Abstract
Antibiotic-producing microorganism can develop strategies to deal with self-toxicity. Cytorhodins X and Y, cosmomycins A and B, and iremycin, are produced as final products from a marine-derived Streptomyces sp. SCSIO 1666. These C-7 reduced metabolites show reduced antimicrobial and comparable cytotoxic activities relative to their C-7 glycosylated counterparts. However, the biosynthetic mechanisms and relevant enzymes that drive C-7 reduction in cytorhodin biosynthesis have not yet been characterized. Here we report the discovery and characterization of a reductase, CytA, that mediates C-7 reduction of this anthracycline scaffold; CytA endows the producer Streptomyces sp. SCSIO 1666 with a means of protecting itself from the effects of its anthracycline products. Additionally, we identified cosmomycins C and D as two intermediates involved in cytorhodin biosynthesis and we also broadened the substrate specificity of CytA to clinically used anthracycline drugs.
Similar content being viewed by others
Introduction
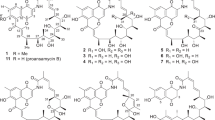
Antibiotic producing bacteria that produce toxic agents can develop and employ resistance mechanisms to survive the effects of their products; several major defensive strategies have come to light and these include: efflux pumps, chemical modifications, prodrugs, compound sequestration, subcellular localization, and target modifications/self-resistant protein variants1. Anthracycline antibiotics are effective agents in the treatment of various cancers despite their cumulative cardiotoxicity2. These compounds consist of a linear tetracyclic polyketide backbone and most are hydroxylated or glycosylated at the C-7 position. Naturally occurring C-7 reduced anthracycline antibiotics are relatively rare (Supplementary Fig. 1). A total of 14 anthracycline antibiotics were purified from marine-derived Streptomyces sp. SCSIO 16663. To our surprise, five C-7 deoxyanthracycline antibiotics, cytorhodins X (1) and Y (2), cosmomycins A (3) and B (4), and iremycin (5)3,4,5,6,7 (Fig. 1) were identified as major products. Previous reports indicated that deoxyanthracyclines generally show reduced toxicity relative to their C-7 glycosylated counterparts8. Compounds 1–10 (Fig. 1) were found to be mycelium-associated whereas fermentation broth supernatants contain less than 5% of the total amount of anthracyclines formed. However, the mechanism that explains the producer’s self-resistance to toxic DNA-intercalating agents remains unknown. Our observations here suggest that possible resistance mechanisms likely exist in Streptomyces sp. SCSIO 1666. Previous investigations into C-7 reduction have mainly focused on exogenous degradation aspects such as: degradation by mammalian tissue preparations9, bacterial bioconversion/degradation10,11, rat liver microsomal NADPH-cytochrome P450 reductase, xanthine oxidase, cytochrome C reductase, and DT-diaphorase-mediated enzymatic reactions in vitro12. Microbial conversions of anthracycline antibiotics by bacterial strains (Aeromonas hydrophila, Citrobacter freundii, and Escherichia coli) suggest that these reactions are NADH-dependent and require anaerobic or semi-anaerobic conditions10,13. Recently, a pair of enzymes, KstA15 and KstA16 that oxidize the C-1 position of aklavinone from the kosinostatin biosynthetic pathway was identified. Additionally, and somewhat surprisingly, the KstA15/KstA16 pair was also found to catalyze elimination of hydroxyl or glycosyl moieties at the aklavinone C-7 position to afford the shunt product 7-deoxyaklavinone under anaerobic conditions14.
a The gene locations of cytA in the cyt gene cluster. b cytA and its homologues locations in their respective gene clusters cyt, cin, acl, and niv from Streptomyces sp. SCSIO 1666, Streptomyces sp. SPB74, Streptomyces galilaeus, and Streptomyces niveus LS2151. c HPLC analysis of fermentation broths from (i) mutant strain ∆cytA; (ii) mutant strain ∆cytA supplemented with 3% XAD-16 resin; (iii) the WT strain Streptomyces sp. SCSIO 1666/ΔtrdA1DH; d Chemical structures of anthracyclines and their aglycones from Streptomyces sp. SCSIO 1666. cytA and its putative homologs are shaded in red; The captured partial cos BGC is shaded in the dotted box. Arrows highlighted in red represented cytA and its homologues; arrows highlighted in blue represented cytB and its homologues.
We have characterized the cytorhodin gene cluster (cyt) and elucidated three glycosyltransferases (GTs) involved in tailoring steps from Streptomyces sp. SCSIO 16663. As indicated, the cyt cluster shows high similarity on the amino acid sequence level (>80% amino acid identity, Supplementary Table 1) to that of cosmomycin D gene cluster (cos). Cosmomycin D is decorated with trisaccharide chains at both C-7 and C-10 positions15 although some key structural differences differentiate the two classes of anthracyclines. The disparity in C-7 oxidation states and substitution patterns between cosmomycin D and cytorhodins X and Y inspired us to hypothesize that an enzymatic process might coordinate formation of the latter. We report here the discovery of CytA, an essential reductase involved in C-7 reduction during the biosynthesis of cytorhodin, a representative anthracycline antibiotic. CytA alone is sufficient to catalyze C-7 reduction under anaerobic conditions and is able to convert clinically employed anthracycline drugs to their inactive, and thereby nontoxic variants. We posit that this unique enzyme and its catalytic chemistry constitute a previously unrecognized resistance mechanism by which Streptomyces sp. SCSIO 1666 defends itself from anthracycline toxicities.
