Abstract
In this study, we performed two-dimensional electrophoresis (2-DE) and matrix-assisted laser desorption/ionisation time of fly mass spectrometry to identify the protein(s) associated with the development of oral squamous cell carcinomas (OSCCs) by comparing patterns of OSCC-derived cell lines with normal oral keratinocytes (NOKs), and found that downregulation of ubiquitous mitochondrial creatine kinase (CKMT1) could be a good candidate. Decreased levels of CKMT1 mRNA and protein were detected in all OSCC-derived cell lines examined (n=9) when compared to those in primary normal oral keratinocytes. Although no sequence variation in the coding region of the CKMT1 gene with the exception of a nonsense mutation in exon 8 was identified in these cell lines, we found a frequent hypermethylation in the CpG island region. CKMT1 expression was restored by experimental demethylation. In addition, when we transfected CKMT1 into the cell lines, they showed an apoptotic phenotype but no invasiveness. In clinical samples, high frequencies of CKMT1 downregulation were detected by immunohistochemistry (19 of 52 (37%)) and quantitative real-time RT–PCR (21 of 50 (42%)). Furthermore, the CKMT1 expression status was significantly correlated with tumour differentiation (P<0.0001). These results suggest that the CKMT1 gene is frequently inactivated during oral carcinogenesis and that an epigenetic mechanism may regulate loss of expression, which may lead to block apoptosis.
Similar content being viewed by others
Main
Oral cancer is a challenging clinical problem and a leading cause of cancer deaths for both men and women, with an estimated 300 000 new cases annually worldwide (Greenlee et al, 2000). Many risk factors have already been identified, including human papilloma virus infection, tobacco use, and alcohol consumption. However, some patients develop oral squamous cell carcinoma (OSCC) without risk factors, suggesting that host susceptibility may play a role. Molecular alterations in a number of oncogenes and tumour-suppressor genes associated with the development of OSCC could be important clues to preventing this disease.
Gene expression profiles using cDNA microarrays have been used to derive a useful molecular-based classification for the diagnosis, treatment, and prevention of several cancers (Alizadeh et al, 2000; Bittner et al, 2000). However, biologic systems comprise protein components resulting from transcriptional and post-transcriptional control, post-translational modifications, and protein shifts among different cellular compartments. These properties cannot be analysed by microarray systems at the RNA level, whereas proteomic analysis allows separation and visualisation of the protein content of a cellular sample (Celis et al, 1996). Considering this, proteome analysis may be a more powerful analytic technology to enhance study of the diagnosis, treatment, and prevention of human diseases, including human cancers (Sanchez et al, 1997; Wolfsberg et al, 2002).
Two-dimensional electrophoresis (2-DE) is the principal tool in proteomics, which can analyse thousands of proteins in one experiment, providing the highest resolution in protein separation. By comprehensively examining the protein expression profiles in normal and diseased samples via 2-DE, proteomics provides information on new biomarkers, disease-associated targets, and pathogenesis (Alaiya et al, 2000; Chambers et al, 2000; Miklos and Maleszka, 2001). This technique has been used extensively to investigate various diseases, including SCC of the oesophagus (Zhang et al, 2003), bladder (Ostergaard et al, 1997; Celis et al, 1999, 2000), lung (Li et al, 2003a, 2003b, 2004), and head and neck (Wu et al, 2002; Drake et al, 2005; Roesch Ely et al, 2005). However, little is known about the state of protein expression profiles in OSCCs. The aim of the present study was to identify the protein(s) associated with the development of OSCCs by comparing the patterns of OSCC-derived cell lines with normal oral keratinocytes (NOKs) using 2-DE and matrix-assisted laser desorption/ionisation time of fly mass spectrometry (MALDI-TOF-mass). We also examined the genetic and epigenetic changes and performed functional analysis of the candidate gene product identified.
Materials and methods
Cells
The nine human OSCCs-derived cell lines used in this study were Ca9-22, Ho-1-N-1, HSC-2, HSC-3, HSC-4, SAS (Human Science Research Resources Bank, Osaka, Japan), OK92 (established from carcinoma of the tongue in our department), Sa3, and H1 (provided by Dr Shigeyuki Fujita, Wakayama Medical University, Wakayama, Japan). All cell lines were maintained at 37°C (humidified atmosphere 5% CO2/95% air) on 150 × 20-mm tissue culture dishes (Nunc, Roskilde, Denmark) and cultured in Dulbecco's modified Eagle's medium F-12 HAM (Sigma Chemical Co, St Louis, MO, USA) with 10% fetal bovine serum (Sigma) and 50 U ml−1 penicillin and streptomycin. Primary cultured NOKs were used as a normal control as described previously (Kang et al, 2000; Endo et al, 2004).
Tissue samples and nucleic acid isolation
Tumours with patient-matched normal oral tissues (when available) were obtained at the time of surgical resection at Chiba University Hospital after informed consent had been obtained from the patients according to a protocol that was approved by the institutional review board of Chiba University. Resected tissues were divided into two parts: one was frozen immediately and stored at −80°C until use, and the other was fixed in 10% buffered formaldehyde solution for pathologic diagnosis. Histopathologic diagnosis of each cancerous tissue was performed according to the International Histological Classification of Tumours (Wahi, 1971) by the Department of Pathology, Chiba University Hospital. Clinicopathologic staging was determined by the TNM Classification of the International Union against Cancer (Hermanek et al, 1987). All patients had OSCC that was histologically confirmed, and tumour samples were checked to ensure the presence of tumour tissue in more than 80% of specimens. Genomic DNA was extracted as described previously (Uzawa et al, 1994); total RNA was prepared using Trizol Reagent (Invitrogen Corp., Carlsbad, CA, USA), according to the manufacturer's protocol.
Two-dimensional polyacrylamide gel electrophoresis
The protein expression profiles of seven OSCCs-derived cell lines (Ca9-22, OK-92, Ho1-N-1, HSC4, SAS, Sa3, and H1) and four NOKs were examined (Figure 2A). When the cultured cells grew into a full monolayer, they were washed three times with cold PBS (Sigma) and treated with lysis buffer containing 30 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 10% glycerol, and protease inhibitor cocktail to generate protein lysates for 10 min at 4°C. The cells were collected with a scraper and centrifuged at 15 000 r.p.m. for 10 min at 4°C. Protein concentrations were determined by the Bradford method. Total protein concentrations were normalised to 1 μg μl−1 for all samples, and the sample aliquots were stored at −20°C until use.
