Abstract
X-linked inhibitor of apoptosis protein (XIAP) is a member of the inhibitor of apoptosis proteins family that selectively binds and inhibits caspase-3, -7 and -9. As such, XIAP is an extremely potent suppressor of apoptosis and an attractive target for cancer treatment. Che-1 is an antiapoptotic agent involved in the control of gene transcription and cell proliferation. Recently, we showed that the checkpoint kinases ATM/ATR and checkpoint kinase 2 physically and functionally interact with Che-1 and promote its phosphorylation and accumulation in response to DNA damage. These Che-1 modifications induce transcription of p53, and Che-1 depletion strongly sensitizes tumor cells to anticancer drugs. Here we show that Che-1 activates XIAP expression in response to DNA damage. This effect is mediated by Che-1 phosphorylation and requires NF-κB. Notably, we found that XIAP expression is necessary for antiapoptotic activity of Che-1 and that in vivo downregulation of Che-1 by small interference RNA strongly enhanced the cytotoxicity of anticancer drugs.
Similar content being viewed by others
Main
Apoptosis is a tightly regulated and selective physiologic process that governs the removal of supernumerary or defective cells.1 Despite a variety of apoptosis-initiating events, apoptosis signaling pathways converge on a common pathway mediated by caspases, a group of intracellular cysteine proteases produced in cells as catalytically inactive zymogens and activated by proteolytic cleavage during apoptosis.2
The enzymatic activity of caspases is subject to inhibition by the conserved IAP (inhibitor of apoptosis) family of proteins.3, 4 Eight distinct mammalian IAPs, including X-linked inhibitor of apoptosis (XIAP), c-IAP1, c-IAP2 and ML-IAP/Livin, have been identified.3, 4 Of these members, XIAP has received the most attention because it is the only member of this family able to directly inhibit both the initiation and execution phases of the caspase cascade crucial to mediate the controlled demise of malignant cells.5 Overexpression studies have clearly demonstrated the cell death suppressing ability of XIAP in cancer cells or in vivo.6 Conversely, downregulation of XIAP expression by multiple approaches (e.g., antisense, RNAi, knockout animals and cell lines, immunodepletion) all led to increased caspase activation and/or cell death under appropriate conditions in vitro or in vivo.7 Thus, the removal of XIAP function, or XIAP itself, by multiple experimental means, supports the notion that XIAP is an apoptosis suppressor whose inhibition or elimination provides increased sensitization to cell death, a fact that can be exploited for cancer therapy.7
Che-1 is a human RNA polymerase II-binding protein highly conserved from yeast to man and involved in the regulation of gene transcription.8, 9, 10, 11, 12, 13 Indeed, Che-1 interacts with retinoblastoma protein (Rb) and affects its growth suppression activity by interfering with the Rb-mediated recruitment of histone deacetylase I on the promoters of E2F1-responsive genes.8, 13 Furthermore, Traube, the mouse hortholog of Che-1, was shown to be essential for proliferation in early embryogenesis.9 Despite this pro-proliferative role of Che-1/Traube/AATF (antiapoptotic transcription factor), it has been described that this protein is involved in the regulation of the apoptosis, exhibiting a strong antiapoptosis activity.11, 14 Furthermore, Che-1 was found downregulated during the apoptotic process by its interaction with mouse double minute 215 and neutrophin receptor-interacting MAGE homolog.16 In agreement with these results, we observed that Che-1 is downregulated in several colon carcinomas17 and that Che-1 overexpression caused cell cycle arrest in human colon carcinoma cell lines through induction of the cyclin-dependent kinase inhibitor p21Waf1 (p21).17 Recently, we showed that DNA damage by different genotoxic agents is associated with Che-1 phosphorylation and extended half-life.18 These post-translational modifications are induced by ataxia telangiectasia mutated protein (ATM) and checkpoint kinase 2 (Chk2), which phosphorylate Che-1 on specific residues and are functionally linked to DNA damage-induced G2/M checkpoint. Moreover, microarray and chromatin immunoprecipitation (ChIP) analyses show that Che-1 activates the transcription of p53 and consequently of several p53 target genes, identifying a new pathway by which ATM and Chk2 modulate p53 levels.18 More importantly, we found that Che-1 inhibition intensifies the cytotoxicity of anticancer drugs that damage the DNA and, in such way, can revert chemoresistance of several tumor cell lines.18
In this report, we describe that in response to genotoxic stress, Che-1 upregulates XIAP expression and this activation is required for Che-1 antiapoptotic activity. Moreover, we show that in vivo depletion of Che-1 is able to sensitize HCT116 tumors to antineoplastic drugs. These findings validate Che-1 as a possible therapeutic target to increase the efficacy of chemotherapy.