Results
Bioinformatics analysis reveals enzymatic candidate for C-7 deoxygenation in the cytorhodin biosynthetic gene cluster (BGC)
Comparisons of mammalian and bacterial metabolic systems responsible for anthracycline degradation mechanisms suggest that naturally occurring 7-deoxyanthracyclines 1–5 are generated by specific biosynthetic oxidoreductases. Additionally, heterologous expression studies with the cosmomycin D (bearing a pendant trisaccharide at C-7, Fig. 1) BGC15, which lacks any discernible CytA homolog, suggested that the enzyme/s driving such chemistry are likely encoded beyond the established cos cluster (Fig. 1a); C-7 reduced cosmomycin analogs are not observed among cos cluster products. This inspired us to closely examine uncharacterized genes proximal to the cyt cluster boundaries. Given that no analogues of the KstA15/KstA16 enzyme pair readily stood out in the cyt cluster and that most cyt genes have been functionally characterized, we focused our attention on a three-gene cassette containing cytABC, the only candidates within the cyt cluster that have thus far eluded assignment. The cytA gene is located immediately adjacent to the FAD-linked redox encoding gene cytB consistent with previously noted organizations (Fig. 1b) for aclacinomycin (acl)16, nivetetracyclate (niv)17, cinerubin (cin)18, and cosmomycin D (cos) BGCs15. Comparative analyses encompassing the cyt as well as acl, niv, and cin clusters reveal that AclJ, NivJ and CinSSBG_00486 are homologues of CytA, and that AknOx, NivO, and CinSSBG_00485 are homologues of CytB. Moreover, CytA (151 aa) shows similarity to AclJ (63% identity), to NivJ (63% identity), and to CinSSBG_00486 (60% identity), respectively. BLAST analyses revealed that CytA, shares amino acid sequence similarities with a family of flavin-dependent reductases (Supplementary Fig. 2) and that CytB is a FAD-linked oxidase whereas CytC is an aldo/keto reductase possibly involved in terminal sugar tailoring. On the basis of these bioinformatics and its genetic organization with respect to the three-gene cassette, we envisioned that CytA may function as a deglycosylase with reductive potential during anthracycline processing. To test this hypothesis, we consequently inactivated cytA in vivo and carried out subsequent metabolomics studies.
In vivo gene inactivation of cytA and resultant metabolite profiles analyses
λ Red-mediated PCR-targeting mutagenesis19,20 was employed to obtain the ∆cytA mutant strain in which cytA was replaced with an apramycin resistance gene cassette. The ∆cytA mutant was verified on the basis of its sensitivity to kanamycin and apramycin resistance along with PCR analyses (Supplementary Fig. 3). Fermentation of the ∆cytA mutant was carried out followed by butanone extractions, solvent removal and resuspension into methanol. The resulting supernatant was then subjected to high-performance liquid chromatography (HPLC) analysis and two new peaks, both having the same UV–Vis profile and representative of 6 and 7 (Fig. 1c) were noted although the overall yield was low. Further liquid chromatography-high-resolution electrospray ionization mass spectrometry (LC-HRESIMS) analyses revealed the molecular formulae of 6 and 7 to be C60H88N2O21 and C60H88N2O22, indicating that 6 and 7 bear one additional trisaccharide chain relative to 3 and 4, respectively. Large-scale fermentation of the ∆cytA mutant strain enabled the isolation of analytically pure 6 and 7 in quantities sufficient for complete structural characterization. 1H and 13C NMR spectroscopic data (Supplementary Table 2) for 6 and 7 were found to be identical to previously reported cosmomycins C and D (Fig. 1d)6, respectively; detailed 2D NMR (COSY, HMQC, and HMBC) analyses further confirmed these structural assignments. Similarly, fermentations with the ∆cytA mutant in the presence of 3% XAD-16 resin to capture putative early precursor metabolites, led to the visualization of two additional peaks 8 and 9 with different HPLC retention times (Fig. 1c). Large-scale fermentations employing XAD-16 resin and subsequent NMR studies enabled us to elucidate structures 8 and 9 as 10-decarbomethoxy-ε-rhodomycin and ε-rhodomycin (Fig. 1d, Supplementary Table 3)21, respectively. That 6–9 all contained the anticipated C-7 glycoside supports our hypothesis that CytA represents a previously unreported reductive deglycosylase and inspired subsequent in vitro experiments as follows.
In vitro biochemical characterization of CytA-catalyzed deglycosylation under anaerobic conditions
To explore the biochemical activity of CytA in vitro, the cytA gene was cloned into the NdeI and EcoRI sites of the pET28a (+) vector and the resulting vector was transformed into E. coli BL21(DE3) using previously described methods22. CytA was overexpressed as an N-terminally His6-tagged soluble protein and purified to homogeneity by Ni affinity chromatography (Fig. 2a). HHpred analysis revealed that CytA is a flavin-dependent protein but the purified protein is colorless. The activity of CytA was first investigated using substrates 6–9 and ε-rhodomycinone (10) under aerobic conditions. Reactions with CytA were conducted at 37 °C in a volume of 100 μL consisting of 50 mM Tris-Cl (pH 8.0), 25 μM substrate, 0.5 μM CytA, and 20 μM NADH. Reactions with boiled CytA served as controls; all reactions were carried out in parallel to ensure reaction consistency. Reactions were terminated by addition of twofold volume of methanol. As expected, CytA failed, under aerobic conditions to modify any of the substrates 6–10. Carrying out the same reactions using rigorously degassed solutions (see “Methods”) afforded very different results. The absence of O2 enabled CytA to readily convert 6 to 3 and 7 to 4 (Fig. 2b) as revealed by HPLC analyses and comparisons with standards (for 3 and 4) as well as LC-HRESIMS analyses for each reaction. Under no conditions were deglycosylated or dehydrated intermediates ever observed in CytA reactions. The absence of any such intermediates supports CytA’s ability to reductively cleave the C-7 C–O bonds of 6 and 7; CytA appears to not require hydrolytic or dehydrative chemistries en route to C-7 reduction. On the heels of this finding we then determined the kinetic parameters of CytA using 6 and 7 as substrates. The resulting kinetic data (Table 1) revealed that CytA possesses high catalytic efficiency with Km of 14.32 and 17.88 μM, and that, of the two substrates evaluated, 6 is preferred. Additionally, CytA showed some enzymatic promiscuity when using 8–10 as substrates (Fig. 2b).