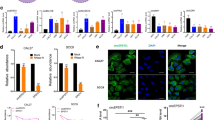
(A) A diagram of the experimental process. For details, see Materials and Methods. Immunofluorescence analyses show the status of CKMT1 protein expression. The CKMT1 proteins of the NOK (B) are surrounded by green fluorescence; fluorescence is not seen in the OSCC-derived cell line (OK92) samples (C). (D) Quantification of mRNA levels in OSCC-derived cell lines by real-time RT–PCR analysis. Significant downregulation of the CKMT1 gene is seen in all cell lines compared to CKMT1 mRNA expression in NOKs. Data are expressed as means±s.d.
For 2-D polyacrylamide gel electrophoresis (PAGE), each protein samples (100 μg) was mixed with sample buffer containing 7 M urea, 2 M thiourea, 4% (w v−1) 3-((3-cholamidopropyl)dimethylammonio)-1-propanesulphonic acid (CHAPS), and 1% (w v−1) DTT. Isoelectric focusing for the first dimension of protein separation was performed with a Multiphore II electrophoresis system (Amersham Biosciences, Piscataway, NJ, USA). Immobiline Dry IPG strips (7 cm, pH 3–10 nonlinear IPG strips; Amersham Biosciences) were rehydrated overnight with the sample/rehydration buffer mixture. The strips were subjected to electrophoresis using a ramping IPG strip (200–5000 V) focusing algorithm. After isoelectric focusing, the gel strips were equilibrated with sodium dodecyl sulohate (SDS) equilibration buffer and electrophoresed in vertical SDS–PAGE slab gels containing 12.5% acrylamide. The gels of the microdissected samples were fixed and silver stained using a Silver Quest silver staining kit (Invitrogen Japan, K.K., Tokyo, Japan), according to the manufacturer's instructions.
Two-dimensional gel analysis and MALDI-TOF-mass spectrometry
The silver stained gel was scanned into Adobe Photoshop 4.0J. (Adobe, San, Jose, CA, USA) with a Umax Powerlook II scanner (Umax, Dallas) and printed. Differences in protein levels were defined as clear visual differences in size, density, or both of the protein spot on the gel. Phoretix 2D Advanced software (Version 5.01, Nonlinear Dynamic, Ltd., Newcastle, UK) was used to estimate the relative differences in spot intensity of a candidate protein. Spots that were consistently and substantially different were selected for MALDI-TOF-mass analysis (Figure 2A). The protein spots were excised from the gels, and in-gel digestion was performed with an enzyme solution containing 50 mM NH4HCO3, 5 mM CaCl2, and 12.5 ng μl−1 trypsin. Aliquots of the purified samples were spotted on matrix crystals of α-cyano-4-hydroxyl-cinnamic acid on a stainless steel target and air dried. Mass determinations were performed on the AXIMA-CFR mass spectrometer (Shimadzu Co. Ltd, Kyoto, Japan). The proteins were identified by the peptide-mass fingerprinting method using Mascot Search on the Web (Matrix Science, Ltd, London, UK).
Immunofluorescence
Cells were grown on glass coverslips, fixed with methanol for 5 min, and permeabilised in 0.01%. Triton X-100 in PBS (pH 7.4) for 10 min. Nonspecific binding was blocked with 5% skim milk in PBS (pH 7.4) for 1 h. Cells were further incubated for 2 h with goat anti-CKMT1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at a dilution of 1:100, washed with PBS, and incubated with rabbit anti-goat secondary antibody labelled with Alexa Fluor 488 (Molecular Probes, Leiden, the Netherlands) for 1 h. Further, a DAPI nuclear stain with (4,6-diamidino-2-phenylindole dihydrochloride) was performed (0.5 μg ml−1). Coverslips were examined by fluorescence microscopy (Figure 2A). The microscope used was a Leica DMIRBE inverted stand equipped with a Leica TCS2-MP confocal system (Leica Laserteknik, Mannheim, Germany) and Coherent Mira tunable pulsed titanium sapphire laser (Coherent Laser Group, Santa Clara, CA, USA).
Immunohistochemistry
Immunohistochemistry (IHC) staining was performed on 4-μm paraffin-embedded specimens (Figure 2A). Briefly, after deparaffinisation and hydration, slides were treated with endogenous peroxidase in 0.3% H2O2 for 30 min and the sections blocked for 2 h at room temperature with 1.5% blocking serum (Santa Cruz Biotechnology) in PBS before reacting with anti-CKMT1 (Santa Cruz Biotechnology) at a dilution of 1 : 500. Sections then were incubated with primary antibody for 30 min at room temperature in a moist chamber. After incubation, the sections were washed three times in PBS buffer and treated with Envision reagent (Dako, Kyoto, Japan), followed by colour development in 3,3′-diaminobenzidine tetrahydrochloride (Dako). Finally, slides were lightly counterstained with haematoxylin and mounted. As a negative control, duplicate sections were immunostained without exposure to primary antibodies. To quantitate CKMT1 protein expression, a scoring method was used in which the mean percentage of positive tumour cells was determined in at least five random fields at × 400 magnification in each section. The intensity of the CKMT1 immunoreaction was scored as follows: 1+, weak; 2+, moderate; and 3+, intense. The percentage of positive tumour cells and the staining intensity were multiplied to produce a CKMT1 IHC staining (CKMT1-IHC) score for each case. Cases with a CKMT1-IHC score less than 62.2 (minimum score of normal tissues) were considered negative. Two independent pathologists without knowledge of or information pertaining to the patients' clinical status scored the cases. A statistically significance difference between the CKMT1-IHC scores and clinicopathologic features was evaluated by the Mann–Whitney's U-test. P<0.05 was considered significant.
mRNA expression analysis
The expression levels of CKMT1 mRNA were examined in the tumours, and paired normal oral tissues from the 50 patients with OSCC, nine OSCC-derived cell lines, and NOKs were analysed (Figure 2A). Control reactions were prepared in parallel without reverse transcriptase. Before cDNA synthesis, residual genomic DNA was removed from the total RNA by DNase I treatment (DNA-free; Ambion, Austin, TX, USA). The primer sequences used for analysis of CKMT1 mRNA expression were 5′-CCTGCTAAGCAAAGATAGCC-3′ (nucleotides 1162–1181), 5′-TAATGCTTGGTGTGGATGAC-3′ (nucleotides 1397–1416). The sequence of specific primers was checked before use to avoid amplification of genomic DNA or pseudogenes using the Primer3 program (http://www-genome.wi.mit.edu/cgibin/primer/primer3_www.cgi). The amplified products were analysed by 3% agarose gel electrophoresis to ascertain the size and purity of the products. To confirm the identity of the PCR products, they were cloned into a pCR 2.1vector (Invitrogen) and sequenced as described previously (Endo et al, 2004).
