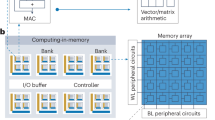
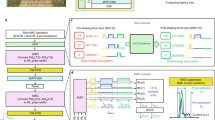
Abstract
As complementary metal–oxide–semiconductor (CMOS) scaling reaches its technological limits, a radical departure from traditional von Neumann systems, which involve separate processing and memory units, is needed in order to extend the performance of today’s computers substantially. In-memory computing is a promising approach in which nanoscale resistive memory devices, organized in a computational memory unit, are used for both processing and memory. However, to reach the numerical accuracy typically required for data analytics and scientific computing, limitations arising from device variability and non-ideal device characteristics need to be addressed. Here we introduce the concept of mixed-precision in-memory computing, which combines a von Neumann machine with a computational memory unit. In this hybrid system, the computational memory unit performs the bulk of a computational task, while the von Neumann machine implements a backward method to iteratively improve the accuracy of the solution. The system therefore benefits from both the high precision of digital computing and the energy/areal efficiency of in-memory computing. We experimentally demonstrate the efficacy of the approach by accurately solving systems of linear equations, in particular, a system of 5,000 equations using 998,752 phase-change memory devices.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Strukov, D. B., Snider, G. S., Stewart, D. R. & Williams, R. S. The missing memristor found. Nature 453, 80–83 (2008).
Chua, L. Resistance switching memories are memristors. Appl. Phys. A 102, 765–783 (2011).
Wong, H.-S. P. & Salahuddin, S. Memory leads the way to better computing. Nat. Nanotech. 10, 191–194 (2015).
Di Ventra, M. & Pershin, Y. V. The parallel approach. Nat. Phys. 9, 200–202 (2013).
Traversa, F. L. & Ventra, M. Di Universal memcomputing machines. IEEE Trans. Neural Netw. Learn. Syst. 26, 2702–2715 (2015).
Sebastian, A. et al. Temporal correlation detection using computational phase-change memory. Nat. Commun. 8, 1115 (2017).
Le Gallo, M., Sebastian, A., Cherubini, G., Giefers, H. & Eleftheriou, E. Compressed sensing recovery using computational memory. In Proc. IEEE Int. Electron Devices Meeting (IEDM) 28.3.1–28.3.4 (IEEE, 2017).
Hu, M. et al. Dot-product engine for neuromorphic computing: programming 1T1M crossbar to accelerate matrix-vector multiplication. In Proc. 53rd Annual Design Automation Conf. (DAC) 19:1–19:6 (ACM, 2016).
Li, C. et al. Analogue signal and image processing with large memristor crossbars. Nat. Electron. 1, 52–59 (2018).
Xu, H. et al. The chemically driven phase transformation in a memristive abacus capable of calculating decimal fractions. Sci. Rep. 3, 1230 (2013).
Ievlev, A. et al. Intermittency, quasiperiodicity and chaos in probe-induced ferroelectric domain switching. Nat. Phys. 10, 59–66 (2014).
Cassinerio, M., Ciocchini, N. & Ielmini, D. Logic computation in phase change materials by threshold and memory switching. Adv. Mater. 25, 5975–5980 (2013).
Sebastian, A., Le Gallo, M. & Krebs, D. Crystal growth within a phase change memory cell. Nat. Commun. 5, 4314 (2014).
Loke, D. et al. Ultrafast phase-change logic device driven by melting processes. Proc. Natl Acad. Sci. USA 111, 13272–13277 (2014).
Wright, C. D., Liu, Y., Kohary, K. I., Aziz, M. M. & Hicken, R. J. Arithmetic and biologically-inspired computing using phase-change materials. Adv. Mater. 23, 3408–3413 (2011).
Hosseini, P., Sebastian, A., Papandreou, N., Wright, C. D. & Bhaskaran, H. Accumulation-based computing using phase-change memories with FET access devices. IEEE Electron. Dev. Lett. 36, 975–977 (2015).
Borghetti, J. et al. ‘Memristive’ switches enable ‘stateful’ logic operations via material implication. Nature 464, 873–876 (2010).
Kvatinsky, S. et al. MAGIC: memristor-aided logic. IEEE Trans. Circ. Syst. II: Express Briefs 61, 895–899 (2014).
Bojnordi, M. N. & Ipek, E. Memristive Boltzmann machine: a hardware accelerator for combinatorial optimization and deep learning. In Proc. IEEE Int. Symp. on High Performance Computer Architecture (HPCA) 1–13 (IEEE, 2016).
Shafiee, A. et al. ISAAC: a convolutional neural network accelerator with in-situ analog arithmetic in crossbars. In Proc. 43rd Int. Symp. on Computer Architecture 14–26 (IEEE, 2016).
Sheridan, P. M. et al. Sparse coding with memristor networks. Nat. Nanotech. 12, 784–789 (2017).
Choi, S., Sheridan, P. & Lu, W. D. Data clustering using memristor networks. Sci. Rep. 5, 10492 (2015).
Ambrogio, S. et al. Statistical fluctuations in HfOx resistive-switching memory: Part I—set/reset variability. IEEE Trans. Electron. Dev. 61, 2912–2919 (2014).
Fantini, A. et al. Intrinsic switching variability in HfO2 RRAM. In 5th IEEE International Memory Workshop 30–33 (IEEE, 2013).