CytA catalyzes reductive deglycosylation of clinically employed anthracycline drugs
In parallel, a set of C-7-O-glycosylated clinically employed anthracyclines including daunorubicin, doxorubicin, idarubicin, epirubicin and pirarubicin (22–26) (Supplementary Fig. 4) were screened for their ability to serve as CytA substrates in vitro. These trials, in particular highlighted the apparent promiscuity of CytA to effect anthracycline deglycosylation chemistry; all five clinical agents were subject to CytA-mediated C-7 reduction/deglycosylatiion albeit with differing efficiencies (Supplementary Figs. 5–7). Enzyme kinetic data for these assays rendered similar Km values (28.47–49.33 μM, Table 1) for all the clinical anthracyclines evaluated. Overall, CytA was found to be 2–3 fold more efficient at processing initial test substrates 6 and 7 than it was for processing 22–26. Notwithstanding, it was surprising to realize that CytA is less efficient (Km value 31.18 μM, kcat 0.14 min−1) at generating ζ-rhodomycinone (11) from 10 given that C-7 hydroxylated aglycones are more sensitive to reduction in sharply contrast to their glycosidic derivatives by exogenous redox enzymes12. Overall, these data clearly demonstrate the ability of CytA to serve as a reductive deglycosidase.
Antibacterial and cytotoxic activity assessments for metabolites 1–13
We carefully evaluated the antibacterial activities of compounds 1–13 (Fig. 1) using a set of five gram-positive bacteria (Table 2) and found that 6 and 7 were more potent antimicrobial agents than their C-7 deglycosylated counterparts 3 and 4. Notably, the removal of the sugar moieties at both C-7 and C-9 positions dramatically reduced antibacterial activities (compounds 10 and 11, MIC values > 32 μg/mL). These findings are consistent with earlier reports in which 6 and 7 were found to be >100-fold more active against Staphylococcus aureus than their reduced congeners 3 and 4, respectively6. This seemed at first to also be the trend for cytotoxicity; cosmomycin C (6) displayed an IC50 of 0.21 μM against MDA-MB-468 cells whereas its C-7 reduced analog cosmomycin B (4) was devoid of activity23. However, in assessing the cytotoxic activities of 1–10 against a panel of tumor cell lines (Table 3) we found that compounds 6 and 7 showed no significant differences in IC50 values relative to their deglycosylated counterparts 3 and 4. At the same time, we noted that cytorhodins X and Y (final biosynthetic products 1 and 2) bearing the C-9α-glycoside, (absent in both 3 and 4), were less active in both antimicrobial and cytotoxicity assays than their accumulated intermediates 3, 4, 6, and 7. This observation strongly suggests that C-7 deglycosylation markedly limits anthracycline bioactivities and that this reaction, especially in light of our findings with CytA, likely constitutes an important self-preservation mechanism for microbial producers of these secondary metabolites.
CytA confers cosmomycins C and D resistance to the producing organism
Our in vitro experiments unequivocally show that CytA catalyzes reductive deglycosylation in cytorhodin biosynthesis. We also noted that the ∆cytA mutant strain accumulated small amounts (80~100-fold lower than WT producer) of the immediate biosynthetic precursors of cosmomycins A (3) and B (4). The in vitro antimicrobial activities of compounds 1–13 against a panel of gram-positive bacteria (Table 2) provided some insight into the possible physiological significance of CytA-catalyzed reductions. Furthermore, we had observed that the ΔcytA mutant strain is clearly impaired relative to the WT strain upon addition of 6 or 7 as potential substrates. To evaluate this effect, we probed the sensitivity of Streptomyces sp. SCSIO 1666 to 6 and 7 in an inhibition zone assay (Fig. 3). A clear inhibition zone (16 mm) using the ΔcytA mutant plate was observed when supplemented with 1 μg of 6 or 7. Alternatively, the WT producer appeared to resist the antimicrobial effects of 6 and 7 and displayed little to no growth inhibition near paper discs impregnated with 6 or 7. That CytA production endows cosmomycin resistance provides strong evidence that the cytA gene directly confers Streptomyces. sp. SCSIO 1666 with the ability to protect itself from the effects by self-resistance to cosmomycins C and D.
SCSIO 1666 toward cosmomycin C (6) and cosmomycin D (7) (1 μg) as determined by growth inhibition zone assays. Compound 10 was inactive against both the WT and ∆cytA mutant strains. This growth inhibition zone assays were performed, using varying concentrations of substrates (1–10 ug) and all showed a similar trend of activity.
