Abstract
Heart Diseases have the highest mortality worldwide, necessitating precise predictive models for early risk assessment. Much existing research has focused on improving model accuracy with single datasets, often neglecting the need for comprehensive evaluation metrics and utilization of different datasets in the same domain (heart disease). This research introduces a heart disease risk prediction approach by harnessing the whale optimization algorithm (WOA) for feature selection and implementing a comprehensive evaluation framework. The study leverages five distinct datasets, including the combined dataset comprising the Cleveland, Long Beach VA, Switzerland, and Hungarian heart disease datasets. The others are the Z-AlizadehSani, Framingham, South African, and Cleveland heart datasets. The WOA-guided feature selection identifies optimal features, subsequently integrated into ten classification models. Comprehensive model evaluation reveals significant improvements across critical performance metrics, including accuracy, precision, recall, F1 score, and the area under the receiver operating characteristic curve. These enhancements consistently outperform state-of-the-art methods using the same dataset, validating the effectiveness of our methodology. The comprehensive evaluation framework provides a robust assessment of the model’s adaptability, underscoring the WOA’s effectiveness in identifying optimal features in multiple datasets in the same domain.
Similar content being viewed by others
Introduction
Heart Disease (HD) is of utmost importance due to the heart’s critical role among other human organs. HD has high death rates worldwide, with approximately 17.9 million people dying from heart conditions in 20191. Heart diseases account for 32% of global deaths, with heart attacks and stroke alone making more than 85% of recorded deaths. Over 75% of cardiovascular deaths in 2019 occurred in underdeveloped nations, accounting for 38% of deaths under 70 years1. Since cardiovascular diseases are fatal, their early detection will enable medical professionals to provide timely healthcare to patients to avert death.
Because of a scarcity of ultra-modern examination tools and medical experts, conventional medical methods for diagnosing heart diseases are challenging, complicated, time-consuming, and exorbitant, making the diagnosis of heart diseases difficult and sometimes unavailable, especially in developing countries2. Machine and deep learning methods have been recently used to analyze clinical data and make predictions3.
Machine learning (ML) provides cost-efficient alternatives where already collected patient data serve as a data mine to perform predictive analysis for diagnostic purposes. To improve the accuracy of ML models, some existing works have focused on using various classifiers or their enhanced forms4,5,6,7. Related works confirm that the feature selection reduces data dimensionality and improves model performance significantly8. Hence, some studies have utilized various methods to improve performance by varying the feature selection methods9,10.
However, some works that utilize feature selection are fraught with redundant features that impact metrics recorded. This is affirmed when wrapper methods are used over filter methods and when embedded methods are used over filter and wrapper methods. It also explains why works, including feature selection, may only record better performance on some datasets if the technique is efficient. In addition, though the researchers do not present the reason some existing works have not reported on specific metrics, studies such as Hicks et al.11 have posited that in a clinical setting, a subset of metrics may give an erroneous outlook of how a model performs and not enabling holistic model performance evaluation. There is an avenue for more scientific work on feature selection methods capable of improving other metrics besides the accuracy metric. This helps to affirm the reliability of the model performance as the unavailability of multiple evaluation metrics is an indication of an unbalanced model not capable of being thoroughly assessed.
This study proposes the use of the whale optimization algorithm (WOA) as a swarm-inspired feature selection algorithm on five (5) heart datasets on ten (10) models (classical ML, ensemble and deep learning models) for the selection of relevant datasets features. The approach contributes to the body of knowledge in the heart disease domain by providing a comprehensive assessment of five different datasets (in the same domain), ten different models and five evaluation metrics. The proposed methodology also validates the robustness of the WOA algorithm on five datasets of variable sizes in the same domain compared to most works, which do not test their methodologies on multiple datasets in the same domain.
Related works
Introduction
Identifying significant features (or redundant features) in a dataset remains a critical activity in modelling12. Feature selection helps to determine relevant subsets from the existing features13 and is recommended for use where one needs to understand the selected features14. Excessive dataset features cause over-fitting, reduce model efficiency, and impair model generalization15. Feature selection (FS) improves the modelling process16. Selected feature selection techniques in literature are mostly grouped into filter, wrapper, hybrid, embedded, and recently, swarm intelligent methods.