Real-time quantitative RT–PCR was performed with a single method using a LightCycler FastStart DNA Master SYBR Green I kit (Roche Diagnostics GmbH, Mannheim, Germany). To prepare the standard curve, 1.5 μg of total RNA from normal oral tissue was reverse transcribed with Superscript reverse transcriptase (Life Technologies, Grand Island, NY, USA) and oligod (T) primer, after which serial dilutions were made corresponding to cDNA transcribed from 300, 30, 3.0, and 0.3 ng of total RNA. The PCR reactions using the LightCycler (Roche Diagnostics) apparatus were carried out in a final volume of 20 μ1 of reaction mixture consisting of 2 μ1 of FirstStart DNA Master SYBR Green I mix (Roche), and 3 mM MgCl2, 0.2 μ1 of the primers, according to the manufacturer's instructions. The reaction mixture was loaded into glass capillary tubes and submitted to an initial denaturation at 95°C for 10 min, followed by 45 rounds of amplification at 95°C (10 s) for denaturation, 56°C (10 s) for annealing, and 72°C for extension, with a temperature slope of 20°C s−1, performed in the LightCycler. The transcript amount for the CKMT1 gene was estimated from the respective standard curves and normalised to the amount of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript determined in corresponding samples.
Mutational analyses
To screen the sequence variations of the CKMT1 gene, PCR-single-strand confirmation polymorphism (SSCP) analysis and DNA sequencing analysis were performed as described previously (Uzawa et al, 1995; Endo et al, 2004) (Figure 2A). Nine sets of oligonucleotide primers as summarised in Table 3 were used to amplify the entire coding region (exons 1–9) of the CKMT1 gene. Sequences for all annotated exons and adjacent intronic sequences were extracted from public (http://www.ncbi.nlm.nih.gov/) draft human genome sequences. Primers for PCR amplification and sequencing were designed using the Primer 3 program as described above.
Methylation analyses
To determine if methylation of a CpG island of the CKMT1 gene contributes to the mRNA expression of CKMT1, DNA samples obtained from the nine OSCC-derived cell lines were analysed using a PCR-based methylation assay (Figure 2A). The DNA samples were precipitated with ethanol, dissolved in distilled water, and further digested with mCpG-sensitive BstUI (1 μg U−1, New England Biolabs, Beverly, MA, USA) at 60°C for 16 h. The BstUI-digested DNAs then were amplified with specific primers for the CKMT1 CpG island (forward: 5′-CAGTGGGTGTCTTTCCCAGT-3′; reverse: 3′-GTTCACTGGCAGCTCGTACA-5′). The PCR reactions were performed in a final volume of 25 μ1 containing 1 μ1 of digested DNA, 2.5 pmol of each specific primer, 50 μ M of dNTPs, 10 mM Tris-HCl buffer (pH 8.3), 50 mM KCl 1.5 mM MgCl2, and 0.5 U of AmpliTaq (Applied Biosystems, Foster City, CA, USA). The amplified PCR products were separated on 3% agarose gel and visualised by ethidium bromide after the run. DNA from the NOKs treated in vitro with Sss I methylase (New England Biolabs) was used as a positive control for methylated alleles. The Sss I-untreated DNA was used as a negative control for methylated genes. To assess reactivation of CKMT1 mRNA expression, demethylating assay was performed using the DNA methyltransferase inhibitor, 5-aza-2′-deoxycytidine (5-aza-2′-dC) (Sigma Chemical Co.), as described previously (Yamamoto et al, 2001) (Figure 2A). The cells were treated with different concentrations (0 and 2 μ M) of 5-aza-2′-dC. On day 5, the cells were washed with PBS and grown for another 10 days without the demethylating chemical. The cells then were harvested, and total RNA was extracted, and the expression of the CKMT1 gene was evaluated by real-time quantitative RT–PCR as described above.
Transiently transfection
Human CKMT1 cDNA was cloned into a pME18SFL3 expression vector for transient transfection experiments (Joshi et al, 2002) (Figure 2A). The nine OSCC-derived cell lines were transfected with pME18SFL3 encoding CKMT1 cDNA using the nonliposomal formulation FuGENE-6 transfection reagent (Roche; Tao et al, 2004). All experiments using these cells were performed 48 h after transfection. Mock transfection of the nine OSCC-derived cell lines cultures with pME18SFL3 expression vector alone was used as a control. Real-time quantitative RT–PCR analyses and immunofluorescence were performed to confirm transfection efficiency.
In vitro invasion assay
The invasive activity of the nine CKMT1-transfected cells and nine mock-transfected cells in vitro were tested using a QCM 96-Well Cell Invasion Assay Kit (Chemicon, Temecula, CA, USA; Wang et al, 1997; Ciolczyk-Wierzbicka et al, 2004) (Figure 2A). The cells were loaded into a chamber coated with ECMatrix at a density of 5 × 105 cells ml−1 in 0.1 ml serum-free Dulbecco's modified Eagle's medium F-12 HAM. The plates with cells were incubated for 16 h at 37°C in a CO2 incubator. Migrating cells on the bottom of the insert were incubated with 150 μ1 of prewarmed cell detachment buffer for 30 min, dissociated from the membrane, and detected by CyQuant GR dye. The fluorescence plate was read using a 480/520 nm filter set.
Apoptosis assay
Apoptosis detection was carried out using a terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate (dUTP)-biotin nick end labeling (TUNEL) method (Tornusciolo et al, 2004) with an In situ Apoptosis Detection Kit (Takara), according to the manufacturer's protocol (Figure 2A). Briefly, the cells were fixed in 4% neutral buffered formalin, dried onto microscope slides, washed with PBS, equilibrated, and incubated with terminal deoxynucleotidyl transferase in a reaction buffer containing digoxigenin dUTP at 37°C for 1 h. The reaction was stopped, the specimens were washed for 30 min, and the slides were incubated with anti-digoxigenin antibody coupled to FITC for 30 min at room temperature and washed three times with PBS before mounting for photomicrography under phase and epifluorescence illumination. Apoptotic index was determined by calculating the percentage of cells that was apoptotic through positive staining. All slides were blindly evaluated by three independent times.