Results and Discussion
Che-1 regulates XIAP expression
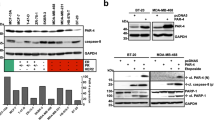
We have previously demonstrated that DNA damage leads to stabilization and accumulation of Che-1, which plays an important role in transcriptional activation of p53 and in maintenance of the G2/M checkpoint.18 Since Che-1 showed a strong antiapoptotic activity,11, 14, 18 and p53 can induce apoptosis in response to stress conditions,19 we assumed that Che-1 could also regulate other pathways in response to genotoxic stress. Interestingly, among the genes induced by Che-1, we identified XIAP, whose antiapoptotic role is well established.3, 4, 5 Microarray data were confirmed by semiquantitative RT-PCR (Figure 1a) and by western blot analysis (Figure 1b). To extend these results in human cells, Che-1 expression was silenced by small interference RNA (siRNA) in HCT116 cells. Figure 1c shows that specific Che-1 depletion significantly inhibited the transcription of XIAP, and this inhibition was associated with a strong reduction of XIAP protein levels (Figure 1d). These data indicate that Che-1 contributes to XIAP regulation.
Che-1 regulates X-linked inhibitor of apoptosis (XIAP) expression. (a) RNA was isolated from NIH3T3 cells uninduced (−Tet) and induced (+Tet) for Che-1 expression. Equal amounts of RNA (RNA input) were analyzed by RT-PCR (25–30 cycles) for expression of indicated genes. The RT control lanes represent RT-PCR in the absence of reverse transcription. (b) Lysates from NIH 3T3 cells stably transfected as in (a) were subjected to western blot analysis with the indicated antibodies. (a and b) HCT116 cells were transiently transfected with siRNA GFP (siRNA control) or siRNA Che-1. (c) Total RNA was isolated and used for RT-PCR analysis to evaluate the expression of the indicated genes. (d) Western blot analysis was performed with the indicated antibodies
Che-1 regulates XIAP in response to DNA damage by NF-κB
Next, we evaluated whether XIAP expression is regulated in response to DNA damage and whether Che-1 is involved in this phenomenon. HCT116 cells treated with doxorubicin (Dox) exhibited an increase of XIAP levels, in particular after Che-1 overexpression (Figure 2a). Consistent with these findings, RT-PCR analysis showed a significant increase of XIAP expression after DNA damage, which was not observed in cells depleted of Che-1 (Figure 2b).