Filter-based feature selection methods
Filter-based methods do not consider the dependence of features on one another; only the intrinsic properties are considered17. Ghosh et al.18 used the Cleveland heart dataset alongside the Decision Tree, K-Nearest Neighbour (KNN), and Random Forest (RF) classifiers, recording the highest accuracy of 93.36% with RF. The most optimal features were selected using relief before being passed to the classifiers for training and testing. Relief selected six of the 13 features, resulting in a significant achievement of accuracy. Narsimhulu et al.19 proposed a Filter Based Feature Selection (FBFS) to detect relevant features and remove the redundant ones. RF recorded the highest accuracy of 95.08%, validating the positive influence of the filter feature selection method on model accuracy. Filter methods can compute the scoring function quickly and efficiently20. Filter methods do not consider feature interdependence and are not dependent on the classifiers21.
Wrapper based feature selection methods
To resolve some of the shortcomings of filter-based methods, the wrapper methods use classifiers and consider feature interdependence, increasing computational time22. Evaluating different feature subsets requires retraining and testing23 compared to the filter feature selection algorithms. El-Sayed employed the Genetic Algorithm (GA) wrapper method on the Cleveland heart disease dataset, obtaining 89.07% and 67.22% for binary and multiclass, respectively. First, GA was used to reduce the attributes from the dataset, and Linear Discriminant Analysis (LDA) was used for classification. MultiLDA and multi-classifiers were then used again after GA for multiclass datasets. In their study, GA used alongside the LDA classifier outperformed KNN, Support Vector Machine (SVM) and Naïve Bayes (NB). Sequential Backward Selection (SBS) is also applied to ascertain more significant features, increasing classifier accuracy while decreasing computational time in the study of Haq et al.10. The Cleveland heart disease database was used in the study, recording an accuracy of 90% with six features selected using SBS for feature selection. The KNN classifier with the SBS method outperforms the KNN classifier used single-handedly. The Recursive Feature Elimination (RFE), another wrapper method, is used in the study of24. Feature selection is performed using the SVM-RFE before using KNN to find the best features and reduce computing time. KNN without FS had an accuracy of 82.65% and 86.33% with feature selection. The Weighted KNN without FS recorded 83.45% and 90.88% with feature selection. The results confirm that with feature selection, the accuracy of the KNN or the weighted KNN experiences a significant improvement. Although wrapper methods perform better than filter methods, they are computationally expensive23.
Embedded based feature selection methods
Embedded methods search for classifier-specific optimal features and keep track of feature interdependence with less complexity than the wrapper method25,26. Embedded methods are less prone to overfitting than wrapper methods and are computationally costlier than the filter method27. For the prediction of HD, the LASSO feature selection was compared to the relief feature selection method by Ghosh et al.28 by using the the Cleveland, Long Beach VA, Switzerland, Hungarian and Statlog datasets. The study achieved the best accuracy of 99.05% with LASSO and the Random Forest Bagging classifier. In a related study, Zhang et al.23 proposed the LinearSVC algorithm, an embedded method with a Deep Neural Network using the Cleveland UCI dataset. The work resulted in an accuracy of 98.56%. The study confirms the advantage of embedded methods of feature selection over the former techniques. The embedded feature approach mitigates the shortcomings of the filter and wrapper methods by engaging with the classifier and accounting for feature dependencies26. Its drawback, however, is its slow performance29.