Phylogenetic analysis of CytA and its homologs
CytA belongs to a family of reductases widely distributed in Bacteria, Archaea and Eukayota. Phylogenetic analysis of CytA revealed its association with incompletely characterized gene products (Supplementary Fig. 8). Notably, most homologous proteins derived from bacteria, especially Streptomyces and Micromonospora, remain largely uncharacterized. CytA also showed similarity with Eukaryota-derived glycoside hydrolase family 74 protein (40.0% identity) from Colletotrichum tofieldiae. The secondary metabolites of these CytA homologues containing microorganisms were largely unexplored. Our work here suggests that CytA and its homologs likely constitute an unidentified self-resistance mechanism associated with anthracycline biosynthetic machineries. As such, these enzymes and their encoding genes may serve as genetic markers for use in genome mining initiatives focused on identifying new anthracyclines from assorted microbial producers (Supplementary Fig. 9).
Discussion
The ability of cytorhodin producers to develop self-defense mechanisms to protect them from the effects of self-made cytotoxins such as glycosides 6 and 7 is clear. Under anaerobic conditions, the CytA-catalyzed reaction with NADH first provides a hydroquinone species which then undergoes C-7 deglycosylation to generate a quinone methide; the quinone methide is then envisioned to quickly rearrange to reduction product 7-deoxyanthracycline (Fig. 2c), in a fashion similar to that observed with previously reported transformation reactions11,24. In the presence of O2, the initially added electron is transferred from the reduced anthracycline to O2 to return the starting quinone; elimination chemistry at the C-7 center is averted (Fig. 2c).
Microbial transformations and/or exogenous redox enzymes identified in previous studies revealed that several anthracycline antibiotics can be converted to their non-toxic 7-deoxyanthracycline congeners (Supplementary Fig. 4). In this study, we discovered an uncharacterized enzyme that drives the deoxy reduction process. Inactivation of cytA in Streptomyces sp. SCSIO 1666 impaired the production of C-7 reduced products. Additionally, in vitro studies with purified CtyA revealed its ability to cleave cosmomycins C and D and a set of clinically used drugs with C-7 sugar moieties. Therefore, we conclude that CytA is responsible for degrading cosmomycins C and D to their less active products cosmomycins A and B in the cytorhodin biosynthetic pathway (Fig. 4). The CytA-catalyzed reaction involves anaerobic one electron reduction of the anthracycline quinone ring, using NADH as a cofactor; the absence of O2 and longevity of the reduced species enables loss of the trisaccharide sugar moiety. This elimination produces a quinone methide intermediate and subsequent tautomerization by C-7 protonation leads to the formation of the 7-deoxyaglycone. This mechanism of anthracycline detoxification appears to complement previously reported soil-bourne bacterial detoxification systems that rely on respiratory electron transport complex I-like multi-enzyme complexes11. We also demonstrated that this enzyme-catalyzed deglycosylation constitutes a mechanism of self-resistance for the antibiotic-producing microbes; that this chemistry has the potential to alter clinically relevant agents was also shown. Hence, to our knowledge, this is the first report of a single enzyme endogenous to the producer that mediates reductive cleavage of anthracycline antibiotics; these studies suggest that CytA plays a key role in both microbial and mammalian mechanisms of anthracycline resistance. Further compounding the importance of these findings is that the CytA homologs AclJ, NivJ, and CinSSBG_00486 are all members of the nitroreductase family which carry out similar chemistries. These enzymes are widespread in Actinobacteria and Mycobacterium tuberculosis but are not yet characterized. Thus, beyond its clear relevance to understanding mechanisms of anthracycline resistance, CytA likely provides us a previously unreported tool by which to further our understanding of nitroreductases that have a clear bearing on human medicine.
Methods
General experimental procedures
Bacterial strains and plasmids used in this study are listed in Supplementary Table 4. E. coli strains were grown on Lysogeny Broth (LB) medium at 28 °C or 37 °C; appropriate antibiotics were added at a final concentration of: 100 μg/mL ampicillin (Amp), 50 μg/mL apramycin (Apr), 50 μg/mL kanamycin (Kan), 25 μg/mL chloroamphenicol (Cml) and 50 μg/mL trimethoprim (TMP) when necessary. Streptomyces sp. SCSIO 1666 was maintained on modified ISP4 medium (M-ISP4, ISP4 medium supplemented with 0.05% yeast extract, 0.1% tryptone and 3% sea salt) plates at 28 °C. Tryptic Soy Broth (TSB) broth was adapted for the spore suspension of Streptomyces sp. SCSIO 1666. M11 medium (soybean powder 1%, glycerin 3%, CaCO3 0.2, 3% crude sea salts, adjusted to pH 7.0) was used for fermentation of Streptomyces sp. SCSIO 1666 and mutant strains.
DNA Sequencing was accomplished at Shanghai Majorbio Bio-pharm Technology Co., Ltd (Shanghai, China). Primers were synthesized at Sangon Biotech Company (Shanghai, China). EasyTaqTM DNA Polymerase, TransStartTM Fast-Pfu DNA Polymerase for polymerase chain reactions (PCR) were purchased from TransGen Biotech Company (Beijing, China). Restriction enzymes and DNA ligase were purchased from Takara Biotechnology Co. Ltd. (Dalian, China). Plasmid, gel extraction and cycle-pure kits were acquired from Omega Bioteck Inc. (GA, USA).
Daunorubicin, doxorubicin, idarubicin, epirubicin and pirarubicin were purchased from Aladdin Biochemical Technology Co., Ltd (Shanghai, China). Other chemical solvents (analytical grade) were all purchased from standard commercial sources.