Che-1 regulates X-linked inhibitor of apoptosis (XIAP) in response to DNA damage by NF-κB. (a) HCT116 cells were transiently transfected with Myc-Che-1 or Myc empty vector and 48 h later treated, where indicated, with 1 μM doxorubicin (Dox) for 2 h. Western blot analysis was performed with the indicated antibodies. (b) HCT116 cells were transiently transfected with siRNA GFP (siRNA control) or siRNA Che-1 and 48 h later treated with 1 μM Dox where indicated. Top: total RNA was isolated and used for RT-PCR analysis to evaluate the expression of the indicated genes. Bottom: western blot analysis performed with the indicated antibodies. (c and d) HCT116 cells were transfected with Myc-Che-1 or Myc-Che-1S4A expression vectors (c) or with Smart-Pool siRNA (siRNA control) or p65 siRNA (siRNA p65) and Myc-Che-1 expression vector (d) and 48 h later treated as in (a). Top: total RNA was isolated and used for RT-PCR analysis to evaluate the expression of the indicated genes. Bottom: western blot analysis was performed with the indicated antibodies. (e) HCT116 cells, whether treated or not with 1 μM Dox for 2 h, were subjected to chromatin immunoprecipitation (ChIP) analysis using the indicated antibodies or control IgGs (IgG). (f and g) HCT116 cells transfected and treated as in (c or d) were subjected to ChIP using the indicated antibodies or control IgGs (IgG)
To verify whether Che-1 activity on XIAP requires its phosphorylation, HCT116 cells were transfected with Myc-Che-1 wild-type or Myc-Che-1S4A mutant, lacking both ATM/ATR (ATM- and Rad3-related protein) and Chk2 phosphorylation residues,18 and treated with Dox. As is shown in Figure 2c, wild-type Che-1 strongly increased XIAP expression, whereas Myc-Che-1S4A mutant did not exert the same effect, thus indicating that XIAP regulation by Che-1 involves its phosphorylation by ATM and Chk2. Since it has been demonstrated that NF-κB regulates the activation of XIAP,20 and Che-1 activates p53 transcription by binding NF-κB subunit p65,18 we tested whether p65 is required for XIAP activation by Che-1. To do this, Che-1 was transfected in HCT116 cells where p65 expression was inhibited by specific siRNA, and RT-PCR and western blot analyses were performed (Figure 2d). These experiments demonstrated that in the absence of p65, Che-1 was not able to activate XIAP transcription (Figure 2d). To confirm these results, we performed ChIP experiments demonstrating that DNA damage increases the presence of endogenous Che-1 to the XIAP promoter (Figure 2e). Similar results were obtained when HCT116 cells were transfected with wild-type Myc-Che-1 (Figure 2f), whereas the mutant Myc-Che-1S4A was only barely detectable on the XIAP promoter both in untreated or Dox-treated cells (Figure 2f). Notably, the amount of Che-1 on the XIAP promoter strongly decreased in the absence of p65 (Figure 2g).
Che-1 was found to activate XIAP transcription in the absence of DNA damage (Figure 1a), but even in this case XIAP transactivation requires Che-1 phosphorylation and binding to NF-κB (Figures 2c and d). Similar results were observed in the regulation of p53 transcription by Che-1,18 and consistent with these results, Che-1 phosphorylation was also slightly detectable even in the absence of Dox treatment,18 probably by intrinsic DNA damage occurring during DNA replication or by other mechanism/s to be characterized. Therefore, it is possible that Che-1 is required for the basal state of p53 and XIAP expression and that this effect is preserved and reinforced in response to DNA damage, supporting the hypothesis of a common antiapoptotic pathway between Che-1 and NF-κB.
Che-1 inhibits apoptosis by XIAP
We have previously shown that Che-1 depletion by siRNA strongly increased Dox cytotoxicity and induced apoptosis in HCT116 cells treated with sublethal doses of chemotherapeutics.18 In addition, Che-1 inhibition reverted chemoresistance in several cancer cell lines.18 Therefore, we asked whether these effects were mediated, at least in part, by inhibition of XIAP. To do this, we explored XIAP levels in HCT116 cells depleted of Che-1 by siRNA and treated with sublethal doses of Dox or cisplatin (DDP). As is shown in Figure 3a, Che-1 inhibition produced a strong decrease of XIAP with a concomitant activation of caspase-3. Similar results were obtained with colon cancer LoVo DX and ovarian cancer c13 cells, resistant to Dox and DDP, respectively (Figure 3a). Notably, Che-1 overexpression did not protect HCT116 XIAP−/− cells (Figure 3b), supporting the requirement of XIAP for Che-1 antiapoptotic activity.