Hybird based feature selection methods
The hybrid methods use two or more methods together for feature selection tasks. The study of30 hybridized Cuckoo Search with the rough set to form the Cuckoo Search with the Rough Set (CSRS) model in their paper. Data from 603 patients are divided into train data for 332 and test data for 271. Cuckoo Search is applied to determine the most optimal features from the training data. Eight features with highly significant values are chosen after 1000 iterations. With optimal features selected, CSRS obtained 93.7% accuracy, outperforming other Cuckoo Search integrated models by 3.37%. Also, a three-phase feature selection approach is proposed by31. In their paper, a three-way feature selection method is proposed for reducing the feature set of the arrhythmia dataset. The three cancer datasets achieved 100% accuracy, and 94.50% was achieved on the arrhythmia dataset. The method selects features with the best accuracy not dependent on the filter methods and the classifiers in the first phase, fusing four techniques: Mutual Information (MI), ReliefF (RFF), Chi-Square (CS), and Xvariance (XV) using three classifiers, KNN, SVM, and NB. The original data set is then subjected to the XGBoost | algorithm, with the top features chosen based on accuracy. In the second phase, the top features in the previous stage are correlated using the Pearson Correlation Coefficient (PCC), and strongly correlated features are dropped, ensuring that the feature subsets have maximum relevance. Again, XGBoost is used to obtain the top features, which are then sent to Phase 3. The optimal feature set is finalized in the third phase, using WOA. Arrhythmia, leukaemia and two others form part of the four datasets used in the trials. The features with the most negligible significance in the dataset had the lowest rankings in information, reliance, and distance. It is also observed that the selected features were reduced by 150 and 1286 times for the arrhythmia and leukemia datasets, respectively. The proposed method enhances the removal of noisy data which are discarded and improves accuracy.
Aside from the authors’ report of limited data, one of the tri-stage’s flaws is the costly computation required to obtain accuracy during the first phase and when WOA is applied.
Arroyo and Delima32 propose a genetically optimized neural network for HD risk prediction. The Cardiovascular disease dataset with 70,000 records and 12 features was employed for ANN modelling. The authors’ work resulted in higher accuracy. However, determining the correct number of layers and neurons takes time and effort. The study of33 also presented techniques based on the KNN, SVM, NB, RF, and a Multilayer Perceptron (MLP) optimized by Particle Swarm Optimization (PSO) merged with Ant Colony Optimization (ACO). The study achieved a maximum of 99.65% accuracy, and the research further enhances the position that with an optimal result set, the accuracy of a study can be significantly improved34. Proposed a hybrid feature selection through Information Gain, Correlation, Chi-Squared, and Relief-F. Using the KNN classifier on a heart failure dataset. The approach recorded 84.61% accuracy. The computational and efficiency challenges inherent in a particular algorithm can be incorporated and reflected in the hybrid algorithm, making the hybrid method very complicated to implement. Therefore, individual algorithms’ challenges must be intentionally handled to improve efficiency and accuracy in hybrid methods.
Swarm intelligent based feature selection methods
Swarm intelligence optimization methods have emerged as robust methods for feature selection in many fields. “Swarm Intelligence (SI) is a type of artificial intelligence that is based on collective behaviours in decentralized and self-organized systems”35. SI is usually inspired by an organized pattern of a random behaviour of a population known as the agents. The intelligent behaviour results in a recognizable pattern helpful in solving optimization problems. Some standard swarm algorithms include the Cuckoo Optimization Algorithm (COA), Ant Colony Optimization (ACO), Bat Algorithm (BA), Grey Wolf Optimization (GWO), Salp Swarm Algorithm (SSA), Marine Predator (MP) and whale optimization algorithm (WOA).