Results
Two-dimensional PAGE
Protein samples were extracted from the OSCC-derived cell lines and concentrations determined by the Bradford method using SmartSpec 3000 (Bio-RAD, Hercules, CA, USA). Total protein extraction rates did not differ significantly between the OSCC-derived cell lines and NOKs. Usually 100 μg of proteins per sample were used for 2-D PAGE separation. The proteins were distributed evenly in the 7-cm gel with isoelectric points in the range of 3–10 and molecular masses of 6–200 kDa. Figure 1A shows typical master gel images for OSCC-derived cell lines and NOK samples. About 450 protein spots in each gel were detected using the Phoretix Two-Dimensional Advanced v5.01 analysis program. Proteins with a >2-fold change in expression were considered significant. Three proteins were upregulated and 27 proteins were suppressed significantly in OSCC-derived cell lines compared with NOKs.
(A) Representative master 2-DE gel images of NOK and OSCC-derived cell line (Ca9-22) samples. After isoelectric focusing for the first dimension of protein separation, the gel strips were electrophoresed in vertical SDS–PAGE slab gels and stained with silver. Close-ups of 2-DE gel images were significantly downregulated in the OSCC-derived cell lines compared with the NOKs. Mw, molecular weight. (B) Results of MALDI-TOF-mass spectrometric analysis of this spot. Amino-acid sequences analysed by peptide mass fingerprinting analysis are redlined on the full-length sequence of the CKMT1 protein.
Protein identification by peptide-mass fingerprinting
Each protein spot was excised and subjected to in-gel tryptic digestion, MALDI-TOF-mass measurement, and database matching. We identified five candidate proteins that were differentially expressed in OSCC cell lines compared with NOKs. Table 1 shows the proteins identified; most of the matched proteins had high sequence coverage, mass accuracy, and MOWSE scores. We identified CKMT1 as a potentially novel molecular marker for OSCCs using this technique (Figure 1B).
Immunofluorescence
We examined the expression level of the CKMT1 proteins based on the results of proteomic analyses by immunofluorescence analysis. Nine OSCCs-derived cell lines and NOKs were examined. The CKMT1 proteins in the NOKs were surrounded by obvious green fluorescence; no fluorescence was seen in all OSCC-derived cell lines. The data were matched to the proteomic study. Figure 2C shows a representative result of significant decrease expression of CKMT1 protein in the OSCC-derived cell line, when compared to its expression in NOK (Figure 2B).
Immunohistochemistry
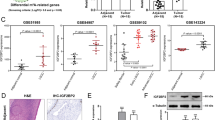
Among 52 OSCCs analysed by IHC staining, 19 had significantly decreased expression of CKMT1 (IHC score <62.2). In contrast, all normal tissues had a strong cytoplasmic immunoreaction for CKMT1. There was a statistically significant difference between the frequency of CKMT1-negative cases and clinicopathologic features (Table 2). All 21 (100%) oral premalignant lesions (OPLs) were CKMT1-positive. Figure 3A–F shows representative results for CKMT1 protein expression in normal oral tissue, OPLs, and primary OSCCs. The CKMT1-IHC scores for normal tissues, OPLs, OSCCs, well, moderately and poorly differentiated type of OSCCs, respectively, ranged from 62.2 to 200 (mean, 116.5), 67.5 to 200 (mean, 120), 0 to 200 (mean, 88.5), 0 to 200 (mean, 92.6), 0 to 123.4 (mean, 56.6), and 0 to 29.6 (mean, 12.6). CKMT1 expression levels in both moderately and poorly differentiated tissues were significantly lower than those in normal oral tissues (P=0.0002 and P=0.0001, respectively). In contrast, we found no significant difference in the CKMT1 IHC scores among normal oral tissue, OPLs, and well-differentiated tumours (P=0.966 and P=0.945, respectively) (Figure 3G).
Immunohistochemistry staining of CKMT1 in normal and tumorous oral tissues. (A) Normal oral tissue exhibits CKMT1 protein expression, which is limited to the cytoplasm of the epithelial cells. (B) CKMT1-positive case of OPL (leukoplakia). (C) CKMT1-positive case of well-differentiated primary OSCC. (D) CKMT1-negative case of well-differentiated primary OSCC. (E) CKMT1-negative case of moderately differentiated primary OSCC. (F) CKMT1-negative case of poorly differentiated primary OSCC. Original magnification × 200. (G) CKMT1 protein expression levels in moderately differentiated and poorly differentiated tumours are significantly lower than in normal oral tissues. No significant difference in the CKMT1 IHC scores is seen among normal oral tissue, OPLs, and well-differentiated tumours. (H) CKMT1 mRNA expression status in primary OSCCs. The relative mRNA expression level in normal tissues and primary OSCCs ranges from 0.1 to 20.1 (mean, 2.02) and 0.02 to 10.6 (mean, 0.86), respectively. There is a significant difference in the CKMT1 mRNA expression levels between negative and positive cases (P=0.0008, Mann–Whitney's U-test).
mRNA expression analysis
Real-time quantitative RT–PCR analysis data were matched to the protein expression values in the 2-DE analyses, immunofluorescence analyses, and immunohistochemistry analyses. mRNA expression of CKMT1 was significantly reduced in tumours of randomly selected CKMT1-negative cases (n=21) compared to the selected CKMT1-positive cases (n=29). mRNA expression levels were normalised to GAPDH. The relative mRNA expression level in the normal tissues and primary OSCCs ranged from 0.1 to 20.1 (mean, 2.02) and 0.02 to 10.6 (mean, 0.86), respectively (Figure 3H). There was no statistically significant difference between the frequency of CKMT1-negative cases and clinicopathologic features (Table 2). A significant decrease in CKMT1 expression was observed in OSCC-derived cell lines compared with NOKs used as controls (Figure 2D). The data are expressed as the mean±s.d. of three independent experiments with samples in triplicate.
Mutational analyses
To investigate the mechanisms responsible for downregulation of CKMT1 gene expression in OSCC-derived cell lines, we analysed genomic DNA by PCR-SSCP and direct sequence analyses. A band shift was detected in four of the nine (44%) OSCC-derived cell lines (Ho-1-N-1, HSC-2, HSC-4, SAS). Sequence variations were observed in exon 8. All sequencing variations were nonsense mutations (AAA changed to AAG and lysine changed to lysine) at codon 407 (data not shown).
