Abstract
The transcription apparatus (TA) is a huge molecular machine. It detects the time-varying concentrations of transcriptional activators and initiates mRNA transcripts at appropriate rates. Based on the general structural organizations of the TA, we propose how the TA dynamically orchestrates transcriptional responses. The activators rapidly cycle in and out of a clamp-like space temporarily formed between the enhancer and the Mediator, with the concentration of activators encoded as their temporal occupancy rate (RTOR) within the space. The entry of activators into this space induces allostery in the Mediator, resulting in a facilitated circumstance for transcriptional reinitiation. The reinitiation rate is much larger than the cycling rate of activators, thereby RTOR guiding the amount of transcripts. Based on this mechanism, stochastic simulations can qualitatively reproduce and interpret multiple features of gene expression, e.g., transcriptional bursting is not mere noise as traditionally believed, but rather the basis of reliable transcriptional responses.
Similar content being viewed by others
Introduction
During cellular signaling, upstream signals may be transmitted into the nucleus, inducing variations in the nuclear concentrations of transcriptional activators. The transcription apparatus (TA), which mainly consists of general transcription factors (GTFs, including TFII-B, -D, -E, -F and -H), RNA polymerase II (Pol II), the Mediator and transcriptional activators, detects such variations and accordingly initiates mRNA transcripts at appropriate rates1,2,3. This process, namely the transcriptional response that exploits the genetic information to modulate cellular functions, is of critical significance to life. There has been a great progress in exploring the mechanism of eukaryotic transcription. The primary structures of the core components of the TA such as Pol II and the Mediator were determined and the basic structural organizations among different components of the TA were also worked out4,5,6. On the other hand, the TA has evolutionarily made use of the stochastic nature of molecular interactions to perform functions. In vivo imaging technologies such as fluorescence recovery after photobleaching (FRAP) provided new insights into this dynamic process7,8,9,10,11. Nevertheless, it is still a great challenge to unravel how the TA dynamically governs transcriptional responses7,9,12,13,14.
Despite gene specificity, the general features of activator-regulated transcription can be summarized as follows4,5,6,12. Transcription begins with activators binding to the enhancer (proximal or distal to the core promoter)4,12, through which specific enzymes are recruited to alter chromatin architecture15. GTFs and Pol II then bind to the matured core promoter, forming the preinitiation complex (PIC). Subsequently, the DNA template strand is positioned into the active center cleft of Pol II, forming the open complex (OPC). After preparations such as the phosphorylation of its carboxy-terminal domain, Pol II gets away into elongation and begins transcribing a full length of mRNA. The complex left behind at the core promoter, termed the scaffold complex (SCF), facilitates transcriptional reinitiation4,12. The bridge between the enhancer-bound activators and the basal transcriptional machinery (composed of GTFs and Pol II at the core promoter) is the Mediator, which is evolutionarily conserved and crucial to almost all Pol II transcription across eukaryotes6,16,17.
To probe the fundamental dynamic mechanism by which the TA orchestrates transcriptional responses, we begin with a minimal model focusing on the essential properties of the TA as mentioned above. This model consists of the core promoter, a proximal enhancer containing one binding site for the activators (distant enhancers can be considered as proximal after DNA looping through the tethering elements9,18) and related proteins (Fig. 1a). Using the theories of probability and statistics, we propose the dynamic mechanism for transcriptional initiation. To further validate this mechanism, we construct a stochastic model of gene expression and the simulation results qualitatively coincide with seven experimental gene-expression profiles. We find that stochastic molecular interactions can be maximally exploited to ensure reliable transcriptional responses to activators.
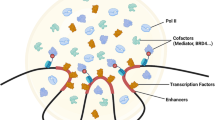
The minimal model of eukaryotic transcriptional initiation and its properties.
