Abstract
Root systems are critical for water and nutrient acquisition by crops. Current methods measuring root biomass and length are slow and labour-intensive for studying root responses to environmental stresses in the field. Here, we report the development of a method that measures changes in the root DNA concentration in soil and detects root responses to drought in controlled environment and field trials. To allow comparison of soil DNA concentrations from different wheat genotypes, we also developed a procedure for correcting genotypic differences in the copy number of the target DNA sequence. The new method eliminates the need for separation of roots from soil and permits large-scale phenotyping of root responses to drought or other environmental and disease stresses in the field.
Similar content being viewed by others
Introduction
Drought (water deficit) represents the most severe constraint to agricultural production and accounts for approximately 70% potential yield loss worldwide1,2. Root systems are critical for uptake of water and nutrients by plants. Effective deployment of genetic variation in root response to drought could have significant implications for world food security3.
Studies on growth of root systems in the field have been restricted by lack of high throughput methods4. Current methods for high throughput phenotyping are based on measurements of seedlings grown in artificial media such as agar5,6,7. However, root growth in artificial media can differ from that of plants grown in heterogeneous soil environments4,8,9,10. Characterising root growth in the field is challenging. Most studies measure changes in root biomass and length, which requires roots to be extracted from soil, separated from the dead roots and roots of other species and then quantified using image analysis, and/or dried to measure root dry weight4. Other methods such as excavation and minirhizotron have also been used11,12.
Cereals such as wheat have fibrous root systems and the fine lateral roots (approximately 0.07–0.15 mm in diameter) account for most of the total root length13. These fine roots fall below the current detection limit for X-ray computed tomography imaging14. They are usually fragile and translucent and difficult to recover for accurate measurements15,16,17, which limits their application to phenotype root growth for field-grown plants4.
A decade ago specific plant DNA sequences were amplified using polymerase chain reaction (PCR)18 for qualitative identification of mixed tree roots isolated from soils19. Recently, quantitative real-time PCR (qPCR) has been used to measure plant DNA concentrations from isolated roots of wild grasses of single or mixed species for estimation of relative abundance of roots of pot-grown plants20,21,22. qPCR has also been used to measure plant DNA concentrations directly from soil for estimation of root dry weight of different forage grasses and root distribution in soil profiles for field-grown plants22,23,24.
Plant DNA in soil samples is largely derived from live roots. The remnant DNA in dead roots is expected to contribute little to the total DNA concentration as over 90% of plant DNA in soils degrades within 7 to 10 days after plant death22,24. Plant DNA concentrations in soils can provide a measure of root response to treatments such as addition of lime to acid soil23 and complete defoliation or a herbicide treatment22, but also of root distribution in soil profiles21,24.
The measurements of plant DNA concentration in soils with qPCR are through amplification of a target DNA sequence. The internal transcribed spacers (ITS1 and ITS2) of the nuclear ribosomal DNA (rDNA) unit are widely used for phylogenetic studies and DNA barcoding as they show high levels of interspecific divergence25,26. ITS2 sequences have been used as a target sequence for qPCR measurements of plant DNA concentration of different forage grass species in soil22,23,24 because of its high specificity at the species level and high copy numbers27 allowing detection of low DNA concentrations22. There have been no reports so far of genotypic differences in root response to environmental stresses and root distribution within crop species, using plant DNA concentrations. In this report, we describe the development of a method for quantitative measurement of wheat DNA concentrations directly in soils. Since there have been reports of interspecies variation in ITS copy numbers25 and intraspecies variation in ITS sequences26,28, we devised a procedure for correcting genotype differences in the copy number of the target DNA sequences. We demonstrate that the DNA-based method provides a new tool for high-throughput measurements of root responses to drought in field trials.
Results
Genotypic variation in copy numbers of ITS2 sequences
Genotypic differences in copy numbers of wheat target sequence ITS2 (TaITS2) among 20 modern Australian spring wheat genotypes were determined using qPCR of TaITS2 and TaPHT1;6A, a single-copy gene, (Supplementary Fig. 1). Ct values of TaITS2 and TaPHT1;6A from root DNA samples were used to calculate TaITS2 copy number per genome. The Ct values of TaPHT1;6A were similar among 20 genotypes (27.68 ± 0.06), whereas the Ct values of TaITS2 among 20 genotypes with sequence-specific primers, ranged from 13.82 ± 0.20 to 16.16 ± 0.18. The genotypic differences in the Ct value of TaITS2 were highly significant (P < 0.001). Calculated copy numbers of TaITS2 varied from approximately 2,900 copies per genome in the variety Seri to 15,000 copies per genome in Young (Fig. 1). The average copy number of TaITS2 in the 20 genotypes was approximately 8800 per genome, comparable to 8200 rDNA repeat units per genome estimated for the two major rDNA loci (Nor1 and Nor2) in wheat by DNA hybridization29. The results indicate that there is genotypic variation in TaITS2 copy numbers among the 20 wheat genotypes.