Usman et al.36 propose the Cuckoo Search Algorithm (CSA) and another variant, the Cuckoo Optimization Algorithm (COA) for FS on the Eric, Hungarian, Stat log, and Z-Alizadeh datasets. Four classifiers are used in their study, namely Support Vector Machine (SVM), Multi-Layer Perceptron (MLP), Naïve Bayes (NB), and Random Forest (RF) classifiers. CSA outperformed COA by selecting fewer features and higher accuracy across all datasets. Other researchers, Al-Tashi et al.37, proposed establishing optimal features to diagnose coronary artery disease using the Grey Wolf Optimization (GWO). The authors first use GWO to determine the significant features. The SVM is then utilized with the optimally selected features as input. The Cleveland UCI dataset was employed, and the model recorded 89.83% accuracy. The other evaluation metrics used are sensitivity, which was recorded at 93%, and 91% for specificity rates. In the study of Al-Tashi et al.37, Grey Wolf Optimization (GWO) finds optimal features in the dataset and then evaluates the fitness function of GWO using SVM. The Cleveland UCI dataset used with the study recorded 89.83%, 93% and 91% in accuracy, sensitivity, and specificity, respectively. By combining GWO and the Naïve Bayes (NB) classifier, a new strategy for detecting cardiac disease is developed38. The features of the heart disease dataset are discretized to increase the accuracy of the classifiers. GWO then automatically selects Naïve Bayes’s weights to maximize NB’s performance, achieving 87.45% accuracy with good values for Sensitivity, F-measure, and G-mean. The Cleveland UCI data was used in the project. The proposed (GWO-NB) method performed better than the standard Naïve Bayes classifier in accuracy from the experiments. It also confirms the effectiveness of GWO on classifiers. To improve the GWO, the paper of Chakraborty et al.39 proposed an enhanced form of GWO for feature selection. Cleveland, Long Beach VA, and Switzerland were utilized in their study. Others include the Hungarian and Statlog datasets. Bagging and boosting techniques produce hybrid classifiers with NB, RF, Decision Tree (DT), K-Nearest Neighbor (KNN), Neural Network, Gradient Boosting, and Adaptive Boosting (AdaBoost). RF achieved the best accuracy of 99.26% with Enhanced-GWO, an improvement of accuracy of 11.90% over the conventional model. The Whale Swarm Algorithm (WSA) proposed by David40 was used to select significant features to determine the presence of cardiovascular disease. Using the Statlog dataset, the study recorded an average selection of 6 features after 100 iterations. Using the Logistic Regression (LR), Random Forest, Support Vector Machine and Gaussian Naive Bayes (GNB) classifiers, the study noticed that the Random Forest outperformed the other classifiers, reporting an accuracy of 85.7%. Another commonly used swarm intelligence algorithm within the heart disease domain is Particle Swarm Optimization (PSO). Shahid and Singh41 in their paper propose a model known as the emotional neural networks (EmNNs) that PSO has hybridized on the Z-Alizadeh Sani dataset. The researchers apply four distinct feature selection algorithms to optimize the performance of the suggested model. The proposed method selected a total of 22 features. The model outperforms all other models used in the study, recording the highest averages for accuracy, precision, sensitivity, specificity, and F1 score at cross-validation, i.e., 88.34%, 92.37%, 91.85%, 78.98%, and 92.12%, respectively, which are comparable to existing methods in the literature. Their work is improved by introducing a novel multi-objective PSO (MOPSO) proposed by Asadi et al.42. Using the Statlog, Cleveland, SPECT, SPECTF, VA Long Beach, and Eric datasets, the study recorded improved accuracy better than when feature selection was not used. Wankhede et al.43 propose a Decision Function-based Chaotic Salp Swarm (DFCSS) method to determine the most significant features after data pre-processing. The relevant attributes are then provided to an enhanced Elman neural network classifier. The experiment demonstrates that the proposed method outperformed existing methods with 98.7% and 98.0% accuracy for CVD and UCI datasets, respectively. The Salp Swarm Algorithm selects valuable features from the UCI Cleveland and heart-failure-clinical-records datasets in Sureja et al.44. The study recorded 98.75% and 98.46% accuracy for the respective datasets. Swarm-based feature selection approaches thus have the potential to enhance classifier performance drastically.
This research explored the use of WOA for feature selection for the following reasons. First, WOA is easy to implement due to the few internal numbers of parameters45, outperforms other swarm algorithms such as the Particle Swarm Optimization (PSO) and gravitational search algorithm (GSA) as recorded in the study of Mirjalili and Lewis46 when used for engineering design problems. It has provided outstanding results for optimization problems in domains such as wireless resource allocation and gold price forecasting (outperforming PSO, Grey Wolf Optimization and genetic algorithm47,48. The study of Ay et al.49 also affirms the creditable performance of WOA compared with cuckoo search (CS), flower pollination algorithm (FPA), and Harris Hawks Optimization (HHO) algorithms, other metaheuristic algorithms. Also, WOA is not widely explored in the heart disease domain; hence, its creditable performance in different fields, simplicity, and excellent output against other metaheuristic methods make it a choice worth selecting for FS in the HD domain that guarantees good results.
Proposed methodology
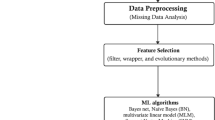
The methodology for this work and the steps used for feature selection, model training, and model evaluation are discussed. Figure 1 depicts the proposed methods employed for this work.