(a) The model. Upstream signals induce variations in the concentration of transcriptional activators that modulate transcriptional initiation through the Mediator. Other proteins such as histones are not depicted in the diagram. (b) The relationships between the transcriptional events. (c) The activators' temporal occupancy rate  is a monotonically increasing function of the number of nuclear activators, na (na is incorporated in aon/aoff). Each dot represents an independent trial with m = 10 (for more cases, see Fig. S1). The black line denotes the average value. (d) The standard deviation of
is a monotonically increasing function of the number of nuclear activators, na (na is incorporated in aon/aoff). Each dot represents an independent trial with m = 10 (for more cases, see Fig. S1). The black line denotes the average value. (d) The standard deviation of  ,
,  , vs. aon/aoff for different m.
, vs. aon/aoff for different m.  decreases with increasing m.
decreases with increasing m.
Results
Mathematical characterization of the dynamics of the TA
The dynamics of the TA are dictated by how its components are spatially and temporally organized on the promoter. Because the TA can assume many distinct configuration states and the state evolution is essentially stochastic, involving numerous molecules and complex interactions, we employ the theories of statistics and probability to investigate the dynamics of the TA. For simplicity, we assume that the concentrations of transcription-associated species such as GTFs remain constant around the model gene and the molecular interactions involving the promoter are in dynamic equilibrium. The term “dynamic equilibrium” does not mean that molecular interactions are all reversible; rather, it just requires that the TA should retrieve its current state after some time. A model gene and all the species around it constitute a system. The above assumptions imply that such a system is in a steady state. Let us consider a statistical ensemble consisting of a large number of such essentially identical systems, with each evolving independently. The number of the systems is large enough so that all possible configuration states of the TA can be covered by this ensemble. That is, each state undergone by an individual gene maps the states of other genes in the ensemble and the proportion of genes in a special state X (e.g., genes with their enhancers bound by activators), P(X), remains constant over time. Equivalently, if an individual gene is observed at any time, the probability that the gene is in the state X is also P(X). In this sense, which state an individual gene is located in is a random event.
For the minimal model (Fig. 1a), we define all configuration states of the TA as a universal set Ω and the various states with the same key features as the following sub-sets, respectively (Fig. 1b). A denotes that the enhancer is bound by an activator. S denotes that the core promoter is bound by the proteins of the SCF. M denotes that a nascent mRNA is in gestation (including the process from PIC formation to Pol II's escape into elongation). J denotes that the enhancer-bound activator is conjectured to the SCF, PIC or OPC through the Mediator. Because eukaryotic transcriptional initiation requires the presence of the SCF on the core promoter4,12, M⊂S. According to the definitions, J⊂AS. In the set MJ, M and A are concurrent, i.e., the enhancer-bound activators can directly affect Pol II's action through the Mediator. Thus, the transcriptional initiation under direct regulation of activators is described by the set MJ, whereas the basal, activator-independent transcriptional initiation is included in the set M−J. The probability of a nascent mRNA in gestation, i.e., the probability that an mRNA is generated, is

where q is a constant representing the basal transcriptional initiation and Aj is a subset of A (see S1 of Supplementary Information for details). In Aj, the enhancer-bound activators are obligate for contacting the SCF-, PIC-, or OPC-joined Mediator. Equation (1) characterizes the relationship between mRNA production and the dynamic properties of the TA.
Encoding the concentration of transcriptional activators
Owing to distinct architectures of promoter chromatin in different transcriptional stages, the enhancer-bound activators may perform various functions, such as promoting histone acetylation and recruiting GTFs4,5,15. Specifically, the set Aj involves the enhancer-bound activators that are responsible for handling the basal transcriptional machinery and controlling transcriptional initiation. Moreover, the activities of these activators are also associated with the encoding of the nuclear concentration of activators, for  is the only factor in equation (1) depending on the concentration of activators. Here, we investigate the dynamics of such activators.
is the only factor in equation (1) depending on the concentration of activators. Here, we investigate the dynamics of such activators.
