol. 34, 1901–1914, 10.1007/s00300-011-1039-5 (2011).
Agogue, H., Brink, M., Dinasquet, J. & Herndl, G. J. Major gradients in putatively nitrifying and non-nitrifying Archaea in the deep North Atlantic. Nature 456, 788–U772, 10.1038/nature07535 (2008).
Lam, P. et al. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proceed. Natl. Acad. Sci. USA 104, 7104–7109, 10.1073/pnas.0611081104 (2007).
Sherr, E. B. & Sherr, B. F. Capacity of herbivorous protists to control initiation and development of mass phytoplankton blooms. Aquat. Microb. Ecol. 57, 253–262, 10.3354/ame01358 (2009).
Huston, A. L. & Deming, J. W. Relationships between microbial extracellular enzymatic activity and suspended and sinking particulate organic matter: seasonal transformations in the North Water. Deep-Sea Res. Pt. II 49, 5211–522, 10.1016/S0967-0645(02)00186-8 (2002).
Monier, A. et al. Upper Arctic Ocean water masses harbor distinct communities of heterotrophic flagellates. Biogeosci. 10, 13, 10.5194/bg-10-4273-2013 (2013).
Sintes, E., Bergauer, K., De Corte, D., Yokokawa, T. & Herndl, G. J. Archaeal amoA gene diversity points to distinct biogeography of ammonia-oxidizing Crenarchaeota in the ocean. Environ. Microbiol. 15, 1647–1658, 10.1111/j.1462-2920.2012.02801.x (2013).
Hallam, S. J. et al. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 4, 520–536, E95 10.1371/journal.pbio.0040095 (2006).
Middelboe, M. & Lundsgaard, C. Microbial activity in the Greenland Sea: role of DOC lability, mineral nutrients and temperature. Aquat. Microb. Ecol. 32, 151–163 (2003).
Darnis, G. & Fortier, L. Zooplankton respiration and the export of carbon at depth in the Amundsen Gulf (Arctic Ocean). J. Geophy. Res.-Oceans 117, C04013 10.1029/2011jc007374 (2012).
Saba, G. K., Steinberg, D. K. & Bronk, D. A. Effects of diet on release of dissolved organic and inorganic nutrients by the copepod Acartia tonsa. Mar. Ecol. Prog. Ser. 386, 147–161, 10.3354/meps08070 (2009).
Forest, A. et al. Carbon biomass, elemental ratios (C:N) and stable isotopic composition (delta(13)C, delta(15)N) of dominant calanoid copepods during the winter-to-summer transition in the Amundsen Gulf (Arctic Ocean). J. Plank. Res. 33, 161–178, 10.1093/plankt/fbq103 (2011).
Deming, J. W. & Fortier, L. Introduction to the special issue on the biology of the circumpolar flaw lead (CFL) in the Amundsen Gulf of the Beaufort Sea (Arctic Ocean). Polar Biol. 34, 1797–1801, 10.1007/s00300-011-1125-8 (2011).
Hamilton, A. K., Lovejoy, C., Galand, P. E. & Ingram, R. G. Water masses and biogeography of picoeukaryote assemblages in a cold hydrographically complex system. Limnol. Oceanogr. 53, 922–935 (2008).
Grasshoff, K., Kremling, K. & Ehrhardt, M. (Eds). Methods of seawater analysis New York Wiley (1999).
Holmes, R. M., Aminot, A., Kérouel, R., Hooker, B. A. & Peterson, B. J. A simple and precise method of measuring ammonium in marine and freshwater ecosytems. Can. J. Fish. Aquat. Sci. 56, 1801–1808 (1999).
Nusch, E. A. Results of interlaboratory tests concerning the determination of chlorophyll-a in water. Z. Wasser Abwass. For.- J. Water Wastewat. Res. 17, 89–94 (1984).
Marie, D., Simon, N. & Vaulot, D. in Algal Culturing Techniques (ed Andersen, R. A.) (Academic Press, 2005).