Datasets
The datasets leveraged in this study are summarized in Table 1. The activities performed on the datasets are discussed in the data pre-processing subsection following.
Dataset visualization before feature selection
For each of the five datasets, four visualization techniques were employed to gain insights into the data’s structure prior to feature selection.
Density-Based Spatial Clustering of Applications with Noise (DBSCAN) is utilized to identify and visualize outliers, to aids the understanding of the data’s distribution and identifying anomalous observations. Correlation Heatmaps was utilized to examine the relationships between features while.
Boxplots was used for to visualize feature distribution. Histograms and density plots give insight on the target variable’s distribution across different classes or values.
The dataset visualizations reveal in the DBSCAN plot a lack of clear, separable clusters, or it could be indicating a need to adjust the algorithm’s parameters for a more meaningful clustering. The correlation heatmap provides insights into potential multicollinearity, which could influence feature selection and model performance. The boxplots on the other hand reveal the presence of outliers and the spread of the data. The target distribution plot shows a fairly balanced dataset. A general observation on the datasets reveal similar observations underpinning the need for the preprocessing tasks and feature selection. The visualizations for the datasets are shown in Figs. 2, 3, 4, 5, 6, 7 and 8 representing the combined, Z-AlizadehSani, Framingham, South African and the Cleveland UCI datasets respectively.
Data pre-processing
Data pre-processing is an essential activity in most ML pipelines. It includes tasks such as data cleaning, transformation, and organizing data before passing it to a model. This step is essential to improve data quality and make it suitable for building accurate and efficient models.
In this work, the target values indicating the existence of HD are coded as 0 or 1 (absent and present, respectively). Other categorical data fields, such as “famhist” in the SA heart dataset with values “absent” and “present”, are coded as 0 for absent and 1 for present. “Male” and “Female” values for all datasets are encoded as 1 and 0, respectively. All other fields with similar values are coded with numerical values.
Data standardization and regularization
The standardization process utilizing the StandardScaler was employed. This involved adjusting the features present in the data to be transformed to possess a zero (0) mean and a one (1) standard deviation. Standardizing the features with StandardScaler has a lower error than MinMaxScaler because StandardScaler scales each feature with zero mean and unit variance, scaling them in an equivalent bell curve and handles missing values preventing overfitting50,51. A Logistic Regression model with an L2 regularization penalty is fitted with the standardized data. Another parameter, “C”, which controls the trade-off between fitting the data well and the simplicity (parsimonious) of the model, is utilized in the logistic regression model. The Logistic Regression classifier fits the model to the scaled train data and the corresponding target. The model’s coefficients, which indicate each feature’s significance in making predictions, are stored in a variable applied to the training data to obtain a new, regularized training dataset. This regularized training data is utilized for model training. Afterwards, the stored coefficient is applied to the test data to regularize the test data and target. Logistic regression with L2 regularization has proven useful in prior studies to promote sparse solutions52,53. The regularized train data is passed to the whale optimization algorithm to select the optimal features for model training modelling and testing. The WOA algorithm is discussed in the adjoining pages.
The whale optimization algorithm (WOA)
WOA is an optimization algorithm that draws inspiration from nature and imitates humpback whales’ hunting techniques developed by Seyedali Mirjalili46. The main components of whale optimization are position updating, encircling prey, and searching for prey. The entire WOA steps are discussed in this section and modified to be utilized for feature selection, which is further explained.
Encircling prey
Encircling uses the first current solution as the target or near to it since the optimal design’s location is initially unknown. The remaining agents then update their locations guided by the location of the best agent. This behaviour is modelled by Eqs. (1) and (2).
where \(t\) is the current iteration, the coefficients \({\vec{\text{A}}}\) and \(\vec{C}\) being vectors and \({\text{X}}^{*}\) being the absolute position vector value for the best solution of the iteration. \({\vec{\text{A}}}\) and \(\vec{C}\) are determined using Eqs. (3) and (4).
where \(\overrightarrow{a}\) linearly reduces from 2 to 0 and \(\overrightarrow{r}\), a randomly generated vector with a value within \(\mathrm{0,1}\) is generated.