Activators move rapidly within the nucleus and the probability of them reaching the enhancer is proportional to their nuclear abundance9. Let us consider a time period, during which the activators involved in the set Aj bind to and then depart from the enhancer for m (m = 1, 2, 3, …) cycles. We define the temporal occupancy rate RTOR of those activators as  , where
, where  and
and  denote the binding and unbinding time of the j-th cycle, respectively. For the fixed number na of activators in the nucleus, we have
denote the binding and unbinding time of the j-th cycle, respectively. For the fixed number na of activators in the nucleus, we have

where aon and aoff are the propensity functions of binding and unbinding, respectively (see
S2
of Supplementary Information for details). aon is a function of na, whereas aoff is independent of na. Equation (2) indicates that as m rises,  converges to a deterministic value, which is a monotonically increasing function of na (Fig. 1c-d and Fig. S1; this is a general property and can be applied to cases where the number of cognate binding sites on the enhancer is greater than one (see equations S13-S18)). This convergence implies that even the time-varying concentration of activators can be encoded by RTOR, provided that the activators cycle on and off the enhancer frequently enough over a time window with their concentration nearly unchanged. Indeed, there are active disassociation mechanisms that guarantee the rapid cycling of activators9,19,20,21,22. The binding time was estimated to be within the range of seconds to tens of seconds9,10. Moreover, it was proved on the endogenous CUP1 gene that such rapid cycling is functional10. Presumably, the RTOR encodes the concentration of transcriptional activators. On the other hand, during the time period where the activators cycle on and off the enhancer for m times, the probability that the enhancer is found bound by such an activator is
converges to a deterministic value, which is a monotonically increasing function of na (Fig. 1c-d and Fig. S1; this is a general property and can be applied to cases where the number of cognate binding sites on the enhancer is greater than one (see equations S13-S18)). This convergence implies that even the time-varying concentration of activators can be encoded by RTOR, provided that the activators cycle on and off the enhancer frequently enough over a time window with their concentration nearly unchanged. Indeed, there are active disassociation mechanisms that guarantee the rapid cycling of activators9,19,20,21,22. The binding time was estimated to be within the range of seconds to tens of seconds9,10. Moreover, it was proved on the endogenous CUP1 gene that such rapid cycling is functional10. Presumably, the RTOR encodes the concentration of transcriptional activators. On the other hand, during the time period where the activators cycle on and off the enhancer for m times, the probability that the enhancer is found bound by such an activator is  . Because the average of
. Because the average of  over the ensemble is also f(na), we have
over the ensemble is also f(na), we have

The constraint conditions ensuring reliable transcriptional responses
Given the stochasticity in the occurrence of transcriptional events, to achieve a reliable transcriptional response requires that the RTOR code, which timely represents the concentration of activators, should be transduced with high fidelity into the amount of transcripts. Ideally, if P(S),  and
and  all equaled 1, the exact information transduction would be accomplished. In the following, we present the conditions under which those three factors can be large enough to ensure reliable transcriptional responses in the presence of random fluctuations (Figs. S2 and S3 provide intuitive explanations for this subsection).
all equaled 1, the exact information transduction would be accomplished. In the following, we present the conditions under which those three factors can be large enough to ensure reliable transcriptional responses in the presence of random fluctuations (Figs. S2 and S3 provide intuitive explanations for this subsection).
Equation (3) implies that the concentration of activators cannot be sufficiently encoded without the persistence of the SCF on the promoter. Thus, the SCF should assemble rapidly when the chromatin architecture allows and be much more stable than the enhancer-bound activators (Condition I). Such stability of the SCF was observed experimentally and the binding time of TBP (TATA-binding protein, the core component of the SCF) on the promoter can be up to 20 min in human cells11. For  , Aj is a precondition of the occurrence of J. Because RTOR is determined by the individual short binding times of activators, J should happen immediately after the occurrence of Aj (Fig. S3). Otherwise, the information about RTOR is largely lost or even falsely utilized to direct transcriptional initiation (note that J is a precondition of M). Therefore, to correctly transfer the RTOR code, the Mediator should act by waiting to bind the cycling activators and transmit the information through allostery23,24,25 (Condition II). This is because other kinds of molecular interactions like free collisions cannot precisely convey the information about the binding time of activators. Such allosterism of the Mediator is supported by the previous work26.
