Abstract
Non-syndromic recessive deafness (NSRD) is the most commonly encountered form of hereditary hearing loss. The majority of NSRD cases in the Mediterranean area are linked to the DFNB1 locus (the connexin 26 GJB2 gene). Unrelated NSRD patients issued from 68 Jordanian families, were tested for mutations of the GJB2 gene by sequencing. Sixteen per cent of the families tested were linked to the DFNB1 locus. The 35delG was the only GJB2 mutation detected in these families. One of these families, presenting with four affected members and not linked to the gene, was subjected to a genome-wide search and was found to be mapped to 9q34.3 with a multipoint lodscore of 3.9. One candidate gene in the interval, coding for the chloride intracellular channel 3, CLIC3, was tested and excluded. The identification of a new NSRD locus, DFNB33, in one Jordanian family, shows the wide genetic heterogeneity that characterizes hearing impairment and the genetic diversity in Middle-Eastern populations.
Similar content being viewed by others
Introduction
Hearing loss is a common pathology that typically illustrates genetic heterogeneity in human populations. It has been a centre of interest in the last few years that witnessed the localisation and the identification of more than 50 responsible genes.1 Genetically transmitted hearing loss represents 50% of congenital deafness and could be either isolated or non-syndromic (70%) or associated with other anomalies (30%). Non-syndromic recessive deafness (NSRD) is the most commonly encountered form of hereditary hearing loss and is generally characterised by a sensorineural defect.1 Nearly thirty loci that harbour a gene responsible for NSRD have been reported till now, and 10 of the corresponding genes have been identified so far (http://www.uia.ac.be/dnalab/hhh/). The connexin 26 gene, GJB2, located on the long arm of chromosome 13 (DFNB1 locus), accounts for the majority of NSRD cases, notably in the Mediterranean area.2
Consanguineous marriages that prevail in countries of the Middle East such as Jordan provide suitable conditions for the occurrence of recessively transmitted diseases, notably NSRD. We report here a new localisation on chromosome 9 of a type of NSRD that affects a Jordanian family, and discuss the frequency of the DFNB1 linked hearing impairment in Jordan.
Patients and methods
Sixty-eight unrelated Jordanian consanguineous families with at least two members having a non-syndromic recessive hearing impairment willingly participated in this study. The patients' hearing loss was prelingual, varied from mild to severe or profound, and was not caused by inflammatory middle ear disease nor specific environmental factors. All cases were examined by otoscopy, and auditory brain stem responses (ABR) were recorded for young affected children. Pure tone audiometry tests for air and bone conduction at frequencies varying from 250 to 8000 Hz were systematically performed.
Screening of the GJB2 gene
EDTA blood samples were obtained from the 68 unrelated patients and DNA was extracted from leucocytes by a previously described technique.3 The patients were tested for mutations in the GJB2 gene to ascertain if their hearing impairment was due to the DFNB1 locus. Systematic sequencing of the sole exon of the GJB2 gene was performed for all patients as previously described4 using the ABI 377 DNA sequencer.
Screening of control chromosomes for the 35delG mutation
Blood samples previously collected from 54 consenting unrelated Jordanian individuals were used as controls. DNA was extracted in order to test, by RFLP, the frequency of the most common mutation of the GJB2 gene, 35delG, in the general Jordanian population. This mutation screening was performed by a previously described RFLP method.5
Genotyping and linkage analysis
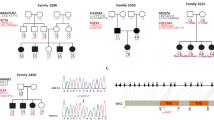
One of the NSRD affected families (Family H) presented with four members having a non-syndromic recessive hearing impairment since their early childhood (Figure 1). Their hearing loss was severe and bilateral, and was not found to be linked to the DFNB1 locus. The family was subsequently subjected to a systematic genome-wide screening in order to determine the implicated NSRD locus. Genotyping and fluorescent microsatellite marker amplification were as previously reported.6,7 Two- point and multipoint lodscores were calculated using respectively the MLINK and the LINKMAP programmes of the LINKAGE package.