Le Gallo, M., Tuma, T., Zipoli, F., Sebastian, A. & Eleftheriou, E. Inherent stochasticity in phase-change memory devices. In Proc. Eur. Solid-State Device Research Conf. (ESSDERC) 373–376 (IEEE, 2016).
Gaba, S., Sheridan, P., Zhou, J., Choi, S. & Lu, W. Stochastic memristive devices for computing and neuromorphic applications. Nanoscale 5, 5872–5878 (2013).
Tuma, T., Pantazi, A., Le Gallo, M., Sebastian, A. & Eleftheriou, E. Stochastic phase-change neurons. Nat. Nanotech. 11, 693–699 (2016).
Bekas, C., Curioni, A. & Fedulova, I. Low cost high performance uncertainty quantification. In Proc. 2nd Workshop on High Performance Computational Finance 8:1–8:8 (ACM, 2009).
Klavík, P., Malossi, A. C. I., Bekas, C. & Curioni, A. Changing computing paradigms towards power efficiency. Phil. Trans. R. Soc. Lond. A 372, 20130278 (2014).
Saad, Y. Iterative Methods for Sparse Linear Systems (Siam, Philadelphia, 2003).
Higham, N. J. Accuracy and Stability of Numerical Algorithms (Siam, Philadelphia, 2002).
Burr, G. W. et al. Recent progress in phase-change memory technology. IEEE J. Emerg. Sel. Top. Circuits Syst. 6, 146–162 (2016).
Koelmans, W. W. Projected phase-change memory devices. Nat. Commun. 6, 8181 (2015).
Sebastian, A., Krebs, D., Le Gallo, M., Pozidis, H. & Eleftheriou, E. A collective relaxation model for resistance drift in phase change memory cells. in International Reliability Physics Symp. (IRPS) MY.5.1–MY.5.6 (IEEE, 2015).
Gallo, M. L., Sebastian, A., Krebs, D., Stanisavljevic, M. & Eleftheriou, E. The complete time/temperature dependence of I–V drift in PCM devices. In Int. Reliability Physics Symp. (IRPS) MY-1-1–MY-1-6 (IEEE, 2016).
Mathew, R., Karantza-Wadsworth, V. & White, E. Role of autophagy in cancer. Nat. Rev. Cancer 7, 961–967 (2007).
Yang, Z. J., Chee, C. E., Huang, S. & Sinicrope, F. A. The role of autophagy in cancer: therapeutic implications. Mol. Cancer Ther. 10, 1533–1541 (2011).
West, J., Bianconi, G., Severini, S. & Teschendorff, A. E. Differential network entropy reveals cancer system hallmarks. Sci. Rep. 2, 802 (2012).
Schramm, G., Kannabiran, N. & König, R. Regulation patterns in signaling networks of cancer. BMC Syst. Biol. 4, 1 (2010).
Hong, S., Chen, X., Jin, L. & Xiong, M. Canonical correlation analysis for RNA-seq co-expression networks. Nucleic Acids Res. 41, e95–e95 (2013).
Feinberg, B., Wang, S. & Ipek, E. Making memristive neural network accelerators reliable. In Proc. IEEE Int. Symp. on High Performance Computer Architecture (HPCA) (IEEE, 2018).
Anzt, H., Heuveline, V. & Rocker, B. in Applied Parallel and Scientific Computing 237–247 (Springer, New York, 2012).
Nandakumar, S. R. et al. Mixed-precision training of deep neural networks using computational memory. Preprint at http://arXiv.org/abs/1712.01192 (2017).
Breitwisch, M. et al. Novel lithography-independent pore phase change memory. In Proc. IEEE Symp. on VLSI Technology 100–101 (IEEE, 2007).
Papandreou, N. et al. Programming algorithms for multilevel phase-change memory. In Proc. Int. Symp. on Circuits and Systems (ISCAS) 329–332 (IEEE, 2011).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Acknowledgements
We thank C. Malossi, M. Rodriguez, C. Hagleitner, L. Kull and T. Toifl for discussions, N. Papandreou, A. Athmanathan and U. Egger for experimental help, T. Delbruck for reviewing the manuscript, and C. Bolliger for help with preparation of the manuscript. A.S. acknowledges funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement number 682675).
Author information
Authors and Affiliations
Contributions
M.L.G., A.S., T.T., C.B., A.C. and E.E. conceived the concept of mixed-precision in-memory computing. M.L.G., A.S. and C.B. designed the research. M.L.G. implemented the mixed-precision in-memory computing system and performed all experiments. M.L.G., R.M. and M.M. performed the research on the RNA expression data. H.G. performed the evaluation of the runtime and energy consumption. All authors contributed to the analysis and interpretation of the results. M.L.G. and A.S. co-wrote the manuscript based on the input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–8
Rights and permissions
About this article
Cite this article
Le Gallo, M., Sebastian, A., Mathis, R. et al. Mixed-precision in-memory computing. Nat Electron 1, 246–253 (2018). https://doi.org/10.1038/s41928-018-0054-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41928-018-0054-8
This article is cited by
-
Solution-processed memristors: performance and reliability
Nature Reviews Materials (2024)
-
Chalcogenide Ovonic Threshold Switching Selector
Nano-Micro Letters (2024)
-
Hardware implementation of memristor-based artificial neural networks
Nature Communications (2024)
-
In-situ electro-responsive through-space coupling enabling foldamers as volatile memory elements
Nature Communications (2023)
-
A memristive neural network based matrix equation solver with high versatility and high energy efficiency
Science China Information Sciences (2023)