Methylation analyses
We analysed the methylation status of the CpG island region of the CKMT1 gene in the 9 OSCC-derived cell lines. CKMT1 methylation was detected in four (44%) of the cell lines (Ho-1-N-1, HSC-2, HSC-3, HSC-4), whereas NOKs had no methylation allele. Figure 4A shows representative results of methylation status. To further study the consequences of loss of expression of CKMT1 in association with CpG island hypermethylation in OSCC-derived cell lines, the cell lines with methylation and transcriptional inactivation of CKMT1 were treated with 5-aza-2′-dC. Significant upregulation (in Ho-1-N-1, HSC-2, HSC-3, HSC-4) of mRNA expression was observed after 5-aza-2′-dC treatment (Figure 4B). The results were matched to methylation states studied in a PCR-based methylation assay. The data are expressed as the mean±s.d. of three independent experiments with samples in triplicate.
(A) Typical results of the methylation assay. CKMT1 methylation is seen in four OSCC cell lines (Ho-1-N-1, HSC-2, HSC-3, and HSC-4), which show reduced CKMT1 mRNA expression. DNA from NOK treated in vitro with SssI methylase is used as a positive control (lane PC) for methylated alleles. SssI-untreated DNA is used as a negative control (lane NC) for methylated genes. M, molecular marker. (B) Quantitative real-time RT–PCR analysis for demethylation assay in CKMT1-methylated OSCC cell lines. Restoration or upregulation of CKMT1 mRNA expression is detected in all CKMT1-methylated OSCC cell lines after treatment with the DNA demethylating agent 5-aza-2′-dC. +, 2 μ M 5-aza-2′-dC treatment; −, 0 μ M 5-aza-2′-dC treatment.
Functional analysis of CKMT1
To confirm transfection efficiency, quantitative real-time RT–PCR analyses and immunofluorescence were carried out. Quantitative real-time RT–PCR showed that CKMT1 was overexpressed 30–5000-fold in CKMT1-transfected OSCC-derived cell lines compared with mock-transfected OSCC-derived cell lines and untreated cell lines (Figure 5A). On immunofluorescence, no significant difference in the CKMT1 expression pattern was observed between mock-transfected OSCC-derived cell lines and untreated cell lines. In contrast, all CKMT1-transfected OSCC-derived cell lines had a strong immunoreaction for CKMT1.
CKMT1 transiently transfected efficiency is confirmed by quantitative real-time RT–PCR analyses and immunofluorescence. (A) Quantitative real-time RT–PCR revealed that CKMT1 is overexpressed 30–5000-fold in all CKMT1 transfectants compared with mock transfectants and untreated cell lines. (B) Representative result of TUNEL assay. More apoptotic cells were clearly seen in HSC-2-CKMT1 transfectants than in mock transfectants.
The invasive activity of the nine CKMT1-transfected cell lines and the nine mock-transfected cell lines were tested using the ECMatrix invasion chamber assay. Transfection of OSCC-derived cell lines with CKMT1 produces no significant decrease in invasive activity (data not shown).
To assess the specific role of CKMT1 downregulation, we evaluated the apoptotic ability between CKMT1-transfected OSCC-derived cell lines and the mock-transfected OSCC-derived cell lines. Cell morphology was analysed using optical microscopy. Morphological change showing apoptotic bodies, such as decrease of cellular and nuclear sizes, were observed in all CKMT1-transfected OSCC-derived cell lines, but not in mock-transfected OSCC-derived cell lines. TUNEL-positive cells were virtually undetected in mock-transfected OSCC-derived cell lines, whereas all the CKMT1-transfected OSCC-derived cell lines had increased numbers of TUNEL-positive cells (Figure 5B). The apoptotic index for CKMT1-transfected OSCC-derived cell lines and mock-transfected OSCC-derived cell lines, respectively, ranged from 26.92 to 76.58 (mean, 43.25), 2.77 to 14.12 (mean, 6.42).
Discussion
We identified CKMT1 as a potentially novel molecular marker for OSCCs. Creatine kinase (CK), originally found in cytosol and the mitochondria of cells, catalyses the reversible exchange of high-energy phosphate between ATP and phosphocreatine (PCr; Wallimann et al, 1992; Wallimann et al, 1998). Cytosolic CKs are dimmers consisting of muscle-type subunits, brain-type subunits, or both. However, two genotypes of mitochondrial CK have been identified, that is, CKMT1 and the sarcomeric mitochondrial CK that are encoded by two different genes on human chromosomes 15 and 5 and expressed in a tissue-specific manner (Stallings et al, 1988; Haas et al, 1989). CKMT1 is coexpressed with the cytosolic brain-type subunits in many cells and tissues with high-energy demand, such as the brain, placenta, kidney, testis, sperm, or endothelial cells. The enzyme is thought to have two main functions in energy metabolism (Wallimann et al, 1992): it buffers the cellular ATP pool by maintaining high cytosolic concentrations of PCr, which can be used in times of high cellular energy demand to regenerate ATP, and it maintains an energy shuttle between subcellular sites of energy supply (oxidative phosphorylation, glycolysis) and sites of energy demand using the easily diffusible creatine (Cr)/PCr. For example, CKMT1, located in the mitochondrial intermembrane space, preferentially catalyses the phosphorylation of Cr to PCr, which then diffuses into the cytosol where it is used in a reverse reaction by cytosolic CK to restore ATP for different, often closely associated ATPases (Schlattner et al, 1998). In addition, CKMT1 has been implicated in the regulation of the Ca2+-induced mitochondrial permeability transition pore (PTP; O'Gorman et al, 1997). PTP is involved in triggering apoptosis by releasing proapoptotic factors into the cytosol (Crompton, 1999). Several studies have shown the aberrant expression of CKMT1 in cancers of various organs, including lung cancer, breast cancer, gastric cancer, colon cancer, and prostate cancer (DeLuca et al, 1981; Tsung, 1983; Kanemitsu et al, 1984; Wallimann et al, 1992), suggesting that CKMT1 is involved in the pathogenesis of various cancers. However, there has been no report regarding CKMT1 expression in oral cancer, which lead us to further analyse this potential protein.