X-linked inhibitor of apoptosis (XIAP) is required for Che-1 antiapoptotic activity. (a) The indicated cells were transiently transfected with siRNA GFP (−) or siRNA Che-1 (+) and 48 h later treated with the indicated doses of Dox or cisplatin (DDP) for 12 h. Western blot analysis was performed with the indicated antibodies. (b) HCT116 and HCT116 XIAP−/− cells were transfected with Myc-Che-1 or Myc empty vector and treated with 1 μM Dox. Cell death was assayed 48 h later. Data are presented as the mean±S.D. from three independent experiments performed in duplicate. In the lower panels, the expression levels of Myc tagged Che-1 were assayed by anti-Myc western blotting
Che-1 is involved in resistance to antineoplastic drugs
Next, we evaluated if in vivo administration of siRNA was able to silence Che-1 expression and to decrease XIAP protein. To do this, mice-bearing HCT116 xenografts were treated with siRNA oligos for Che-1 or for negative control siRNA (green fluorescent protein (GFP)). At day 15 after tumor cells injection (5 days after completion of siRNA treatment), western blot analysis was performed on excised tumors. As reported in Figure 4a, an almost complete inhibition of Che-1 expression was evident in tumors treated with siRNA for Che-1, and this effect was associated with a marked reduction of XIAP expression. The downregulation of Che-1 correlated with a significant reduction of growth of tumors (Figure 4b), as about 30% inhibition of tumor weight (P=0.02), calculated at the nadir of the effect, was evident in mice treated with siRNA. More interestingly, this treatment sensitized HCT116 tumors to antineoplastic drugs; in fact, while this tumor exhibited resistance to DDP treatment (only 15% reduction of tumor weight versus untreated, P>0.05), the combination with Che-1 depletion elicited a marked antitumor efficacy, being about 60% reduction in tumor weight that was significantly different (P<0.001) compared to untreated groups and to groups treated with siRNA or DDP given alone. It is interesting to note that the maximum inhibitory effect on the growth of tumor is evident just a few days after the end of treatment thus confirming the relationship between the downregulation of Che-1 and sensitization of tumors to DDP antineoplastic activity (Figure 4b). Figure 4c shows representative tumor-bearing mice or excised tumors, confirming the marked decrease of tumor mass in mice treated with the siRNA for Che-1/DDP combination compared to untreated mice or to mice treated with each drug alone. The treatment with control siRNA did not increase the response to DDP (data not shown).
Che-1 is involved in resistance to antineoplastic drugs. (A) Western blot analysis of Che-1 and X-linked inhibitor of apoptosis (XIAP) protein expression on tumors treated with siGFP (siRNA Control) or siRNA Che-1. Analysis was performed 5 days after the end of siRNA treatment. (B) Percent of tumor weight in the different treated groups compared to untreated tumors was evaluated at different days after the injection of tumor cells. Points are means (bars, S.D.). Each group included six mice. Groups are as follows: Δ, DDP; •, siChe-1; •, siChe-1/DDP. The graphic is representative of two different experiments with similar results. (C) Representative mice (up) or excised tumors (down) from untreated group (a), or from groups treated with DDP (b), siChe-1 (c) alone or in combination (d), 20 days after the injection of tumor cells. A millimetric scale is also shown
These data conclusively show that (a) Che-1 activates XIAP transcription; (b) this activity is increased in response to DNA damage, is mediated by ATM and Chk2, and requires NF-κB; (c) XIAP is requested for Che-1 antiapoptotic activity; (d) in vivo Che-1 downregulation by siRNA sensitizes cancer cells to chemotherapy. This latter observation that Che-1 inhibition not only induces apoptosis, which inhibits p53 and p21 expression,18 but also depletes XIAP cellular levels, strongly reinforcing the notion that Che-1 can be considered a valid target for design of drugs that can enhance the efficacy of antitumor agents.