Enzymatic reactions were carried out under anaerobic conditions in an anaerobic workstation (Don Whitley Scientific A45) containing a gas phase of N2/H2/CO2 (80:10: 10%, by vol.). 1H, 13C, and 2D (COSY, HMQC, HMBC and NOESY) NMR spectra were recorded at 25 °C with an Avance 700 MHz spectrometer instruments (Bruker). High-resolution mass spectra were obtained with a Maxis quadrupole-time-of-flight mass spectrometer (Bruker).
Genomic library screening and annotation of open reading frames
The genomic library of Streptomyces sp. SCSIO 1666 using the SuperCos1 vector has been constructed previously3,25. Secondary metabolite BGC were detected and analyzed using online antiSMASH software (http://antismash.secondarymetabolites.org/). The ORFs were determined by application of the FramePlot 4.0 beta program (http://nocardia.nih.go.jp/fp4/). Protein sequences were compared with BLAST programs (http://blast.ncbi.nlm.nih.gov/) and also analyzed by HHpred method (https://toolkit.tuebingen.mpg.de/#/tools/hhpred).
Inactivation of cytA in Streptomyces sp. SCSIO 1666 to afford the ∆cytA mutant
The λ Red-mediated PCR-targeting method according to standard procedures19,26 was performed to inactivate the target gene cytA. Primers designed for inactivation are listed in Supplementary Table 5. A general procedure is described as follows. First, cosmid p11D5 which containing the partical cyt BGC was transformed into E. coli BW25113/pIJ790 for gene inactivations. Then, the gene disruption cassette aac(3)IV-oriT was amplified using a fragment from plasmid pIJ773 that was digested with Eco RI and Hind III. PCR products of the aac(3)IV-oriT cassette for the disrupted gene were then transformed into E. coli BW25113/pIJ790 containing cosmid p11D5 for λ Red-mediated recombination to yield the recombinant cosmid. The recombinant cosmid was transformed into E. coli ET12567/pUZ8002 and suffered from conjugation with Streptomyces sp. SCSIO 1666 wild-type (WT) strain. Double crossover mutants were selected on the basis of the KanSAprR phenotype and then further confirmed by PCR (Supplementary Fig. 3) using primers listed in Supplementary Table 5. Finally, the ∆cytA mutant strain of Streptomyces sp. SCSIO 1666 was generated.
Analysis of production profiles of the Streptomyces sp. SCSIO 1666 WT and mutant strains
Unless otherwise stated, the solvent system of analytical HPLC consisted of solvent A (0.1% TFA in ddH2O) and solvent B (100% CH3CN). To analyze the metabolite profiles of Streptomyces sp. SCSIO 1666 and its mutants, analytical HPLC was performed on an Agilent 1260 HPLC system (Agilent Technologies Inc., USA) equipped with a binary pump and a diode array detector using a Phenomenex Prodigy ODS column S5 (150 × 4.60 mm, 5 μm) with UV detection at 254 and 500 nm. The samples were eluted with a linear gradient of 5 to 80% solvent B over 20 min, followed by 80 to 100% solvent B in 1 min, and then eluted with 100% solvent B in 5 min, at a flow rate of 1.0 mL/min.
The solvent system used for semi-preparative HPLC consisted of solvent A (50 mM ammonium acetate, pH 8.0) and solvent B (100% CH3CN). Semi-preparative HPLC was accomplished with a Hitachi Model D2000 Elite Chromatography Data Station (Hitachi, Japan) equipped with the Hitachi pump and diode array detector, using an YMC-Pack ODS C8 column (YMC, 250 × 10 mm, 5 μm). Samples were eluted at 2.5 mL/min with a linear gradient from 40 to 90% solvent B for 20 min, 90 to 100% solvent B for 2 min, followed by holding at 100% B for 5 min, and then elution with 40% solvent B for 3 min; UV detection was at wavelengths of 254 nm and 500 nm.
Large-scale fermentation, isolation and structural elucidation of intermediates 6–9 from ∆cytA mutant strain
To characterize intermediates and related compounds from the ∆cytA mutant strain, a two-step large-scale fermentation strategy was employed. A general procedure is described. First, a suitable portion of spore from solid M-ISP4 medium plate was used to inoculate 50 mL TSB medium in a 250-mL flask as a seed culture; the flask was cultured at 28 °C and 200 rpm for 36-48 h. Then, the seed culture (50 mL) was transferred to 200 mL of fermentation medium (soybean 10 g/L, glycerol 30 g/L, CaCO3 2 g/L, crude sea salt 30 g/L, adjusted to pH 7.0 ~ 7.2) in a 1000 mL flask; the flask was cultured at 28 °C and 200 rpm for an additional 7–9 days. A total of 30 L culture was carried out. After incubation, the culture broth was harvested and centrifuged to yield a supernatant and a mycelium cake. The supernatant was extracted with an equal volume of butanone three times and the combined butanone fractions then evaporated to dryness; the mycelium was extracted with 1.5 L acetone three times and organics then evaporated to dryness; the two organic extracts were then combined to yield a residue. The combined residue was then subjected to normal phase silica gel column chromatography, eluted with a gradient elution of CHCl3/MeOH mixture from 100/0, 98/2, 96/4, 94/6, 92/8, 90/10, 80/20, and 50/50 to yield eight fractions (Fr. A1-Fr. A8). The fraction containing the targeted compound on the basis of HPLC analysis was dissolved in MeOH and passed through a 0.22 μm syringe filter followed by semi-preparative HPLC purification to give compounds 6 (3 mg) and 7 (1.8 mg).