CLIC3 gene screening
The whole genomic sequence of CLIC3 was amplified for one affected member of Family H, using the following pair of primers: CLIC3-5F (5′-CAGGATGCCTGGGTCTGCTTCC-3′) and CLIC3-5R (5′-GGTTTGGGGATGGGAAGTGATCC-3′). The middle part of the gene, consisting of 656 base pairs, was further amplified with primers CLIC3-a (5′-GAGGACGCGCGCAAGACAGA-3′) and CLIC3-b (5′-GTATTTGAACTCTTTCTCCTGCATCG-3′). The amplified fragments were sequenced using the ABI 377 DNA sequencer.
Results and Discussion
In the Jordanian families involved in this study, sequencing of the GJB2 single exon revealed only the 35delG mutation, found at the homozygous state in 11 patients out of the 68 tested. It was not detected at the heterozygous state, and no other mutations of the gene were identified in any of the patients. Subsequently, only 16.2% of the families were linked to the DFNB1 locus. This frequency is lower than reported in other Mediterranean countries such as Italy and Spain,8 Israel9 and Lebanon,10 where 49%, 41.3% and 33% of NSRD respectively were due to GJB2 mutations. The 35delG mutation, a frameshift mutation that generates a premature stop codon, was found in 100% of DFNB1-linked cases, whereas it accounts for 70% of the GJB2 mutations in families of different origins,4 88% in Italy and 55% in Spain,8 55% in Australia,11 94% in Lebanon.10
In fact, the 35delG mutation is thought to occur in a hot spot site for mutations,4,12,13 although a recent article also suggested that its frequency could be secondary to a founder effect.14 As the number of Jordanian NSRD families linked to the DFNB1 locus was small, testing 108 control chromosomes was necessary to assess the frequency of this mutation in the general Jordanian population. None of these chromosomes bore the 35delG mutation, an indication of its relatively low frequency in this population, which is comparable to its prevalence in some other populations such as African Americans, Asian Americans, Egyptian and English populations.14 Therefore, its rare allelic frequency in the general Jordanian population and the subsequently rare DFNB1 involvement in Jordanian NSRD families (16%) are not in favour of a common founder for the 35delG GJB2 mutation. Indeed, the absence of a common founder for this mutation could be expected since Jordan has always been a crossroad in the region and was conquered successively by various populations throughout history.
The genome-wide search performed for the Jordanian Family H unlinked to DFNB1 revealed a 6.3 cM homozygosity region on the telomeric side of chromosome 9 (9q34.3) between the microsatellite polymorphic markers D9S1826 and D9S1838 (Figure 1). The highest lodscore was obtained with the D9S905 marker and was equivalent to 3.38 at θ=0. A maximum lodscore of 3.9 was obtained in the multipoint linkage analysis with microsatellites in the telomeric region of chromosome 9. This is the first family for which a deafness locus has been mapped to 9q34.3, thereby defining a new NSRD locus, DFNB33. This new localisation differed from NSRD localisations previously found in neighbouring Lebanon, where at least 13 implicated loci have been identified.10 Its discovery raises a solid argument for the presence of unknown isolates in the Middle-East countries.
The 9q34.3 region contains various plausible candidate genes for this pathology, such as the one coding for the chloride intracellular channel 3, CLIC3. This latter is predominantly localised in the nucleus and stimulates chloride conductance when expressed in cells.15 The CLIC3 gene is a small gene composed of five exons. Screening of its genomic sequence in one patient did not reveal any sequence variant, whether a mutation or a polymorphism, which allowed exclusion of CLIC3 as a potential candidate gene.
In conclusion, we have identified a new NSRD locus, DFNB33, mapped to 9q34.3, and showed the low frequency of DFNB1 involvment in Jordanian NSRD families, and of the 35delG in the Jordanian population at large. Screening more affected families and testing more control individuals for 35delG will contribute to more valid conclusions, and probably new NSRD localisations.