Li, W. K. W., McLaughlin, F. A., Lovejoy, C. & Carmack, E. C. Smallest Algae Thrive As the Arctic Ocean Freshens. Science 326, 539–539, 10.1126/science.1179798 (2009).
Aljanabi, S. M. & Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucl. Acids Res. 25, 4692–4693, 10.1093/nar/25.22.4692 (1997).
Harding, T., Jungblut, A. D., Lovejoy, C. & Vincent, W. F. Microbes in high arctic snow and implications for the cold biosphere. Appl. Environ. Microbiol. 77, 3234–3243, 10.1128/aem.02611-10 (2011).
Church, M. J., Short, C. M., Jenkins, B. D., Karl, D. M. & Zehr, J. P. Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl. Environ. Microbiol. 71, 5362–5370, 10.1128/aem.71.9.5362-5370.2005 (2005).
Zhu, F., Massana, R., Not, F., Marie, D. & Vaulot, D. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol. Ecol. 52, 79–92, 10.1016/j.femsec.2004.10.006 (2005).
Diez, B., Pedros-Alio, C. & Massana, R. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 67, 2932–2941, 10.1128/AEM.67.7.2932-2941.2001 (2001).
Pouliot, J., Galand, P. E., Lovejoy, C. & Vincent, W. F. Vertical structure of archaeal communities and the distribution of ammonia monooxygenase A gene variants in two meromictic High Arctic lakes. Environ. Microbiol. 11, 687–699, 10.1111/j.1462-2920.2008.01846.x (2009).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res. 25, 3389–3402 (1997).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599, 10.1093/molbev/msm092 (2007).
Schloss, P. D. & Handelsman, J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71, 1501–1506, 10.1128/aem.71.3.1501-1506.2005 (2005).
Hammer, Ø., Harper, D. A. T. & Ryan, P. D. PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4, 9 (2001). http://palaeo-electronica.org/2001_1/past/issue1_01.htm (Date of access:04/03/2014)
Acknowledgements
This work is an ArcticNet contribution, with additional funds from the Natural Science and Engineering Research Council (NSERC) of Canada, including a Discovery and other grants to CL. Financial support from Fonds de recherché du Quebec- nature et technologies (FRQNT) to Québec Océan is gratefully acknowledged. We thank W.K.W. Li for flow cytometry data and Mary Thaler for cell counts, advice on statistics and comments on the manuscript. We would also like to thank and the Canadian Coast Guard crew of the CCGS Amundsen and scientists who helped with sampling over the numerous missions.
Author information
Authors and Affiliations
Contributions
C.L. and P.E.G. wrote the main manuscript text and E.P., M.P., J.E.T. contributed technical sections and reviewed the manuscript. E.P. carried out the light experiment, Baffin Bay sampling and clone library construction, M.P. optimized and carried out qPCR protocols and J.E.T. contributed nutrient data.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information S1
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images in this article are included in the article's Creative Commons license, unless indicated otherwise in the image credit; if the image is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the image. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Pedneault, E., Galand, P., Potvin, M. et al. Archaeal amoA and ureC genes and their transcriptional activity in the Arctic Ocean. Sci Rep 4, 4661 (2014). https://doi.org/10.1038/srep04661
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep04661
This article is cited by
-
Vertical segregation and phylogenetic characterization of archaea and archaeal ammonia monooxygenase gene in the water column of the western Arctic Ocean
Extremophiles (2023)
-
Transcriptomic Insights into Archaeal Nitrification in the Amundsen Sea Polynya, Antarctica
Journal of Microbiology (2023)
-
Community structure, distribution pattern, and influencing factors of soil Archaea in the construction area of a large-scale photovoltaic power station
International Microbiology (2022)
-
Need for focus on microbial species following ice melt and changing freshwater regimes in a Janus Arctic Gateway
Scientific Reports (2018)
-
Distinctive Soil Archaeal Communities in Different Variants of Tropical Equatorial Forest
Microbial Ecology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.