Genetic variation in copy numbers of TaITS2 per genome among 20 wheat genotypes.
Total root DNA was isolated from primary roots of one month-old plants grown in a potting mix and used to determined copy numbers of TaITS2 per genome using quantitative real-time PCR. Ct values of TaITS2 and TaPHT1;6A using sequence-specific primers and probes were determined for all samples. The copy number of TaITS2 per genome was calculated by 2ΔCt (ΔCt = TaPHT1;6A Ct – TaITS2 Ct). Predicted means of four biological replicates are presented.
As other plant species closely related to wheat (Triticum aestivum) could be present in the field, such as T. urartu, T. monococcum, Aegilops speltoides, A. tauschii and Secale cereal, amplification refractory mutation system (ARMS)30 primers were designed for TaITS2 with deliberate 3′ mis-matches to eliminate DNA amplification from these related plant species. The ARMS primers were tested for efficacy in assessment of genotypic variation in TaITS2. The mismatches in the ARMS primers increased Ct values of TaITS2 for all 20 wheat genotypes, but there was a highly significant correlation (r2 = 0.986) in copy numbers of TaITS2 per genome between ARMS and sequence-specific primers (Supplementary Fig. 2). This result further confirms the genotypic variation in TaITS2 copy numbers among the 20 wheat genotypes and indicates that the ARMS primers are suitable for amplification of TaITS2. The ARMS primers were therefore used in DNA quantification for all soil samples.
For genotype comparison, it was necessary to correct the differences in copy numbers of TaITS2 among 20 genotypes used in the experiments. The copy number of TaITS2 in Krichauff was selected as the reference genotype because it was close to the average copy number of TaITS2 per genome (Supplementary Table 1). A scaling factor was calculated for each genotype to correct the difference in the copy number of TaITS2 (Supplementary Table 1).
Genotypic variation in root responses to drought in controlled and field environments
Root responses to drought were initially studied in pot-grown plants in a controlled environment. As root length density (RLD) is often used to measure root growth in pot and field-grown plants31, both RLD and root DNA density (RDD) were examined in two wheat cultivars, Kukri and Gladius for root response to stable and moderate drought (−0.4 MPa soil water potential). The drought treatment started after seedling establishment and ended at flowering. Genotypic differences in shoot dry weight at flowering were significant in the well-watered plants between Kukri and Gladius, but not in the drought-treated plants (Fig. 2a). The drought treatment reduced shoot growth by 35% in Gladius and 53% for Kukri relative to the well watered controls (Fig. 2a). Differences in RLD were similar to shoot dry weight for the genotypes and water treatments (Fig. 2b). Kukri had significantly higher RLD than Gladius in the well-watered plants, but no difference in RLD was observed between the two genotypes exposed to the drought treatment (Fig. 2b). The reduction in RLD by the drought treatment was 46% in Gladius and 60% in Kukri (Fig. 2b). In contrast, Gladius had significantly higher overall RDD than Kukri regardless of the water treatments (Fig. 2c). The drought treatment significantly reduced RDD by approximately 30% in both genotypes relative to that of the well watered treatment (Fig. 2c). These results indicate that RDD detects root responses to the drought as RLD and reveals the difference between RDD and RLD.
Effect of stable and moderate drought on shoot dry weight, root length density and root DNA density on wheat plants grown in pots under a controlled environment.
Effect of stable and moderate drought on shoot dry weight (a), root length density (b) and root DNA density (c) of two wheat cultivars, Kukri and Gladius. Soil moisture tension in the stable and moderate treatment was maintained at −0.4 MPa for four weeks after sowing until flowering. Well watered (WW) plants were the control. Plants were grown in a controlled climate environment. Means of five replicates are represented. There are significant differences in shoot dry weight and RLD for interactions of genotype × drought (P < 0.05). The differences in RDD for the genotypes and drought treatments were significant (P < 0.006), but the interaction of genotype × drought was not significant (P = 0.56). Vertical lines indicate the least significant difference at P = 0.05.