For purification of compounds 8 and 9, the fermentation broth was supplemented with 3% XAD-16 resin during the early stage fermentation. After 7–9 days, the resin was harvested and extracted with 2 L of acetone three times. The solvent was evaporated to dryness to give the crude residue, which was suspended with ddH2O (300 mL), and then extracted with ethyl acetate. The combined organic phase was evaporated to dryness. A similar purification procedure was carried out to finally obtain pure compounds 8 (30 mg) and 9 (28 mg).
Physicochemical properties and structure Elucidation of compounds 6–9
Compound 6 had a molecular formula of C60H88N2O21, as determined by HRESIMS. The MS, 1H and 13C NMR spectroscopic data of 6 are fully consistent with cosmomycin C in the literature27. Thus, the structure of 6 was established.
Compound 6: red powder; 1H (700 MHz, CDCl3) and 13C NMR (175 MHz, CDCl3), 1H and 13C NMR data, see Supplementary Table 2; (+)-HRESIMS m/z [M + H]+ 1173.5964 (calcd. for C60H88N2O21, 1173.5952); NMR spectra, see Supplementary Figs. 10–15.
Compound 7 had a molecular formula of C60H88N2O22 as indicated by HRESIMS, corresponding to one more hydroxyl group attached to 6. Analysis of the 1H, COSY, HSQC, and HMBC NMR data of 7 allowed full assignment of the NMR signals of 7. 1H and 13C NMR spectroscopic data of 7 are fully consistent with cosmomycin D data previously reported in the literature27,28. Thus, the structure of 7 was established.
Compound 7: red powder; 1H (700 MHz, CDCl3) and 13C NMR (175 MHz, CDCl3), 1H and 13C NMR data, see Supplementary Table 2; (+)-HRESIMS m/z [M + H]+ 1189.5916 (calcd. for C60H88N2O22, 1189.5901); NMR spectra, see Supplementary Figs. 16–21.
Compound 8 had a molecular formula of C28H33NO9, as determined by HRESIMS, indicating the absence of one -COOCH3 group relative to compound 9. Analysis of the 1H, COSY, HSQC, and HMBC NMR data of 8 (Supplementary Figs. 22–27) allowed full assignment of the NMR signals of 8. Detailed 1H and DEPT NMR data revealed one more methylene group (δΗ 3.23, d, J = 19.0 Hz; 2.53, d, J = 19.0 Hz, δC 36.6) was observed in compound 8. HMBC correlations from H-10 to C-6a, C-8, C-9, and C-10a further confirmed this methylene group at C-10 position. Thus, the structure of 8 was established as a previously unreported compound and named as 10-decarbomethoxy-ε-rhodomycin.
Compound 8: red powder; 1H (700 MHz, CDCl3) and 13C NMR (175 MHz, CDCl3), 1H and 13C NMR data, see Supplementary Table 3; (+)-HRESIMS m/z [M + H]+ 528.2249 (calcd. for C28H34NO9, 528.2228), see Supplementary Fig. 6f; NMR spectra, see Supplementary Figs. 22–27.
Compound 9 had a molecular formula of C30H35NO11, as determined by HRESIMS. The MS, 1H and 13C NMR spectroscopic data of 9 are fully consistent with those of previously reported ε-rhodomycin29. Thus, the structure of 9 was established.
Compound 9: red powder; 1H (700 MHz, CDCl3) and 13C NMR (175 MHz, CDCl3), 1H and 13C NMR data, see Supplementary Table 3; (+)-HRESIMS m/z [M+H]+ 586.2451 (calcd. for C30H36NO11, 586.2283); NMR spectra, see Supplementary Figs. 28–33.
Overexpression and purification of CytA
Overexpression of CytA in E. coli BL21(DE3) is described below. The cytA gene was PCR-amplified from cosmid p115D with primer pairs of CytA-expF (GGAATTCCATATGGACGAACAGGACGAGGTGCT, the underlined sequences represent NdeI site) and CytA-expR (CCGGAATTCTCATCCCCGTGCGGGCCCCT, the underlined sequences represent EcoRI site). The PCR products of cytA were recovered from an agarose gel using a gel extraction kit. The corresponding fragments were then excised with NdeI/EcoRI and cloned into the same site of pET/28a(+) vector to yield plasmid pET28a(+)/cytA, which were transformed into E. coli BL21(DE3) to yield strains E. coli BL21(DE3)/pET28a(+)/cytA for protein expression.
E. coli BL21(DE3)/pET28a(+)/cytA was cultured at 37 °C and 200 rpm to OD600 = 0.6. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was then added to a final concentration of 0.1 mM to induce the expression of cytA. After cultivation at 16 °C for an additional 20 h, the cells were collected by centrifugation, washed with 50 mM Tris-HCl buffer (pH 8.0) twice, resuspended in the binding buffer (50 mM Tris-HCl, 500 mM NaCl, and 5 mM imidazole, pH 8.0), sonicated (0 °C) and centrifuged. The supernatant was loaded onto a 1 mL Ni affinity column packed by Ni-NTA His•Bind Resin, washed with 3 mL washing buffer I (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 30 mM imidazole, 10% glycerol) and 3 mL washing buffer II (50 mM Tris-HCl buffer, pH 8.0, 500 mM NaCl, 50 mM imidazole, 10% glycerol), eluted by 2.5 mL elution buffer I (50 mM Tris-HCl buffer, pH 8.0, 500 mM NaCl, 250 mM imidazole, 10% glycerol) and 1 mL elution buffer II (50 mM Tris-HCl buffer, pH 8.0, 500 mM NaCl, 1 M imidazole, 10% glycerol). The fractions that were eluted by elution buffer I containing CytA were then desalted with a PD-10 desalting column, concentrated by filtration on a 3 K Amicon Ultra-15 centrifugal filters, and ultimately dissolved in a storage buffer (10% glycerol, 50 mM Tris-HCl buffer, pH 8.0) and stored at −80 °C for further experiments. All protein purification steps were conducted at 4 °C. The concentration of purified CytA was determined using Bio-Rad protein assay dye according to the manufacturer’s protocol.