Materials and Methods
Cell culture, transfections and analysis
HCT116 and HCT116 XIAP−/− (kindly provided by Dr. Bunz, Johns Hopkins University, Baltimore) human colon carcinoma cell lines and NIH3T3 expressing tetracycline (Tet)-conditional human Che-1 13 were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS). Dox-resistant LoVo Dx cells and DDP-resistant c13 cells were grown in RPMI-1640 plus 10% FBS. Transfection was carried out by Lipofectamine and Plus reagent (Invitrogen). Transfection efficiency ranged between 30 and 50% of cells. Dox was purchased from Sigma, whereas clinical-grade DDP (Cisplatino Teva) was obtained from Teva Pharma.
Plasmids and antibodies
The Myc-tagged Che-1 and its mutant Che-1S4A have been described previously.18 The following rabbit polyclonal antibodies were used: anti-Che-1,8 p65 (sc-6243, Santa Cruz), XIAP and cleaved caspase-3 (Cell Signaling Technology). Mouse monoclonal antibodies anti-Myc 9e10 (Invitrogen) and α-tubulin (sc-8035, Santa Cruz) were also used.
Western blot analysis
Cells were lysated in lysis buffer (50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 10% glycerol, 0.5 mM EDTA, 0.5 mM EGTA, 100 mM NaF, 3 mM Na3VO4, 10 nM okadaic acid, 1 mM PMSF, 10 μg/ml leupeptin, 1% NP40). Western blots were prepared by standard procedures and immunoreactivity was detected by ECL (Amersham).
Transcriptional profiling experiments
Microarray analysis was performed utilizing messenger RNA extracted from NIH3T3 cells conditionally expressing human Che-1 cDNA in a Tet-regulated manner as already described.18
RT-PCR analysis
For semiquantitative RT-PCR analysis, cells were harvested and total RNA was isolated using TRIZOL reagent (Invitrogen). RT-PCR was performed using a Platinum quantitative RT-PCR kit (Invitrogen) following the manufacturer's instructions. PCR products were separated onto 1.5% agarose gel. The following primers were employed: human XIAP forward, 5′-CAACACTGGCACGAGCAGGG-3′; human XIAP reverse, 5′-CATGGCAGGGTTCCTCGGGT-3′; human glyceraldehyde-3-phosphate dehydrogenase (Gapdh) forward, 5′CCAAGGTCATCCATGACAAC-3′; human Gapdh reverse, 5′-TTACTCCTTGGAGGCCATG T-3′; human Che-1 forward, 5′-CCGGAATTCGTTTCGCAGTGCTCTGAA-3′; human Che-1 reverse, 5′-CCGCTCGAGATGCCATTTCTGTAGTGT-3′.
Chromatin immunoprecipitation assay
ChIP assays were performed using anti-Che-1 or anti-Myc antibodies as described.18 Immunoprecipitations with no specific immunoglobulins (Santa Cruz) were performed as negative control. The following primers were employed to amplify XIAP promoter: forward, 5′-TAATGTCGAGCTCAGTATTT-3′; reverse, 5′-GCAGGTACACAAGTTTAGAT-3′.
siRNA
The modified (siStable) 22-nucleotide siRNA duplexes corresponding to nucleotides 1472–1493 of the human Che-1 sequence and to nucleotides 122–143 of the negative control green fluorescent protein (GFP) sequence were synthesized by Dharmacon. RNA interference was performed as described previously.13 SiRNA-mediated interference experiments of p65 expression were performed by transfecting SMART pool specific or nonspecific control pool double-stranded RNA oligonucleotides (Dharmacon) using Lipofectamine Plus (Invitrogen).