Next, root responses to a Mediterranean-type drought scenario32 was measured using RDD. Plants of the two wheat cultivars, Kukri and Gladius were grown in field soil under a rain-out shelter and exposed to two cycles of drought during flowering (Fig. 3a). The cyclic drought reduced soil moisture potential to −0.5 MPa (Fig. 3a), leading to a significant reduction of RDD in both cultivars (at least 64% relatively compared to well watered plants) in the top 10-cm soil layer (Fig. 3b). There was no effect of the cyclic drought on RDD of either cultivar below 10-cm depth (Fig. 3b). This indicates that RDD in the top 10-cm soil is particularly sensitive to the cyclic drought. RDD of both genotypes in the well watered and cyclic drought plots was significantly lower in 20–30 cm layer than in the top soil layers (Fig. 3b). Furthermore, RDD of Gladius across the three depths and the drought treatments was significantly higher than that of Kukri, revealing the genotypic variation in response to the cyclic drought between the two genotypes.
Effects of cyclic drought and continuous drought on root DNA density (RDD) in soil profile.
(a) Soil moisture tension was measured at 15 cm and 35 cm depth. Two cycles of the drought treatment (Drought) were applied to the plants grown in a rain-out shelter at 55 and 65 days after seed sowing, respectively. Well watered (WW) plants were the control. (b) Effects of the cyclic drought on RDD of two wheat genotypes, Kukri and Gladius in 0–30 cm depth. Four soil cores (2.5 cm in diameter) were collected at 70 days after seed sowing from between rows for each plot. Means and standard errors of four replicates are presented for RDD. There are highly significant differences in RDD for cultivars, cyclic drought and depth and for interactions of cyclic drought × depth × genotypes (P < 0.003). (c) RDD of two wheat genotypes, Krichauff and Berkut in 0–40 cm depth at flowering. Five soil cores (2.5 cm in diameter) were collected from between rows of plots at Roseworthy, South Australia in 2007. Means and standard deviations of three replicates are presented for RDD. There are highly significant differences in RDD for cultivars and depth (P < 0.001). Ct values of TaITS2 were determined for all soil samples using quantitative real-time PCR with ARMS primers and the TaITS2 probe. RDD presented was standardised with the scaling factor for each genotype.
The vertical distribution of roots from 0 to 40 cm depth was also measured using RDD at flowering for two wheat cultivars, Krichauff and Berkut during a severe drought in 2007. RDD of the two cultivars was the greatest in the 0–10 cm layer (Fig. 3c), similar to the well-watered plants (Fig. 3b). There was a sharp decline in RDD of both cultivars below 10-cm depth and little change from 10 to 40 cm depths (Fig. 3c). However, Berkut had a significantly higher RDD than Krichauff across all four depth measurements (Fig. 3c). These results reveal that there is the genotypic difference in vertical distribution of live roots under a severe drought condition.
High-throughput screen for genetic variation in root response to drought
To test whether the DNA-based method is suitable for studying genetic variation in root response to drought, a large trial consisting of twenty modern wheat genotypes with diverse genetic background was conducted at five locations in South Australia in 2008. The growing-season rainfall in 2008 was well below the yearly average and each location varied in accumulated rainfall of the growing season (Supplementary Table 2). On average, there was an approximately two-fold variation of RDD in the top 10-cm soil at flowering across the 20 genotypes for the five locations (Fig. 4a). Genetic variance at each location was highly significant and the heritability at each site was greater than 0.5 (Supplementary Table 3), indicating a strong genetic component underlying RDD. The genetic correlations between sites were also highly significant (Supplementary Table 3).
Genetic variation of root DNA density (RDD) in field-grown wheat genotypes.