, Aj is a precondition of the occurrence of J. Because RTOR is determined by the individual short binding times of activators, J should happen immediately after the occurrence of Aj (Fig. S3). Otherwise, the information about RTOR is largely lost or even falsely utilized to direct transcriptional initiation (note that J is a precondition of M). Therefore, to correctly transfer the RTOR code, the Mediator should act by waiting to bind the cycling activators and transmit the information through allostery23,24,25 (Condition II). This is because other kinds of molecular interactions like free collisions cannot precisely convey the information about the binding time of activators. Such allosterism of the Mediator is supported by the previous work26.  determines how the information about RTOR inherited by J is converted to guide the amount of transcripts. Because RTOR depends on the intermittent binding of activators, a large
determines how the information about RTOR inherited by J is converted to guide the amount of transcripts. Because RTOR depends on the intermittent binding of activators, a large  requires that during the short binding periods, transcripts should be produced at a rather rapid rate (Fig. S2) (Condition III). This feature is also verified by computational estimates of the experimental data (see
S3
of Supplementary Information). Therefore, all the three conditions can be satisfied naturally.
requires that during the short binding periods, transcripts should be produced at a rather rapid rate (Fig. S2) (Condition III). This feature is also verified by computational estimates of the experimental data (see
S3
of Supplementary Information). Therefore, all the three conditions can be satisfied naturally.
The dynamic mechanism of activator-regulated transcription
The above three constraint conditions together determine how the TA operates. There repetitively arises a state in which a relatively stable clamp-like space is formed between the Mediator and the enhancer (Fig. 2; according to Conditions I and II). As the SCF was shown experimentally to be not very stable11, this space is temporarily constructed. The clamp-like space attracts free activators and then rapidly peels them off, with RTOR decided by the activators' concentration (according to equations (2-3)). Once one activator molecule gets into this space, there arises allostery in the Mediator, resulting in a facilitated circumstance for the GTFs and other related proteins to perform their functions. Consequently, Pol IIs can initiate/reinitiate transcription very rapidly (according to Conditions II and III), with the RTOR governing the quantity of transcripts.
Illustration of the dynamic mechanism for the TA orchestrating a reliable transcriptional response.
There repetitively arises a state where a relatively stable clamp-like space is formed between the Mediator and the enhancer. Transcriptional activators cycle in and out of this space rapidly. Only when this space is occupied by activators do Pol IIs initiate/reinitiate transcription at a faster rate than the cycling rate of the activators.
This mechanism suggests that the molecular interactions involving the promoter obey elegant dynamic principles as follows. Whereas the clamp-like space is temporarily formed, it is much more stable than the activators settled in it. The activators can cycle in and out of the space for many times even during short episodes when their concentration nearly remains unchanged; thus, the concentration of activators can be represented by RTOR timely. Because the Mediator transmits the information via allostery and the rate of transcriptional reinitiation is much larger than the cycling rate of activators, the RTOR code is effectively employed to direct mRNA synthesis. In a word, the clamp-like space is the structural basis for reliable transcriptional responses. Rather than being an obstacle, the stochastic nature of the molecular interactions is fully utilized to induce transcription reliably; this largely depends on different extents of the stability of the components of the TA, which span several orders of magnitude. The above arguments are supported by experimental data and the typical time scales are as follows: the half lifetime of the clamp-like space is about 5 min11, the occupancy time of activators in the space is within the range of seconds to tens of seconds10, allostery usually occurs within the time of milliseconds to no more than 1 second23,24,25 and it just takes several seconds to reinitiate a transcript (see S3 of Supplementary Information).
Validation of the mechanism by numerical simulations
To further verify the proposed dynamic mechanism, we build a simplified stochastic model of gene transcription with physiologically realistic parameters (see Fig. S4 and S4 in Supplementary Information for details). This model depicts the key state transitions of the TA and also simply describes the related chromatin dynamics, thereby capable of charactering the transcriptional response to transcriptional activators. In the following, the “input” and “output” denote the nuclear concentration of activators and the amount of gene products, mRNA or protein, respectively.