In vivo experiments and statistical analysis
Male CD-1 nude (nu/nu) mice (6–8 weeks old and 22–24 g in body weight) were purchased from Charles River Laboratories (Calco, Italy). All procedures involving animals and their care were conducted in conformity with institutional guidelines, which are in compliance with national (DL no. 8. GU, July 1994) and international laws (EEC Council Directive 86/609, OJ L 358. 1, December 12, 1987; Guide for Care and use of Laboratory Animals, United States National Research Council, 1996). Each experimental group included six mice.
Mice were injected subcutaneously with a cell suspension of 106 HCT116 cells per mouse. Six days after cell implant, a tumor mass of about 100 mg was evident in all injected mice. Starting from this time, the mice were treated with intratumoral injection of modified double-strand RNA oligos (siStable) for siRNA Che-1 or for the negative control siRNA (GFP), at 0.1 mg per mouse per day for four consecutive days. Clinical-grade DDP was given intraperitoneally at 10 mg/kg in a single bolus (10% lethal dose, LD10). In the combination experiments, the mice received siRNA at days 6–9 and DDP at day 10 after tumor cell implant. As controls, tumor-bearing mice received the same volume of diluents used to dissolve DDP and siRNA for the same period of combination treatment.
Tumor weight was calculated from caliper measurements according to the method of Geran et al.21, 22 Antitumor efficacy of treatment was assessed by using percent tumor weight in treated mice compared to untreated ones (T/C%). To evaluate protein levels in xenografts, 100 mg of mechanically disaggregated control and treated tumors were solubilized in lysis buffer and analyzed by western blotting. The statistical difference of tumor weight among the different groups was determined by Student's t-test assuming unequal variances. Differences were considered statistically significant when P<0.05.
Abbreviations
- AATF:
-
antiapoptotic transcription factor
- ATM:
-
ataxia telangiectasia mutated protein
- ATR:
-
ATM- and Rad3-related protein
- ChIP:
-
chromatin immunoprecipitation
- Chk2:
-
checkpoint kinase 2
- DDP:
-
cisplatin
- Dox:
-
doxorubicin
- GFP:
-
green fluorescent protein
- Rb:
-
retinoblastoma protein
- siRNA:
-
small interference RNA
- Tet:
-
tetracycline
- XIAP:
-
X-linked inhibitor of apoptosis
References
Jin Z, El-Deiry WS . Overview of cell death signaling pathways. Cancer Biol Ther 2005; 4: 139–163.
Salvesen GS, Dixit VM . Caspases: intracellular signalling by proteolysis. Cell 1997; 91: 443–446.
Deveraux QL, Reed JC . IAP family proteins-suppressors of apoptosis. Genes Dev 1999; 13: 239–252.
Salvesen GS, Duckett CS . IAP proteins: blocking the road to death's door. Nat Rev Mol Cell Biol 2002; 3: 401–410.
Eckelman BP, Salvesen GS, Scott FL . Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep 2006; 7: 988–994.
LaCasse EC, Baird S, Korneluk RG, MacKenzie AE . The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene 1998; 17: 3247–3259.
Schimmer AD, Dalili S, Batey RA, Riedl SJ . Targeting XIAP for the treatment of malignancy. Cell Death Differ 2006; 13: 179–188.
Fanciulli M, Bruno T, Di Padova M, De Angelis R, Iezzi S, Iacobini C et al Identification of a novel partner of RNA polymerase II subunit 11, Che-1, which interacts with and affects the growth suppression function of Rb. FASEB J 2000; 14: 904–912.
Thomas T, Voss AK, Petrou P, Gruss P . The murine gene, Traube, is essential for the growth of preimplantation embryos. Dev Biol 2000; 227: 324–342.
Lindfors K, Halttunen T, Huotari P, Nupponen N, Vihinen M, Visakorpi T et al Identification of novel transcription factor-like gene from human intestinal cells. Biochem Biophys Res Commun 2000; 276: 660–666.