(a) Genetic variation of RDD in top 10-cm soil among 20 wheat genotypes at five field sites. Ten representative soil cores (one cm in diameter × 10 cm in depth) from between rows were collected at flowering for each plot. Predicted means of four replicates are presented for RDD. (b) Genotypic variation of RRD in soil profile among seven wheat genotypes. Five representative soil cores (2.5 cm in diameter and 45 cm in depth) were collected at flowering from between rows for each plot at one field site, Karoonda. The 45-cm soil cores were separated into three 15-cm sections (0–15 cm, 15–30 cm and 30–45 cm) and the corresponding sections of five soil cores were combined for root DNA analysis. Natural logarithm transformed data were used for statistical analysis and means (n = 3) are presented for RDD. There are significant differences in RDD for genotype and depth (P < 0.001) and for interactions of genotype × depth (P < 0.034). Vertical lines indicate the least significant difference (LSD0.05) for interactions of genotype × depth. Ct values of TaITS2 were determined for all soil samples using quantitative real-time PCR with ARMS primers and the TaITS2 probe. RDD was standardised with the scaling factor for each genotype.
Genotypic variation in RDD was also examined for root vertical distribution at flowering at one site, Karoonda. Seven of the 20 genotypes were assessed to 45-cm depth (Fig. 4b). No significant differences in RDD across three depths were observed for Gladius and Berkut, whereas significant differences were seen between the 0–15 cm layer and those deeper in the soil profile for the other five cultivars (Fig. 4b). These results indicate that different wheat genotypes have distinct root responses to severe drought in terms of RDD distribution in the soil profile.
Discussion
Current methods based on root biomass and length can only provide low-throughput measurements for root growth in field-grown crops, which restricts studies on genetic diversity of root systems in the field4. Quantitative PCR allows measurements of plant DNA concentration in soil and can detect short-term and long-term changes of root systems in responses to stresses such as drought (Fig. 2c, 3b, 3c). Overall, these data indicate that plant DNA concentration in soil can be used to assess root responses to stresses, which may not be obtainable from root biomass or length. Plant DNA concentrations in soils can also be used to detect wheat root distribution in soil profiles (Fig. 3c, 4c). The vertical distribution of RDD are generally analogous to that measured with root biomass or length in field-grown wheat plants33,34. In addition, the plant DNA concentration in soil can be used for estimation of root biomass and length with careful calibration21,22,23,24. However, the proportion of live root cells in soil changes with different field sites, soil depths and ages of plants23. The calibration is needed for each situation as it is unlikely that there exists a close correlation between plant DNA concentration and root biomass or length in different soils.
Two types of target sequences (inter simple sequence repeat and ITS) have been used to quantify plant DNA concentration by qPCR20,21,22,23. ITS2 sequences were used in this study as they are highly specific at the species level and are in high copy numbers27, which allows detection of low DNA concentrations in soil22. However, potential differences in the copy number of the ITS2 sequences between genotypes need to be quantified and corrected for genotype comparison. Such correction allows comparison in plant DNA concentration in soil between wheat genotypes (Fig. 2c, 3b, 3c, 4a, 4b). There was a strong genetic correlation in RDD across multiple field sites (Supplementary Table 3) indicating that RDD is a heritable trait. In addition, the results from the field trials demonstrate that the DNA-based method can provide high-throughput assessment of root responses to drought in the field. These results show that RDD could assist in the characterisation of genetic diversity in root system architecture and root responses to water and nutrient supply in field-grown crops.
The association of RDD with drought tolerance in yield has been observed for some of the genotypes (Fig. 4a). The southern Australian or Mediterranean-type environment is characterised by late season drought and low soil moisture and poor quality. In this environment deep rooting offers few advantages since there is little useful moisture at depth. In much of southern Australia, groundwater is saline or there are toxic levels of boron at depth. Genotypes that perform well in these environments35, Excalibur, RAC875, Yitpi and Wyalkatchem had higher RDD in the top 10 cm soil, while less well adapted genotypes, such as Correll, Drysdale, Kukri and Young had lower RDD. However, two drought tolerance cultivars, Gladius and Mace show relatively low RDD (Fig. 4a). It may be premature to draw definitive correlation between RDD and yield. Yield is determined by a complex interaction of a wide range of environmental parameters over the full developmental phases of plant growth. Consequently, further analyses are required to separate effect of root traits in terms of RDD on yield from other traits. As more data are accumulated on changes in RDD across different germplasm pools and different environments, the significance of RDD relative to other measures of stress tolerance should become clearer. In addition, future studies should compare RDD with results from X-ray computed tomography14 and other in situ techniques for live roots, such as a green fluorescent protein reporter10.