First, we explore the temporal evolution of the number of cellular mRNAs at constant input levels (Fig. 3a). Notably, mRNAs are produced in a burst-like manner, consistent with the prevailing view14,27,28,29,30,31,32. For low-level inputs, one allele is transcribed while the other is silent in a diploid cell and thus the bursting phenomenon is apparent. For high-level inputs, however, both the two alleles burst frequently so that the sum becomes almost constant. This suggests that the phenotype of persistent elevated transcriptional responses may be observed at high input levels14.
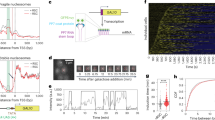
Transcriptional responses to activators based on the proposed dynamic mechanism.
The input equals aon/aoff, which is positively related to the nuclear concentration of activators. (a) Temporal evolution of the number of cellular mRNAs in a single diploid cell with different input levels. mRNAs produced by two alleles are shown separately in red and black. The transcriptional burst becomes dense with increasing the input strength. (b) The average input/output relationship in individual diploid cells. The maximal outputs are normalized to 1. Error bars denote the standard deviation of the output, SDout. The inset shows the ratio of SDout to the mean output vs. the input. Because the abundance of mRNAs or proteins also depends on their degradation/inactivation rates, which are modulated by cellular signaling, the rate of mRNA production more directly reflects the dynamics of the TA (see also Fig. S9, where the production rate of proteins is also shown44,45). (c) The curves of the SDout vs. the input. These curves nearly remain bell-shaped even at various degradation rates of mRNAs or proteins (see also Fig. S10). (d) The distribution of mRNA levels across a cell population for different input levels. The bin size is 10. (e) The state evolution of a promoter in response to a periodically varying input. G1 denotes the enhancer is bound by an activator. SCF denotes the core promoter is bound by the SCF. OPC denotes the core promoter is in the OPC state. The curves describe the input, the corresponding states of the promoter and the production of mRNAs (from top to bottom), respectively. (f) ChIP simulation of the transcriptional response. The input and the symbols are the same as in panel (e). TATAn and Pol II denote that the core promoter is bound by histones and Pol II, respectively.
A recent experimental analysis precluded the possibility that the chromatin environment plays a central role in shaping transcriptional bursting32. Here, we demonstrate that a burst of transcripts originates from persistent reinitiation by Pol IIs when the clamp-like space is occupied by the activators (Fig. 4). That is, the initiation of mRNA is itself burst-like. The bursting is not mere noise; instead, it is a direct manifestation of the RTOR code, which represents the concentration of activators and guides mRNA production.
The essence of transcriptional burst.
Shown is a microscopic view of a transcriptional burst. ‘CA’ denotes that an activator is in the clamp-like space. ‘OPC’ denotes that the transcription machinery is in the OPC state (a zoom-in panel is also displayed). When an activator molecule is present in the clamp-like space, rapid transcriptional reinitiation results in a burst of mRNAs.
Second, we investigate the average input/output relationship of transcriptional response. The average output resembles a Hill function of the input, which is widely used in systems biology to model gene expression3,33,34 (Fig. 3b). The curve of the standard deviation SDout of the output versus the input is approximately bell-shaped (Fig. 3c). The intensity of the intrinsic noise, defined as the ratio of SDout to the mean output35, is inversely correlated with the input (the inset of Fig. 3b). Moreover, the above features are insensitive to the slight fluctuations in the input (i.e., extrinsic noise) (Fig. S5), suggesting the robustness of the transcriptional response to noise. All these results are in good agreement with the experimental measurements in both Saccharomyces cerevisiae36 and Drosophila embryos37. Particularly, the left side of the SDout curve is lower than its right side; this feature is almost quantitatively in accordance with the experimental data37 (see S5 of Supplementary Information for further discussions). By contrast, deviations from the dynamic principles proposed above (including the circumstances under which the activators' cycling is slow, the scaffold complex/clamp-like space is not stable, or/and the rate of transcriptional reinitiation is low) would reduce the capability of the TA to respond reliably to the input (Fig. S6).