References
Resendes BL, Williamson RE, Morton CC . Review article. At the speed of sound: Gene discovery in the auditory system Am J Hum Genet 2001 69: 923–935
Zelante L, Gasparini P, Estivill X et al. Connexin26 mutations associated with the most common form of non-syndromic neurosensory autosomal recessive deafness (DFNB1) in Mediterraneans Hum Mol Genet 1997 6: 1605–1609
Miller SA, Dynes DD, Polesky F . A simple salting out procedure for extracting DNA from human nucleated cells Nucleic Acids Res 1988 16: 1215
Denoyelle F, Weil D, Maw MA et al. Prelingual deafness: high prevalence of a 30delG mutation in the connexin 26 gene Hum Mol Genet 1997 6: 2173–2177
Storm K, Willocx S, Flothmann K, Van Camp G . Determination of the carrier frequency of the common GJB2 (connexin-26) 35delG mutation in the Belgian population using an easy and reliable screening method Hum mutat 1999 14: 263–266
Dib C, Fauré S, Fizames S et al. A comprehensive genetic map of the human genome based on 5,264 microsatellites Nature 1996 380: 152–154
Gyapay G, Ginot F, Nguyen S, Vignal A, Weissenbach J . Genotyping procedures in linkage mapping Methods 1996 9: 91–97
Estivill X, Fortina P, Surrey S et al. Connexin-26 mutations in sporadic and inherited sensorineural deafness Lancet 1998 351: 394–398
Sobe T, Vreugde S, Shahin H et al. The prevalence and expression of inherited connexin 26 mutations associated with nonsyndromic hearing loss in the Israeli population Hum Genet 2000 106: 50–57
Mustapha M, Salem N, Delague V et al. Autosomal recessive non-syndromic hearing loss in the Lebanese population: prevalence of the 30delG mutation and report of two novel mutations in the connexin 26 (GJB2) gene J Med Genet 2001 38: E36
Wilcox SA, Saunders K, Osborn AH et al. High frequency hearing loss correlated with mutations in the GJB2 gene Hum Genet 2000 106: 399–405
Kelley PM, Harris DJ, Comer BC et al. Novel mutations in the connexin 26 gene (GJB2) that cause autosomal recessive (DFNB1) hearing loss Am J Hum Genet 1998 62: 792–799
Morell RJ, Kim HJ, Hood LJ et al. Mutations in the connexin 26 gene (GJB2) among Ashkenazi Jews with nonsyndromic recessive deafness N Engl J Med 1998 339: 1500–1505
Van Laer L, Coucke P, Mueller RF et al. A common founder for the 35delG GJB2 gene mutation in connexin 26 hearing impairment J Med Genet 2001 38: 515–518
Qian Z, Okuhara D, Abe MK, Rosner MR . Molecular cloning and characterization of a mitogen-activated protein kinase-associated intracellular chloride channel J Biol Chem 1999 274: 1621–1627
Acknowledgements
We thank Mr Arnaud Lemainque, and Mrs Delphine Torchard-Pagnez and Delphine Bacq for their help, and Dr Salim Adib for his review. This work was supported by a Saint Joseph University grant for Scientific Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Medlej-Hashim, M., Mustapha, M., Chouery, E. et al. Non-syndromic recessive deafness in Jordan: mapping of a new locus to chromosome 9q34.3 and prevalence of DFNB1 mutations. Eur J Hum Genet 10, 391–394 (2002). https://doi.org/10.1038/sj.ejhg.5200813
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200813
Keywords
This article is cited by
-
Genetic etiology of hereditary hearing loss in the Gulf Cooperation Council countries
Human Genetics (2022)
-
Prevalence of GJB2 (CX26) gene mutations in south Iranian patients with autosomal recessive nonsyndromic sensorineural hearing loss
Molecular Biology Reports (2012)
-
DFNB79: reincarnation of a nonsyndromic deafness locus on chromosome 9q34.3
European Journal of Human Genetics (2010)
-
Re-assigning the DFNB33 locus to chromosome 10p11.23–q21.1
European Journal of Human Genetics (2009)