In summary, the measurement of wheat root DNA in soil by qPCR can detect root responses to drought with high throughput. It is expected that this method will provide a new tool to complement other techniques to characterise root responses to abiotic and biotic stresses in field conditions. The application of this new technique will promote exploration of genetic diversity in root systems for improvement of water and nutrient acquisition4,36. This new technique should be readily applicable to crops other than wheat and diverse field environments and natural ecosystems.
Methods
Determination of genetic variation in TaITS2 copy numbers among 20 wheat genotypes
Twenty wheat (Triticum aestivum L.) genotypes were grown in pots (15 cm in diameter × 15 cm in height) which contained 2 kg of University of California potting mix with complete plant nutrients. One pre-germinated seed per pot was grown in a growth room with 15/10°C (day/night) temperature and 12 hr day/night cycle for one month. Light density was 300 μmol m−2 s−1 at the plant level. At harvest, pots were soaked briefly in water and the entire root system was carefully washed free of potting mix. Excess water on roots was blotted on laboratory tissues. Primary roots with laterals were collected, immediately frozen in liquid nitrogen and freeze-dried.
Total DNA from freeze-dried roots was isolated as described for soil DNA extraction (see below). DNA concentrations of samples were determined using QuantiFluor™ dsDNA System (Promega, Madison, WI, USA) and diluted to 2.5 ng μL−1. Quantitative real-time PCR using TaqMan® Minor Groove Binder (MGB) probes (Applied Biosystems, Foster City, CA, USA) was performed on ABI PRISM® 7900HT as described22. For TaITS2, the Ct value was determined using the TaqMan probe (6FAM 5′-CGGCATCTGGTCCCT-3′) with two different primer pairs. TaITS2 sequence-specific primer pair consisted of forward primer (5′-CCAAAACACGCTCCCAACCA-3′) and reverse primer (5′-TCGGTCCACCGTCCCTTGA-3′). The ARMS primer pair for TaITS2 consisted of forward primer (5′-CCAAAACACGCTCCCAAACA-3′) and reverse primer (5′-TCGGTCCACCGTCCCTAGA-3′). The ARMS primers were designed with 3′ mis-matches for increased specificity to discriminate T. aestivum closely-related species. For single copy gene, TaPHT1;6A located on chromosome 5AL (Supplementary Fig. 1), Ct values were determined using forward primer (5′-CGGACGTCGTCGGACACT-3′) and reverse primer (5′-AAACCATGAAAAGCATCCATACAC-3′) with the TaqMan® MGB probe (6FAM 5′–CCAACACCTGCATAGTA-3′ on the reverse strand). For each wheat genotype, the copy number of TaITS2 per genome was calculated by 2ΔCt (ΔCt = TaPHT1;6A Ct – TaITS2 Ct).
Measurements of root DNA density in soil
Soil cores were dried overnight at 45°C in a dehydration oven and DNA was extracted by the commercial service, Root Disease Testing Service (SARDI, Adelaide, SA, Australia). This DNA extraction method shows consistency with two other methods available for root DNA isolation and measurements23. Quantitative real-time PCR was used to determine Ct values of soil samples using the ARMS primer pair of TaITS2 with the TaqMan probe of TaITS2 as described above22. The Ct values could not be directly compared in different batches of samples. Therefore, the Ct values were converted to pg root DNA g−1 soil using standard calibration curves obtained from 10-fold serial dilution of Krichauff root DNA, ranging from 200 to 0.002 pg μl−1 for TaITS2 and from 20,000, to 2.0 pg μl−1 for TaPHT1;6A measurements. The root DNA of Krichauff was isolated from 7 day-old, sterile roots.
Plant accelerator experiment
Two genotypes (Gladius and Kukri) were used in the experiment. Plants were grown in white plastic pots (14 cm in diameter × 19 cm in height) containing 2.5 kg of soil mixture (equal volume of clay-loam soil and coco-peat mix) with complete fertilizers. Three seeds per pot were sown on 21st February, 2011 and thinned to one plant per pot two weeks after sowing. The plants were treated with two watering regimes from 28 days after sowing. In the well-watered regime, soil water content was maintained at 20% by watering every second day; equivalent to a soil water potential of approximately −0.05 MPa. Soil water potential was estimated by weighing pots every two days. For the moderate drought regime, watering was stopped at 28 days after sowing. When soil water potential reached to −0.4 MPa, the pots were rewatered. This mean level of soil water potential was maintained until the end of the experiment by watering every second day.