The input/output relationship observed in Drosophila embryos was believed to be realized by maximally utilizing the limit of molecular interactions37,38,39. The properties of such microscopic interactions are integrated to be macroscopically manifested as SDout. The SDout curve is still bell-shaped overall compared with the SDin curve (cf. Fig. 1d). That is, the signature of the RTOR code can be directly transmitted to the output. This confirms that the temporal occupancy rate of activators is really exploited to regulate transcription and the Mediator transmits the information through allostery. On the other hand, the SDout curve is asymmetrical, with the right side being higher than the left. The reason is obvious. When the input is very high, the enhancer is bound by the activators almost all the time and thus the fluctuations mainly reflect the dynamic properties of the SCF and the transcriptional reinitiation by Pol IIs. Our further simulations show that the right side of the SDout curve drops as the stability of the SCF or the rate of transcriptional reinitiation is increased; only when increasing their strength beyond the physiological ranges can the curve become symmetrical (Fig. S6F). This also verifies that both P(S) and  are indeed large enough. Therefore, the properties of SDout should conclusively prove the microscopic transcriptional mechanism.
are indeed large enough. Therefore, the properties of SDout should conclusively prove the microscopic transcriptional mechanism.
Third, we probe the distribution of mRNA levels across a large cell population exposed to the same input (Fig. 3d). For small inputs, the bursting phenomenon is especially obvious and most cells have no or few mRNAs. This is consistent with the experimental observation27,30,31. But the distribution gradually becomes normal as the input increases. For aon/aoff >1, the distribution becomes sharper with increasing the input. These results await experimental identification.
Fourth, we simulate the transcriptional response to a periodically varying input. The microscopic process on a promoter is rather dynamic and stochastic, with different components of the TA exhibiting distinct stabilities (Fig. 3e). However, the amount of mRNAs can follow the input. These results are in good agreement with the results revealed by FRAP, i.e., the TA is a highly dynamic apparatus8,10,11,22. On the other hand, chromatin immunoprecipitation assays (ChIP) simulations, which characterize the temporal evolution of the distribution of different states of the promoter among a cell population, reveal a strong regularity in the distributions (Fig. 3f). The patterns of both the activators binding to the enhancer and the SCF and Pol II binding to the promoter follow the input. mRNA transcripts are produced in phase with the input, whereas histones occupy the promoter in a reversed phase. All these results are well consistent with the experimental findings22,40. Therefore, the observed discrepancies between the results from FRAP and ChIP experiments may originate from different resolutions involved in the measurements. ChIP measurements integrate molecular interactions both temporally and over the cell population, whereas FRAP more tightly reflects the instantaneous interactions. Moreover, the transcriptional response to the time-varying input is robust to extrinsic noise but sensitive to composite input signals and deviations from the dynamic principles (such as the cases with low cycling rate of activators, the unstable scaffold complex/clamp-like space, or/and low rate of transcriptional reinitiation) would reduce the response capability (see Figs. S7 and S8).
Discussion
Based on the general features of the eukaryotic TA, we have proposed the fundamental dynamic mechanism by which the TA orchestrates reliable transcriptional responses to cellular signals. Although our work is built on the general architecture of the TA, different profiles of gene expression can be accounted for simultaneously. This implies that the eukaryotic TA likely shares the same basic mechanism in mediating transcriptional responses, just as the same set of GTFs is involved. We have shown that the TA is an elegant apparatus; the stabilities of its components are widely differentiated such that the stochastic nature of molecular interactions is employed to achieve reliable transcriptional responses. The activators' cycling in and out of the clamp-like space modulates the amount of mRNA transcripts initiated through the Mediator's allostery. The concentration of activators are represented by the statistical quantity RTOR, which essentially leads to burst-like mRNA production. Thus, the transcriptional bursting is not only the phenotype but also the basis of reliable transcriptional responses.
Traditionally, it was believed that activators regulate gene transcription through recruiting proteins such as GTFs and Pol II41. Here, we argue that this is mainly the process of TA assembly. Our results demonstrate that it is through controlling the circumstance where Pol IIs reinitiate transcription that activators mediate the responses to upstream signals; the clamp-like space between the enhancer and the Mediator is the structural basis for RTOR to guide the amount of mRNA transcripts. It is worthy to note that the transcriptional mechanism proposed here can be viewed as a complicated realistic version of the clustering model13,27,42,43. In that model, a promoter is in an "off" or "on" state and only when the promoter is in the "on" state, the gene is transcribed actively. Here, we further show that Pol IIs reinitiate transcripts rapidly only when the clamp-like space is occupied by activators. Finally, it is worth mentioning that it is still difficult to experimentally elucidate the dynamic mechanisms of molecular machines, whereas our approach employing statistics and probability theory is powerful in this field.