In the present study, we showed the state of CKMT1 gene mutation and its mRNA/protein expression in OSCC-derived cell lines and NOKs by SSCP/DNA sequencing, quantitative real-time RT–PCR, and immunofluorescence. Our data indicate that CKMT1 expression decreases or is lost in OSCC-derived cell lines but is abundant in NOKs. When DNA from OSCC-derived cell lines was analysed, we found nonsense mutations in the coding sequence of the CKMT1 gene. In contrast to mutational analyses, quantitative real-time RT–PCR analysis of CKMT1 mRNA showed frequent downregulation of the gene in all cell lines, indicating that other mechanisms, such as post-transcriptional modification and upregulated degradation, may be involved in CKMT1 gene silencing. In this context, with accumulating knowledge of the mechanisms of inactivation of tumour-suppressor genes, abnormal methylation at the promoters of tumour-suppressor genes is another mechanism that suppresses gene activity (Jones and Laird, 1999). In oral tumours, the promoters of several tumour-suppressor genes, that is, p16, p15, p14, and E-cadherin, are highly methylated in addition to a rare gene mutation in human OSCCs (Saito et al, 1998; Miracca et al, 1999; Shintani et al, 2001). In the present study, we found an association between methylation and suppression of CKMT1 gene expression in OSCC-derived cell lines. Treatment of OSCC-derived cell lines showed that CKMT1 methylation with a demethylating agent restored or significantly upregulated CKMT1 expression. These findings indicate that reduced expression of the CKMT1 gene may be linked to human oral carcinogenesis and/or progression of cancer due to epigenetic silencing. Thus, we suggest that CKMT1 could be a class II tumour-suppressor gene in that it is structurally intact in sequence but underexpressed or unexpressed due to downregulation or silencing in transcription or translation (Jones and Laird, 1999). These data show that proteomic analyses coupled with genetic and epigenetic analyses may be an essential strategy to discover novel molecular targets in the development of OSCCs.
To confirm the correlation between invasiveness and reduction of the CKMT1 protein, we examined the invasive activity of CKMT1-transfected OSCC-derived cell lines and mock-transfected OSCC-derived cell lines by in vitro invasion assay. The results showed that transfection of OSCC-derived cell lines with CKMT1 produces no substantial difference in invasive activity. We suggest that CKMT1 is unrelated to the invasiveness of the cells and suppresses or promotes metastasis of OSCCs.
To assess the specific role of CKMT1 downregulation, we evaluated apoptotic ability. The results showed that all CKMT1-transfected OSCC-derived cell lines had increased TUNEL-positive cells compared with mock-transfected OSCC-derived cell lines. Apoptosis can be mediated via several pathways, one of which involves the engagement of the so-called death receptors belonging to the tumour necrosis factor receptor superfamily. A cascade of proteolytic digestion occurs, involving caspases, resulting in cell death. However, caspase-8 may digest the Bid protein to yield a truncated form that can induce mitochondrial damage, eventually leading to cell death. In addition, many agents induce cellular stress, which can also lead to mitochondrial perturbation and ultimately cell death. A mechanism causing the mitochondrial dysfunction mentioned previously has been proposed that consists of mitochondrial membrane permeability transition, dissipation of the inner membrane potential, osmotic swelling of the matrix, rupture of the outer mitochondrial membrane, release of cytochrome c and other apoptogenic proteins from the mitochondria, and formation of the caspase-3 activation complex, the apoptosome (Debatin et al, 2002; Newmeyer and Ferguson-Miller, 2003). Permeability transition involves the opening of a PTP. The main components of this pore are adenine nucleotide translocator (ANT) and cyclophilin D in the inner membrane of the mitochondria and a voltage-dependent anion channel and a peripheral benzodiazepine receptor (VDAC) in the outer mitochondrial membrane. The PTP is formed in regions of contact between the inner and outer mitochondrial membranes. Prolonged opening of the PTP leads to the previously mentioned effects, exposes cytosol to the contents of the mitochondria, and culminates in cell death (Debatin et al, 2002; Newmeyer and Ferguson-Miller, 2003). Interestingly, mitochondrial permeability transition can lead to both apoptosis and necrosis (Lemasters et al, 1998). CKMT1 is located in the intermembrane space and also interacts with VDAC–ANT complexes (Marzo et al, 1998). We suggest that CKMT1 might induce apoptosis through specialised systems such as the PTP in OSCCs.
We showed the status of CKMT1 expression in clinical tissue samples obtained from primary OSCCs and the corresponding normal oral mucosa by quantitative real-time RT–PCR and IHC staining; high frequencies of CKMT1 downregulation were detected. Moreover, no difference in protein expression was observed in OPLs. In addition, we found a significant correlation between CKMT1 expression status and the clinicopathologic features. Most of the primary OSCCs diagnosed as moderately or poorly differentiated tumours showed downregulation of CKMT1 protein expression, and a statistically significant difference was observed between the CKMT1-reduced primary OSCCs and the histopathologic type (P<0.0001). Recently, a relation between mitochondrial homeostasis and tumour differentiation was reported (Krueger, 1995; Morgan et al, 2004). For example, the VDAC is mainly found on the outer mitochondrial membrane of cells as described previously. The VDAC plays a role in several cellular functions including haem synthesis, steroidogenesis, DNA synthesis, cell growth and differentiation, and apoptosis (Krueger, 1995). In cutaneous neoplasms and other skin diseases, a heterogeneous pattern of VDAC expression at lower intensity was seen depending on the tumour type and the degree of differentiation (Morgan et al, 2004). The VDAC expression was greatest in well-differentiated tumours, synonymous with the VDAC expression gradient seen in normal skin, and least in poorly differentiated and infiltrative tumours (Morgan et al, 2004). Although we have not examined the relationship between CKMT1 expression and oral cancer cell differentiation in vitro, above-mentioned evidences and our data suggest that change of mitochondrial homeostasis, in part, causes tumour differentiation. Furthermore, the potential role of CKMT1 in the regulation of both differentiation and apoptosis could be involved in the development of OSCC.
Based upon our data, we have concluded that downregulation of CKMT1 is, in part, involved in the oral carcinogenesis and that an epigenetic mechanism may regulate loss of its expression, which leads to block apoptosis through specialised systems such as mitochondrial permeability transition pores in oral cancer cells. Further investigations with greater number of clinical samples including precancerous lesions are needed to improve our ability to diagnose, prevent, and treat this neoplasm.