Plants were grown in the glasshouse (phenotyping platform of “The Plant Accelerator”, Adelaide, Australia). Air temperature and relative humidity were 18° day/24°C night and 50–90% humidity. Plants were harvested at 52 days after sowing, 24 days after being exposed to the two water regimes. Shoot dry weight was recorded after 3 days at 85°C. All soil in the pot was carefully removed and split into four quarters, two for analysis of RDD and the other two for root length density.
Root DNA density in a rain-out shelter experiment
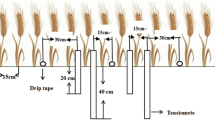
Plants of two wheat cultivars, Kukri and Gladius were grown in small experimental plots (20 × 60 cm, 8 plants per row, two rows per cultivar) in a rain-out shelter at the Waite Campus, Adelaide, Australia. The plots were fertilized twice with 8 g Aquasol m−2 (Hortico, Clayton, VIC, Australia) at 37 and 50 days after sowing. Seeds were sown on 3rd September, 2009. Two cycles of drought treatments were applied to plants at 55 and 65 days after sowing. Two moisture sensors were embedded at 15-cm and 35-cm depth, respectively to monitor soil moisture potential. Four soil cores (30 cm long × 2.5 cm wide) were taken at 70 days after sowing from between rows in each plot for each wheat cultivar. The 30-cm soil cores were divided into three 10-cm sections and the corresponding sections of four soil cores from each plot were combined for analysis of root DNA density.
Root DNA density in field experiments
Two field experiments were conducted. The first was at Roseworthy (South Australia) in 2007. The distribution of plant DNA in the soil profiles was examined for two wheat mapping parents, Berkut and Krichauff. Five cores (40 cm long × 2.5 cm wide) were taken from between rows of three plots per cultivar and then divided into 10-cm sections for analysis of root DNA density. The second multiple genotype experiment was conducted at five sites in South Australia in 2008. The growing-seasonal rainfall in 2008 was the lowest in decades. Rainfall data were obtained from weather stations closest to the trial sites (Supplementary Table 2). Twenty-one, hexaploid semi-dwarf wheat genotypes from diverse genetic background and widely used in breeding programs around Australia were included. The trial design was randomized split block design (12 ranges × 7 rows) and four replicates. Plots were 3.20 × 1.25 m, row spacing 0.25 m and managed as per district practice. Seeding rate was 200 seeds per square metre. Soil cores (10 cm long × one cm wide) were collected from the top soil at flowering. Twenty cores were taken from between rows and combined to make a composite sample per plot for plant DNA analysis. As mixed seeds were found for one of 21 genotypes, RDD data of 20 genotypes were presented.
For the genotypic variation in the distribution of roots in the soil profile, soil cores were taken at Karoonda site from between rows of seven wheat genotypes with three replicates. Five soil cores (45 cm long × 2.5 cm wide) per plot were collected at flowering four days after the top soil sampling. The 45-cm cores were separated into three 15-cm sections and the corresponding sections of five cores were combined to make a composite sample for plant DNA analysis.
Root length measurements
Soil cores were soaked in water. Roots were carefully washed free of soil particles and plant debris was removed from roots. The roots were spread out in root position tray (Reagent Instruments Inc., Canada) and scanned at 600 dpi. Scanning parameters were set as those described37. Length of roots was determined using the WinRHIZOPro Image Analysis software (Reagent Instruments Inc., Canada).
Statistical analyses
Experiments were analysed using the R (R Development Core Team, 2011, URL http://www.R-project.org/) package ASReml-R38. In the statistical analyses of TaITS2 copy number per genome, shoot dry weight, RLD and RDD from the pot experiment in controlled environment, RDD from the rain-out shelter experiment, the first field experiment and deep soil cores in the second field experiment, genotypes were fitted as fixed effects, along with the other treatments of interest, with an analysis of variance used to test the significance of effects. For RDD of the 0–10 cm soil cores from the second field experiment, a linear mixed model was fitted with genotype as a random effect, using a factor analytic model39 to model genetic correlations in RDD across five sites. An autoregressive process across ranges and rows was used to account for spatial variation at each site. Predicted means were calculated for each genotype by site combination.
References
Boyer, J. S. Plant productivity and environment. Science 218, 443–448 (1982).