Methods
Mathematical derivations
The relationships between the transcriptional events depend on the basic knowledge that eukaryotic transcriptional initiation requires the SCF4,12 and only the enhancer-bound activators rather than free activators can regulate transcription. Equation (1) was derived using the probability theory. Equation (2) was derived by employing the Gillespie theory46,47. The implications of equations (1–3) and the constraint conditions were also intuitively illuminated in Figs. S2–S3. All mathematical derivations are detailed in Supplementary Information.
Stochastic simulations
The stochastic model was constructed based on the proposed dynamic mechanism of activator-regulated eukaryotic transcription and related experiments. The Gillespie algorithm47 was used to perform simulations. The very detailed description of the model, the parameter values and the methods of numerical simulations are presented in Supplementary Information.
References
Brivanlou, A. H. & Darnell, J. E., Jr Signal transduction and the control of gene expression. Science 295, 813–818 (2002).
Weake, V. M. & Workman, J. L. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 11, 426–437 (2010).
Zhang, X. P., Liu, F., Cheng, Z. & Wang, W. Cell fate decision mediated by p53 pulses. Proc. Natl. Acad. Sci. U. S. A. 106, 12245–12250 (2009).
Hahn, S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11, 394–403 (2004).
Thomas, M. C. & Chiang, C. M. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol 41, 105–178 (2006).
Kornberg, R. D. The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. U. S. A. 104, 12955–12961 (2007).
Darzacq, X. et al. Imaging transcription in living cells. Annu. Rev. Biophys. 38, 173–196 (2009).
Darzacq, X. et al. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 14, 796–806 (2007).
Hager, G. L., McNally, J. G. & Misteli, T. Transcription dynamics. Mol. Cell 35, 741–753 (2009).
Karpova, T. S. et al. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science 319, 466–469 (2008).
Chen, D., Hinkley, C. S., Henry, R. W. & Huang, S. TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol. Biol. Cell 13, 276–284 (2002).
Fuda, N. J., Ardehali, M. B. & Lis, J. T. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461, 186–192 (2009).
Pedraza, J. M. & Paulsson, J. Effects of molecular memory and bursting on fluctuations in gene expression. Science 319, 339–343 (2008).
Raj, A. & van Oudenaarden, A. Nature, nature, or chance: stochastic gene expression and its consequences. Cell 135, 216–226 (2008).
Li, B., Carey, M. & Workman, J. L. The role of chromatin during transcription. Cell 128, 707–719 (2007).
Malik, S. & Roeder, R. G. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30, 256–263 (2005).
Kornberg, R. D. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30, 235–239 (2005).
Levine, M. & Tjian, R. Transcription regulation and animal diversity. Nature 424, 147–151 (2003).
Freeman, B. C. & Yamamoto, K. R. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science 296, 2232–2235 (2002).
Reid, G. et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ER alpha on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 11, 695–707 (2003).
Stavreva, D. A., Muller, W. G., Hager, G. L., Smith, C. L. & McNally, J. G. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol. Cell. Biol. 24, 2682–2697 (2004).
Bosisio, D. et al. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kappa B-dependent gene activity. EMBO J. 25, 798–810 (2006).
Henzler-Wildman, K. & Kern, D. Dynamic personalities of proteins. Nature 450, 964–972 (2007).
Li, W. . Wolynes, P. G. & Takada, S. Frustration, specific sequence dependence and nonlinearity in large-amplitude fluctuations of allosteric proteins. Proc. Natl. Acad. Sci. U. S. A. 108, 3504–3509 (2011).
Hyeon, C., Jennings, P. A., Adams, J. A. & Onuchic, J. N. Ligand-induced global transitions in the catalytic domain of protein kinase A. Proc. Natl. Acad. Sci. U. S. A. 106, 3023–3028 (2009).