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Alaiya AA, Franzén B, Auer G, Linder S (2000) Cancer proteomics: from identification of novel markers to creation of artificial learning models for tumor classification. Electrophoresis 21: 1210–1217
Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson Jr J, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503–511
Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, Sampas N, Dougherty E, Wang E, Marincola F, Gooden C, Lueders J, Glatfelter A, Pollock P, Carpten J, Gillanders E, Leja D, Dietrich K, Beaudry C, Berens M, Alberts D, Sondak V (2000) Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature 406: 536–540
Celis JE, Gromov P, Ostergaard M, Madsen P, Honore B, Dejgaard K, Olsen E, Vorum H, Kristensen DB, Gromova I, Haunso A, Van Damme J, Puype M, Vandekerckhove J, Rasmussen HH (1996) Human 2-D PAGE databases for proteome analysis in health and disease:. http://biobase.dk/cgi-bin/celis. FEBS Lett 398: 129–134
Celis JE, Ostergaard M, Rasmussen HH, Gromov P, Gromova I, Varmark H, Palsdottir H, Magnusson N, Andersen I, Basse B, Lauridsen JB, Ratz G, Wolf H, Orntoft TF, Celis P, Celis A (1999) A comprehensive protein resource for the study of bladder cancer:. http://biobase.dk/cgi-bin/celis. Electrophoresis 20: 300–309
Celis JE, Wolf H, Ostergaard M (2000) Bladder squamous cell carcinoma biomarkers derived from proteomics. Electrophoresis 21: 2115–2121
Chambers G, Lawrie L, Cash P, Murray GI (2000) Proteomics: a new approach to the study of disease. J Pathol 192: 280–288
Ciolczyk-Wierzbicka D, Amoresano A, Casbarra A, Hoja-Lukowicz D, Litynska A, Laidler P (2004) The structure of the oligosaccharides of N-cadherin from human melanoma cell lines. Glycoconj J 20: 483–492
Crompton M (1999) The mitochondrial permeability transition pore and its role in cell death. Biochem J 341: 233–249
Debatin KM, Poncet D, Kroemer G (2002) Chemotherapy: targeting the mitochondrial cell death pathway. Oncogene 21: 8786–8803
DeLuca M, Hall N, Rice R, Kaplan NO (1981) Creatine kinase isozymes in human tumors. Biochem Biophys Res Commun 99: 189–195
Drake RR, Cazare LH, Semmes OJ, Wadsworth JT (2005) Serum, salivary and tissue proteomics for discovery of biomarkers for head and neck cancers. Expert Rev Mol Diagn 5: 93–100
Endo Y, Uzawa K, Mochida Y, Shiiba M, Bukawa H, Yokoe H, Tanzawa H (2004) Sarcoendoplasmic reticulum Ca(2+) ATPase type 2 downregulated in human oral squamous cell carcinoma. Int J Cancer 110 (2): 225–231
Greenlee RT, Murray T, Bolden S, Wingo PA (2000) Cancer statistics, 2000. CA Cancer J Clin 50: 7–33
Haas RC, Korenfeld C, Zhang ZF, Perryman B, Roman D, Strauss A W (1989) Isolation and characterization of the gene and cDNA encoding human mitochondrial creatine kinase. J Biol Chem 264: 2890–2897
Hermanek P, Sobin LH (eds) (1987) TNM Classification of Malignant Tumours, 4th edn, pp 16–18. Berlin: Springer-Verlag
Jones PA, Laird PW (1999) Cancer epigenetics comes of age. Nat Genet 21: 163–167
Joshi BH, Kawakami K, Leland P, Puri RK (2002) Heterogeneity in interleukin-13 receptor expression and subunit structure in squamous cell carcinoma of head and neck: differential sensitivity to chimeric fusion proteins comprised of interleukin-13 and a mutated form of Pseudomonas exotoxin. Clin Cancer Res 8: 1948–1956
Kanemitsu F, Kawanishi I, Mizushima J, Okigaki T (1984) Mitochondrial creatine kinase as a tumor-associated marker. Clin Chim Acta 138: 175–183
Kang MK, Bibb C, Baluda MA, Rey O, Park NH (2000) In vitro replication and differentiation of normal human oral keratinocytes. Exp Cell Res 258 (2): 288–297
Krueger KE (1995) Molecular and functional properties of mitochondrial benzodiazepine receptors. Biochim Biophys Acta 1241: 453–470
Lemasters JJ, Nieminen AL, Qian T, Trost LC, Elmore SP, Nishimura Y, Crowe RA, Cascio WE, Bradham CA, Brenner DA, Herman B (1998) The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim Biophys Acta 1366: 177–196
Li C, Chen Z, Xiao Z, Wu X, Zhan X, Zhang X, Li M, Li J, Feng X, Liang S, Chen P, Xie JY (2003a) Comparative proteomics analysis of human lung squamous carcinoma. Biochem Biophys Res Commun 309: 253–260
Li C, Chen ZC, Xiao ZQ, Wu XY, Zhan XQ, Li MY, Feng XP, Zhang XP, Li JL, Chen P, Liang SP (2004) Differential analysis of two-dimension gel electrophoresis profiles of human lung squamous carcinoma and tumor-adjacent tissue. Ai Zheng 23: 28–35
Li C, Zhan X, Li M, Wu X, Li F, Li J, Xiao Z, Chen Z, Feng X, Chen P, Xie J, Liang S (2003b) Proteomic comparison of two-dimensional gel electrophoresis profiles from human lung squamous carcinoma and normal bronchial epithelial tissues. Genomics Proteomics Bioinformatics 1: 58–67
Marzo I, Brenner C, Zamzami N, Susin SA, Beutner G, Brdiczka D, Remy R, Xie ZH, Reed JC, Kroemer G (1998) The permeability transition pore complex: a target for apoptosis regulation by caspases and bcl-2-related proteins. J. Exp. Med 187: 1261–1271
Miklos GLG, Maleszka R (2001) Integrating molecular medicine with functional proteomics: realities and expectations. Proteomics 1: 30–41
Miracca EC, Kowalski LP, Nagai MA (1999) High prevalence of p16 genetic alterations in head and neck tumours. Br J Cancer 81: 677–683
Morgan J, Oseroff AR, Cheney RT (2004) Expression of the peripheral benzodiazepine receptor is decreased in skin cancers in comparison with normal skin. Br J Dermatol 151: 846–856
Newmeyer DD, Ferguson-Miller S (2003) Mitochondria: releasing power for life and unleashing the machineries of death. Cell 112: 481–490
O'Gorman E, Beutner G, Dolder M, Koretsky AP, Brdiczka D, Wallimann T (1997) The role of creatine kinase in inhibition of mitochondrial permeability transition. FEBS Lett 414: 253–257