In vitro biochemical analysis of CytA
To validate the in vitro biochemical activities of CytA, we overexpressed and purified this enzyme as a soluble N-terminally His6-tagged protein from E. coli (Fig. 2a). Tris-Cl buffer was made O2 free by boiling, followed by cooling under N2 flow; NADH stock solution (dissolved in Tris-Cl), substrate stock solution (dissolved in DMSO), and enzyme solution (dissolved in Tris-Cl supplemented with 10% glycerin) were subjected to O2 eliminating process in the anaerobic workstations for at least two cycles (40 min for one cycle). CytA enzymatic activity was tested in 50 μL volume containing 50 mM Tris-Cl (pH 8.0), 25 μM substrate, 0.5 μM CytA, and 20 μM NADH, at 37 °C for 30 min. The reaction was quenched with the addition to 2 volumes MeOH. After centrifugation to remove protein, supernatants were analyzed with Agilent HPLC equipment using a Phenomenex Prodigy ODS column S5 (150 × 4.60 mm, 5 μ) and UV detection at 254 and 500 nm. Samples were eluted with a gradient elution of phase A containing 0.1% TFA in ddH2O and phase B containing 100% acetonitrile, at a flow rate of 1 mL/min.
Kinetic analyses of CytA with putative substrates 6–10, daunorubicin, doxorubicin, idarubicin, epirubicin and pirarubicin, involved a reaction volume of 20 μL consisting of 50 mM Tris-Cl (pH 8.0), 0.5 μM CytA, 20 μM NADH, and substrate concentrations of 4.5–450 μM.
The kinetics of CytA were measured by determination of the initial velocity for the formation of deglycosylation products with different concentrations of substrates (4.5 μM–450 μM). All assays were performed in triplicate and Km and Vmax values were calculated on the basis of curve fitting to the Michaelis-Menten equation: v = Vmax *[S]/(Km + [S]) (Supplementary Fig. 7).
Antibacterial activities
The antibacterial activities of compounds 1–13 were assessed using twofold serial dilutions of antibacterial agents in MH broth, according to previously reported standard methods provided by Clinical and Laboratory Standards Institute (CLSI)30. These compounds were tested for their antibacterial activities against Bacillus subtilis BS01, Staphyloccocus aureus ATCC 29213, Staphyloccocus aureus 745524, Bacillus thuringiensis BT01, and Enterococcus faecalis ATCC 29212 using a broth dilution method. Each of the reported MIC values in Table 2 is the lowest concentration of antimicrobial agent that completely inhibits growth of the organism in microdilution wells as detected by unaided eyes.
Cytotoxic activity assessments for compounds 1–10
Compounds 1–10 were evaluated for cytotoxic activity using seven human tumor cell lines, A549, HeLa, HepG2, RKO, MCF-7, MDA-MB-231, MDA-MB-468, and two normal cell lines L02, Huvec-12 using MTT method31. All data were obtained in triplicate and are presented as means.
Statistics and reproducibility
For kinetics of CytA, Origin 9.0 (OriginLab Co.) was used to analyze on the basis of curve fitting to the Michaelis-Menten equation. Experiments were performed in triplicate independently. For antibacterial activity, experiments were performed in triplicate using 96-well plates.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Sequence data that support the findings of this study has been deposited in GenBank with accession codes MF773975 for cytorhodin gene cluster. Source data are in Supplementary Data 1. The authors declare that all other relevant data supporting the findings of this study are available within the article and its Supplementary Information files and from the corresponding author upon reasonable request.
References
Almabruk, K. H., Dinh, L. K. & Philmus, B. Self-resistance of natural product producers: past, present, and future focusing on self-resistant protein variants. ACS Chem. Biol. 13, 1426–1437 (2018).
McGowan, J. V. et al. Anthracycline chemotherapy and cardiotoxicity. Cardiovasc. Drugs Ther. 31, 63–75 (2017).
Gui, C., Mo, X., Gu, Y. C. & Ju, J. Elucidating the sugar tailoring steps in the cytorhodin biosynthetic pathway. Org. Lett. 19, 5617–5620 (2017).
Hedtmann, U. et al. The new cytotoxic antibiotic cytorhodin X, an unusual anthracyclinone-9α-glycoside. J. Antibiot. 45, 1373–1375 (1992).
Speitling, M., Gruen-Wollny, I., Fiebig, H. H. & Laatsch, H. Rhodomycinone-derivatives. PCT Int. Appl. WO 2001096352 A1 20011220 (2001).
Morioka, H. et al. Production and isolation of cosmomycins A, B, C and D: new differentiation inducers of friend cell F5-5. Agric. Biol. Chem. 49, 1951–1958 (1985).
Ihn, W., Schlegel, B., Fleck, W. F. & Sedmera, P. New anthracycline antibiotics produced by interspecific recombinants of streptomycetes. J. Antibiot. 33, 1457–1461 (1980).
Tsuji, T. et al. Differentiation inducing activity of anthracycline compounds in friend leukemia cells. Agric. Biol. Chem. 50, 1697–1701 (1986).
Rueckert, P. W., Wiley, P. F., Mcgovren, J. P. & Marshall, V. P. Mammalian and microbial cell-free conversion of anthracycline antibiotics and analogs. J. Antibiot. 32, 141–147 (1979).