Salekdeh, G. H., Reynolds, M., Bennett, J. & Boyer, J. Conceptual framework for drought phenotyping during molecular breeding. Trends Plant Sci. 14, 488–496 (2009).
Gewin, V. Food: An underground revolution. Nature 466, 552–553 (2010).
Gregory, P. J. et al. Root phenomics of crops: opportunities and challenges. Funct. Plant Biol. 36, 922–929 (2009).
Bengough, A. G. et al. Gel observation chamber for rapid screening of root traits in cereal seedlings. Plant Soil 262, 63–70 (2004).
Iyer-Pascuzzi, A. S. et al. Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol. 152, 1148–1157 (2010).
Clark, R. T. et al. Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol. 156, 455–465 (2011).
Whitmore, A. P. & Whalley, W. R. Physical effects of soil drying on roots and crop growth. J. Exp. Bot. 60, 2845–2857 (2009).
Rich, S. M. & Watt, M. Soil conditions and cereal root system architecture: review and considerations for linking Darwin and Weaver. J. Exp. Bot. 64, 1193–1208 (2013).
Chapman, N., Miller, A. J., Lindsey, K. & Whalley, W. R. Roots, water and nutrient acquisition: let's get physical. Trends Plant Sci. 17, 701–710 (2012).
Zhu, J., Brown, K. M. & Lynch, J. P. Root cortical aerenchyma improves the drought tolerance of maize (Zea mays L.). Plant Cell Environ. 33, 740–749 (2010).
Trachsel, S., Kaeppler, S. M., Brown, K. M. & Lynch, J. P. Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 341, 75–87 (2011).
Manske, G. G. B. & Vlek, P. L. G. Plant roots: the hidden half 3rd edn. pp 249-259. Marcel Dekker Inc. New York, USA (2002).
Flavel, R. J. et al. Non-destructive quantification of cereal roots in soil using high-resolution X-ray tomography. J. Exp. Bot. 63, 2503–2511 (2012).
Bengough, A. G., Castrignano, A., Pages, L. & van Noordwijk, M. Sampling strategies, scaling and statistics. Root Methods: A Handbook, 147–173 (2000).
Pierret, A., Moran, C. J. & Doussan, C. Conventional detection methodology is limiting our ability to understand the roles and functions of fine roots. New Phytol. 166, 967–980 (2005).
Watt, M., Magee, L. J. & McCully, M. E. Types, structure and potential for axial water flow in the deepest roots of field-grown cereals. New Phytol. 178, 135–146 (2008).
Mullis, K. B. & Faloona, F. A. Specific synthesis of DNA invitro via a polymerase-catalyzed chain-reaction. Methods in Enzymol. 155, 335–350 (1987).
Jackson, R. B., Moore, L. A., Hoffmann, W. A., Pockman, W. T. & Linder, C. R. Ecosystem rooting depth determined with caves and DNA. Proc. Natl. Acad. Sci. USA 96, 11387–11392 (1999).
Mommer, L., Wagemaker, C. A. M., De Kroon, H. & Ouborg, N. J. Unravelling below-ground plant distributions: a real-time polymerase chain reaction method for quantifying species proportions in mixed root samples. Mol. Ecol. Res. 8, 947–953 (2008).
Mommer, L. et al. Unveiling below-ground species abundance in a biodiversity experiment: a test of vertical niche differentiation among grassland species. J Ecol. 98, 1117–1127 (2010).
Riley, I. T., Wiebkin, S., Hartley, D. & McKay, A. C. Quantification of roots and seeds in soil with real-time PCR. Plant Soil 331, 151–163 (2010).
Haling, R. E. et al. Direct measurement of roots in soil for single and mixed species using a quantitative DNA-based method. Plant Soil 348, 123–137 (2011).
Haling, R. E., Simpson, R. J., Culvenor, R. A., Lambers, H. & Richardson, A. E. Field application of a DNA-based assay to the measurement of roots of perennial grasses. Plant Soil 358, 176–192 (2012).
Alvarez, I. & Wendel, J. F. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29, 417–434 (2003).
Song, J. et al. Extensive pyrosequencing reveals frequent intra-genomic variations of internal transcribed spacer regions of nuclear ribosomal DNA. PloS one 7, e43971–e43971 (2012).
Prokopowich, C. D., Gregory, T. R. & Crease, T. J. The correlation between rDNA copy number and genome size in eukaryotes. Genome 46, 48–50 (2003).