Ostergaard M, Rasmussen HH, Nielsen HV, Vorum H, Orntoft TF, Wolf H, Celis JE (1997) Proteome profiling of bladder squamous cell carcinomas: identification of markers that define their degree of differentiation. Cancer Res 57: 4111–4117
Roesch Ely M, Nees M, Karsai S, Magele I, Bogumil R, Vorderwulbecke S, Ruess A, Dietz A, Schnolzer M, Bosch FX (2005) Transcript and proteome analysis reveals reduced expression of calgranulins in head and neck squamous cell carcinoma. Eur J Cell Biol 84: 431–444
Saito Y, Takazawa H, Uzawa K, Tanzawa H, Sato K (1998) Reduced expression of E-cadherin in oral squamous cell carcinoma: relationship with DNA methylation of 5′CpG island. Int J Oncol 12: 293–298
Sanchez JC, Rouge V, Pisteur M, Ravier F, Tonella L, Moosmayer M, Wilkins MR, Hochstrasser DF (1997) Improved and simplified in-gel sample application using reswelling of dry immobilized pH gradients. Electrophoresis 18: 324–327
Schlattner U, Forstner M, Eder M, Stachowiak O, Fritz-Wolf K, Wallimann T (1998) Functional aspects of the X-ray structure of mitochondrial creatine kinase: a molecular physiology approach. Mol Cell Biochem 184: 125–140
Shintani S, Nakahara Y, Mihara M, Ueyama Y, Matsumura T (2001) Inactivation of the p14ARF, p15INK4B and p16INK4A genes is a frequent event in human oral squamous cell carcinomas. Oral Oncol 37: 498–504
Stallings RL, Olson E, Strauss AW, Thompson LH, Bachinski LL, Siciliano MJ (1988) Human creatine kinase genes on chromosomes 15 and 19, and proximity of the gene for the muscle form to the genes for apolipoprotein C2 and excision repair. Am J Hum Genet 43: 144–151
Tao DD, Jiang M, Feng YD, Wu JH, Gong JP (2004) Expression of estrogen receptor in different breast cancer cell lines transfected with estrogen receptor plasmid HEGO5 mediated by FuGENE6. Ai Zheng 23: 1166–1170
Tornusciolo DR, Schmidt RE, Roth KA (2004) Associations among beta-TrCP, an E3 ubiquitin ligase receptor, beta-catenin, and NF-kappaB in colorectal cancer. J Natl Cancer Inst 96: 1161–1170
Tsung SH (1983) Creatine kinase activity and isoenzyme pattern in various normal tissues and neoplasms. Clin Chem 29: 2040–2043
Uzawa K, Suzuki H, Yokoe H, Tanzawa H, Sato K (1995) Mutational state of p16/CDKN2 and VHL genes in squamous-cell carcinoma of the oral cavity. Int J Oncol 7: 895–899
Uzawa K, Yoshida H, Suzuki H, Tanzawa H, Shimazaki J, Seino S, Sato K (1994) Abnormalities of the adenomatous polyposis coli gene in human oral squamous-cell carcinoma. Int J Cancer 58: 814–817
Wahi PN (1971) Histological typing of oral and oropharyngeal tumours. In International Histological Classification of Tumours Cohen B, Luthra UK, Torloni H (eds), 4th edn, pp 9–28. Geneva: WHO
Wallimann T, Dolder M, Schlattner U, Eder M, Homemann T, O'Gorman E, Ruck A, Brdiczka D (1998) Some new aspects of creatine kinase (CK): compartmentation, structure, function and regulation for cellular and mitochondrial bioenergetics and physiology. Biofactors 8: 229–234
Wallimann T, Wyss M, Brdiczka D, Nicolay K, Eppenberger HM (1992) Intracellular compartmentation, structure and function of creatine kinase isoenzymes in tissues with high and fluctuating energy demands: the ‘phosphocreatine circuit’ for cellular energy homeostasis. Biochem J 281: 21–40
Wang M, Liu YE, Greene J, Sheng S, Fuchs A, Rosen EM, Shi YE (1997) Suppression of breast cancer growth and metastasis by a serpin myoepithelium-derived serine proteinase inhibitor expressed in the mammary myoepithelial cells. Oncogene 14: 2767–2774
Wolfsberg TG, Wetterstrand KA, Guyer MS, Collins FS, Baxevanis AD (2002) A user's guide to the human genome. Nat Genet 32: 1–79
Wu W, Tang X, Hu W, Lotan R, Hong WK, Mao L (2002) Identification and validation of metastasis-associated proteins in head and neck cancer cell lines by two-dimensional electrophoresis and mass spectrometry. Clin Exp Metast 19: 319–326
Yamamoto N, Uzawa K, Yakushiji T, Shibahara T, Noma H, Tanzawa H (2001) Analysis of the ANA gene as a candidate for the chromosome 21q oral cancer susceptibility locus. Br J Cancer 84: 754–759
Zhang LY, Ying WT, Mao YS, He HZ, Liu Y, Wang HX, Liu F, Wang K, Zhang DC, Wang Y, Wu M, Qian XH, Zhao XH (2003) Loss of clusterin both in serum and tissue correlates with the tumorigenesis of esophageal squamous cell carcinoma via proteomics approaches. World J Gastroenterol 9: 650–654
Acknowledgements
We thank Dr Yasuo Iwadate (Department of Neurological Surgery, Graduate School of Medicine, Chiba University) for helpful comments on the experiments. We also thank Lynda C Charters for editing this manuscript. This work was supported by Research Grant (No. 16209059) from the Ministry of Education, Science and Culture, Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Onda, T., Uzawa, K., Endo, Y. et al. Ubiquitous mitochondrial creatine kinase downregulated in oral squamous cell carcinoma. Br J Cancer 94, 698–709 (2006). https://doi.org/10.1038/sj.bjc.6602986
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6602986
Keywords
This article is cited by
-
Mitochondrial creatine kinase 1 regulates the cell cycle in non-small cell lung cancer via activation of cyclin-dependent kinase 4
Respiratory Research (2023)
-
Ubiquitous mitochondrial creatine kinase promotes the progression of gastric cancer through a JNK-MAPK/JUN/HK2 axis regulated glycolysis
Gastric Cancer (2023)
-
An integrative pan-cancer analysis of molecular characteristics and oncogenic role of mitochondrial creatine kinase 1A (CKMT1A) in human tumors
Scientific Reports (2022)
-
Mitochondrial creatine kinase 1 in non-small cell lung cancer progression and hypoxia adaptation
Respiratory Research (2021)
-
Structure and function of resistance arteries from BB-creatine kinase and ubiquitous Mt-creatine kinase double knockout mice
Amino Acids (2020)