Meyer, K. D., Lin, S. C., Bernecky, C., Gao, Y. & Taatjes, D. J. p53 activates transcription by directing structural shifts in Mediator. Nat. Struct. Mol. Biol. 17, 753–760 (2010).
Raj, A., Peskin, C. S., Tranchina, D., Vargas, D. Y. & Tyagi, S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4, e309 (2006).
Blake, W. J. et al. Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell 24, 853–865 (2006).
Chubb, J. R., Trcek, T., Shenoy, S. M. & Singer, R. H. Transcriptional pulsing of a developmental gene. Curr. Biol. 16, 1018–1025 (2006).
Zenklusen, D., Larson, D. R. & Singer, R. H. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 15, 1263–1271 (2008).
Kaufmann, B. B. & van Oudenaarden, A. Stochastic gene expression: from single molecules to the proteome. Curr. Opin. Genet. Dev. 17, 107–112 (2007).
Suter, D. M. et al. Mammalian genes are transcribed with widely different bursting kinetics. Science 332, 472–474 (2011).
Brandman, O., Ferrett, J. E., Jr, Li, R. & Meyer, T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science 310, 496–498 (2005).
Sneppen, K., Krishna, S. & Semsey, S. Simplified models of biological networks. Annu. Rev. Biophys. 39, 43–59 (2010).
Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002).
Blake, W. J., Kaern, M., Cantor, C. R. & Collins, J. J. Noise in eukaryotic gene expression. Nature 422, 633–637 (2003).
Gregor, T., Tank, D. W., Wieschaus, E. F. & Bialek, W. Probing the limits to positional information. Cell 130, 153–164 (2007).
Tkacik, G., Callan, C. G., Jr. & Bialek, W. Information flow and optimization in transcriptional regulation. Proc. Natl. Acad. Sci. U. S. A. 105, 12265–12270 (2008).
Okabe-Oho, Y., Murakami, H., Oho, S. & Sasai, M. Stable, precise and reproducible patterning of bicoid and hunchback molecules in the early Drosophila embryo. PLoS Comput. Biol. 5, e1000486 (2009).
Stavreva, D. A. et al. Ultradian hormone stimulation induces glucocorticoid receptor-mediated pulses of gene transcription. Nat. Cell Biol. 11, 1093–1102 (2009).
Watson, J. D. et al. Molecular Biology of the Gene (6th edition, CSHL press, New York, 2008), pp. 597–600.
Peccoud, J. & Ycart, B. Markovian modeling of gene-product synthesis. Theor. Popul. Biol. 48, 222–234 (1995).
Feng, H., Hensel, Z., Xiao, J. & Wang, J. Analytical calculation of protein production distributions in models of clustered protein expression. Phys. Rev. E 85, 031904 (2012).
Yu, J., Xiao, J., Ren, X., Lao, K. & Xie, X. S. Probing gene expression in live cells, one protein molecule at a time. Science 311, 1600–1603 (2006).
Hensel, Z. et al. Stochastic properties of transcription factor expression revealed by single molecule noise analysis. Nat. Struct. Mol. Biol., accepted (2012).
Gillespie, D. T. A general method for numerically simulating stochastic time evolution of coupled chemical reactions. J. Comput. Phys. 22, 403–434 (1976).
Gillespie, D. T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81, 2340–2361 (1977).
Acknowledgements
This work was supported by the NNSF of China (10834002 and 11175084), the NSF of Jiangsu Province (BK2009008), the Program for New Century Excellent Talents in Universities (NCET-08-0269) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Contributions
Y.W., F.L. and W.W. designed the research; Y.W. performed computations; Y.W., F.L. and W.W. wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplymentary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareALike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Wang, Y., Liu, F. & Wang, W. Dynamic Mechanism for the Transcription Apparatus Orchestrating Reliable Responses to Activators. Sci Rep 2, 422 (2012). https://doi.org/10.1038/srep00422
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00422
This article is cited by
-
Mathematical approaches in studying bicoid gene
Quantitative Biology (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.