Marshall, V. P., Reisender, E. A., Reineke, L. M., Johnson, J. H. & Wiley, P. F. Reductive microbial conversion of anthracycline antibiotics. Biochemistry 15, 4139–4145 (1976).
Westman, E. L. et al. Bacterial inactivation of the anticancer drug doxorubicin. Chem. Biol. 19, 1255–1264 (2012).
Yoshimoto, A., Tobe, H., Johdo, O., Ishikura, T. & Takeuchi, T. Structure-sensitivity relationship of anthracycline antibiotics to C7-reduction by redox enzymes. Jpn. J. Antibiot. 44, 287–295 (1991).
Gille, L. & Nohl, H. Analyses of the molecular mechanism of adriamycin-induced cardiotoxicity. Free Radic. Biol. Med. 23, 775–782 (1997).
Zhang, Z. et al. Hydroxyl regioisomerization of anthracycline catalyzed by a four-enzyme cascade. Proc. Natl Acad. Sci. USA 114, 1554–1559 (2017).
Larson, C. B., Crüsemann, M. & Moore, B. S. PCR-independent method of transformation-associated recombination reveals the cosmomycin biosynthetic gene cluster in an ocean streptomycete. J. Nat. Prod. 80, 1200–1204 (2017).
Alexeev, I., Sultana, A., Mäntsälä, P., Niemi, J. & Schneider, G. Aclacinomycin oxidoreductase (AknOx) from the biosynthetic pathway of the antibiotic aclacinomycin is an unusual flavoenzyme with a dual active site. Proc. Natl Acad. Sci. USA 104, 6170–6175 (2007).
Chen, C. et al. Nivetetracyclates A and B: novel compounds isolated from Streptomyces niveus. Org. Lett. 15, 5762–5765 (2013).
Kersten, R. D. et al. Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules. Proc. Natl Acad. Sci. USA 110, E4407–E4416 (2013).
Gust, B., Challis, G. L., Fowler, K., Kieser, T. & Chater, K. F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl Acad. Sci. USA 100, 1541–1546 (2003).
Mo, X. et al. Δ11, 12 Double bond formation in tirandamycin biosynthesis is atypically catalyzed by TrdE, a glycoside hydrolase family enzyme. J. Am. Chem. Soc. 134, 2844–2847 (2012).
Yoshimoto, A. et al. Anthracycline metabolites from baumycin-producing Streptomyces sp. D788. II. New anthracycline metabolites produced by a blocked mutant strain RPM-5. J. Antibiot. 46, 56–64 (1993).
Gui, C. et al. Discovery of a new family of Dieckmann cyclases essential to tetramic acid and pyridone-based natural products biosynthesis. Org. Lett. 17, 628–631 (2015).
Kim, J. et al. Cosmomycin C inhibits signal transducer and activator of transcription 3 (STAT3) pathways in MDA-MB-468 breast cancer cell. Bioorg. Med. Chem. 19, 7582–7589 (2011).
Kleyer, D. L. & Koch, T. H. Spectroscopic observation of the tautomer of 7-deoxydaunomycinone from elimination of daunosamine from daunomycin hydroquinone. J. Am. Chem. Soc. 105, 2504–2505 (1983).
Mo, X. et al. Cloning and characterization of the biosynthetic gene cluster of the bacterial RNA polymerase inhibitor tirandamycin from marine-derived Streptomyces sp. SCSIO1666. Biochem. Biophys. Res. Commun. 406, 341–347 (2011).
Gust, B. et al. λ Red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 54, 107–128 (2004).
Ando, T. et al. Cosmomycin D, a new anthracycline antibiotic. Agric. Biol. Chem. 49, 259–262 (1985).
Johdo, O. et al. Anthracycline metabolites from Streptomyces violaceus A262. I. Isolation of antibiotic-blocked mutants from Streptomyces violaceus A262. J. Antibiot. 44, 1110–1120 (1991).
Johdo, O. et al. Anthracycline metabolites from Streptomyces violaceus A262. II. New anthracycline epelmycins produced by a blocked mutant strain SU2-730. J. Antibiot. 44, 1121–1129 (1991).
Wikler, M. A. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; Approved Standard–Eighth Edition (2003).
Mosmann, T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63 (1983).
Acknowledgements
This study was supported in part by the China NSF (81425022, U1501223, and 41476133), CAS (XDA11030403), Guangdong NSF (2016A030312014), and the Syngenta Ph.D. Fellowship awarded to C. Gui. Additionally, we thank the analytical facility center (Ms. Aijun Sun, Dr. Zhihui Xiao and Mr. Chuanrong Li) of the South China Sea Institute of Oceanology for recording NMR and MS data.
Author information
Authors and Affiliations
Contributions
J.J. and C.G. designed research; C.G., J.C., Q.X., and S.Z. performed the research; C.G., X.M., H.Z., J.M., Q.L., Y.G., and J.J. analyzed data; C.G. and J.J. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gui, C., Chen, J., Xie, Q. et al. CytA, a reductase in the cytorhodin biosynthesis pathway, inactivates anthracycline drugs in Streptomyces. Commun Biol 2, 454 (2019). https://doi.org/10.1038/s42003-019-0699-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-019-0699-5
This article is cited by
-
Reductive inactivation of the hemiaminal pharmacophore for resistance against tetrahydroisoquinoline antibiotics
Nature Communications (2021)
-
Diversity, function and assembly of mangrove root-associated microbial communities at a continuous fine-scale
npj Biofilms and Microbiomes (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.