Simon, U. K. et al. Accession-Specific Haplotypes of the Internal Transcribed Spacer Region in Arabidopsis thaliana-A Means for Barcoding Populations. Mol. Biol. Evol. 29, 2231–2239 (2012).
Luo, M. C., Yang, Z. L. & Dvorak, J. Position effects of ribosomal RNA multigene loci on meiotic recombination in wheat. Genetics 149, 1105–1113 (1998).
Newton, C. R. et al. Analysis of any point mutation in DNA - The amplification refractory mutation system (ARMS). Nucl. Acids Res. 17, 2503–2516 (1989).
Wojciechowski, T., Gooding, M. J., Ramsay, L. & Gregory, P. J. The effects of dwarfing genes on seedling root growth of wheat. J. Exp. Bot. 60, 2565–2573 (2009).
Fischer, R. A. & Turner, N. C. Plant productivity in arid and semi-arid zones. Ann. Rev. Plant Physiol. Plant Mol. Biol. 29, 277–317 (1978).
Sheng, Q. & Hunt, L. A. Shoot and root dry-weight and soil-water in wheat, tricale and rye. Can. J. Plant Sci. 71, 41–49 (1991).
Gregory, P. J. & Eastham, J. Growth of shoots and roots and interception of radiation by wheat and lupin crops on a shallow, duplex soil in response to time of sowing. Aust. J. Agric. Res. 47, 427–447 (1996).
Fleury, D., Jefferies, S., Kuchel, H. & Langridge, P. Genetic and genomic tools to improve drought tolerance in wheat. J. Exp. Bot. 61, 3211–3222 (2010).
Coudert, Y., Perin, C., Courtois, B., Khong, N. G. & Gantet, P. Genetic control of root development in rice, the model cereal. Trends Plant Sci. 15, 219–226 (2010).
Genc, Y., Huang, C. Y. & Langridge, P. A study of the role of root morphological traits in growth of barley in zinc-deficient soil. J. Exp. Bot. 58, 2775–2784 (2007).
Butler, D., Cullis, B., Gilmour, A. & Gogel, B. ASReml-R Reference Manual. VSN International Ltd. Hempstead UK (2009).
Smith, A., Cullis, B. & Thompson, R. Analyzing variety by environment data using multiplicative mixed models and adjustments for spatial field trend. Biometrics 57, 1138–1147 (2001).
Acknowledgements
The advice and encouragement of Drs K. Ophel-Keller and D. Giblot-Ducray, initial statistical analyses of field-trial results by A. Smith and technical support provided by Y. Li, H. Zhou, R. Burns, I. Dadej, I. Dumitrescu, D. Pounsett and A. Thomas is gratefully acknowledged. This work was supported by the Grains Research and Development Corporation, the Australian Research Council, the South Australian Government, the University of Adelaide, Australian Grain Technologies, CSIRO Ecosystem Sciences, South Australian Grain Industry Trust Fund and the project DROPS, DROught tolerant yielding PlantS from funding of the European Community's Seventh Framework Programme under the grant agreement FP7-244374. http://www.drops-project.eu.
Author information
Authors and Affiliations
Contributions
C.Y.H., P.L. and A.C.M. conceived of the approach and co-wrote the paper. H.K. designed and J.E and S.H. conducted field trials. B.P. conducted the experiments in controlled environment and in the rain-out shelter. P.E. conducted statistical analyses, H. performed qPCR data analyses, D.M.H. designed ARMS primers.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Huang, C., Kuchel, H., Edwards, J. et al. A DNA-based method for studying root responses to drought in field-grown wheat genotypes. Sci Rep 3, 3194 (2013). https://doi.org/10.1038/srep03194
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep03194
This article is cited by
-
Interspecific competition among catch crops modifies vertical root biomass distribution and nitrate scavenging in soils
Scientific Reports (2019)
-
DNA based diagnostic for the quantification of sugarcane root DNA in the field
Scientific Reports (2018)
-
Transcriptomic signature of drought response in pearl millet (Pennisetum glaucum (L.) and development of web-genomic resources
Scientific Reports (2018)
-
Using root DNA to assess responses to phosphorus by surface roots in wheat and barley
Plant and Soil (2017)
-
Dynamic root responses to drought and rewatering in two wheat (Triticum aestivum) genotypes
Plant and Soil (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.