Page G, Lodige I, Kogel D, Scheidtmann KH . AATF, a novel transcription factor that interacts with Dlk/ZIP kinase and interferes with apoptosis. FEBS Letts 1999; 462: 187–191.
Burgdorf S, Leister P, Scheidtmann KH . TSG101 interacts with AATF and enhances AR-mediated transcription by promoting its mono-ubiquitination. J Biol Chem 2004; 279: 17524–17534.
Bruno T, De Angelis R, De Nicola F, Barbato C, Di Padova M, Corbi N et al Che-1 affects cell growth by interfering with the recruitment of HDAC1 by Rb. Cancer Cell 2002; 2: 387–399.
Guo Q, Xie J . AATF inhibits aberrant production of amyloid beta peptide 1–42 by interacting directly with Par-4. J Biol Chem 2004; 279: 4596–4603.
De Nicola F, Bruno T, Iezzi S, Di Padova M, Floridi F, Passananti C et al The prolyl isomerase Pin1 affects Che-1 stability in response to apoptotic DNA damage. J Biol Chem 2007; 282: 19685–19691.
Di Certo MG, Corbi N, Bruno T, Iezzi S, De Nicola F, Desantis A et al NRAGE associates with the anti-apoptotic factor Che-1 and regulates its degradation to induce cell death. J Cell Sci 2007; 120: 1852–1858.
Di Padova M, Bruno T, De Nicola F, Iezzi S, D'Angelo C, Gallo R et al Che-1 arrests human colon carcinoma cell proliferation by displacing HDAC1 from the p21WAF1/CIP1 promoter. J Biol Chem 2003; 78: 36496–36504.
Bruno T, De Nicola F, Iezzi S, Lecis D, D'Angelo C, Di Padova M et al Che-1 phosphorylation by ATM and Chk2 kinases activates p53 transcription and the G2/M checkpoint. Cancer Cell 2006; 10: 479–486.
Vousden KH, Lu X . Live or let die: the cell's response to p53. Nat Rev Cancer 2002; 2: 594–604.
Tang G, Minemoto Y, Dibling B, Purcell NH, Li Z, Karin M et al Inhibition of JNK activation through NF-kappa-B target genes. Nature 2001; 414: 313–317.
Geran RI, Greenberg NH, Macdonald MM, Shumacher AM, Abbott BJ . Protocols for screening chemical agents and natural products against animal tumors and other biological systems. Ed 3 Cancer Chemother Rep 1972; 3: 1–88.
Schimmer AD . Inhibitor of apoptosis proteins: translating basic knowledge into clinical practice. Cancer Res 2004; 64: 7183–7190.
Acknowledgements
We thank Dr. F Bunz for sharing XIAP−/− HCT116 cells. This work was supported by the Italian Association for Cancer Research (AIRC) and Regional Grant-Lazio (AIRC). Simona Iezzi is the recipient of an FIRC fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by M Oren
Rights and permissions
About this article
Cite this article
Bruno, T., Iezzi, S., De Nicola, F. et al. Che-1 activates XIAP expression in response to DNA damage. Cell Death Differ 15, 515–520 (2008). https://doi.org/10.1038/sj.cdd.4402284
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4402284
Keywords
This article is cited by
-
Polymerase iota (POLI) confers radioresistance of esophageal squamous cell carcinoma by regulating RAD51 stability and facilitating homologous recombination
Cell Death Discovery (2023)
-
XIAP as a multifaceted molecule in Cellular Signaling
Apoptosis (2022)
-
AATF Competitively Interacts with Nuclear AIF and Inhibits Parthanatos of Neurons in dMCAO/R and OGD/R Models
Journal of Molecular Neuroscience (2022)
-
METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner
Experimental & Molecular Medicine (2021)
-
AATF suppresses apoptosis, promotes proliferation and is critical for Kras-driven lung cancer
Oncogene (2018)