Bubble-net attacking method (exploitation phase)
Bubble-net attack (exploitation) uses encircling and spiral updating techniques to control the whale mechanisms in WOA. The shrinking encircling mechanism is governed by Eqs. (2) and (3), with the varying range of vector A relying on vector a. Equation (3) can compute this range. The vector \(a\) diminishes from 2 to 0 across numerous rounds, affecting vector A. When vector A lies within [− 1, 1], the agent’s following location is between the present and target positions. The spiral-based position update procedure begins by computing distances between the whale’s present position and the positions of the targets. Based on this range, the spiral motion mimicking the humpback whale’s swimming pattern is generated. The modelled pattern is explained in Eq. (5).
The logarithmic spiral is defined by the constant b, and the value of l is chosen at random from [− 1, 1]. There is a 50% likelihood of using shrinking or the spiral technique to attack the prey successfully. Equation (6) defines the equation.
\(p\) is a number between 0 and 1.
Search for prey
The last process to be discussed in the WOA process is how the target is searched. Humpback whales disperse and explore the search space at random to identify the target, as Eq. (3) describes. When the value of vector \(A\) exceeds \(1\) or falls below -1, it prompts the whales to spread out and perform a random search. The goal of this phase is to integrate exploratory abilities into the WOA. During this stage, the following location of the agents is determined at random, irrespective of the existing best solution’s value. Equations (7) and (8) explain the search process further.
Vector (\(X_{r}\)) is randomly positioned and drawn from the selected population. WOA begins with randomly generated solutions and iteratively refines the optimal solution using either random search agents or the best-performing one. The technique is based on three key parameters: vector a, vector A, and vector p. Vector a, which diminishes through 2 to 0, is utilized to maintain a suitable equilibrium between exploration and exploitation. Vector A determines whether to use a random or the best search agent for position updating. Suppose vector A is more significant than one (1); a randomly generated search agent is used to effect an update on the position. However, the current best solution is used if it is less than one. This assists the WOA in maintaining the proper equilibrium between exploring and exploiting. p facilitates the search agents to vary between cyclic and spiral movement, increasing their adaptability.
Feature selection using WOA
The proposed modified WOA determines the best features using the whale optimization algorithm with input from the preprocessed data. The initial location of the whales (indicating feature selection or non-selection) is generated randomly as binary values. The algorithm then updates each whale’s position in each iteration, shrinking the search space with each iteration. The new position is determined by combining the best position found thus far with random numbers. If the new position has better fitness, it becomes the best. The algorithm then returns the indices of the selected features, considered the best feature. The returned indices are passed to the next step to aid the prediction process. Each whale represents a potential solution in the context of feature selection, with a binary vector encoding the presence or lack of features. The algorithm updates each whale’s location based on a linearly decreasing search space coefficient and the best position over a predetermined number of iterations (10, 20, 30, 40, 50 to 100) and agents (10, 20, 30, 40, 50 to 100).
Ten agents are paired against ten iterations for the WOA. This is repeated in additions of 10 (for agents and iterations). By training a Logistic Regression classifier penalized by an L1 regularization on the chosen feature sets, the fitness function is utilized to compute the performance of the provided subset of features. Each iteration updates the whale position. If the new position improves fitness, the position and fitness are updated accordingly. The maximized prediction performance on the validation with a minimized number of features is considered the optimal feature for that iteration. The best position, therefore, reflects the ideal subset of features so far as the algorithm has run through the required number of iterations. The Scaled training data and its target, the number of whales and iterations are the input factors used by WOA for feature selection. The entire process is captured in Algorithm 1.
Fitness function
Our study presents a novel approach to the fitness function in the whale optimization algorithm (WOA). Central to this method is integrating a logistic regression model with L1 regularization, a choice motivated by the model’s inherent capacity for feature selection and sparsity52.
The logistic regression model, known for its effectiveness in binary classification problems, is enhanced with L1 regularization. This regularization technique introduces a penalty term equivalent to the absolute value of the magnitude of the coefficients. The primary advantage of incorporating L1 regularization is its tendency to produce sparse solutions, inherently performing feature selection by driving the coefficients of less significant features to zero54,55. This aspect is particularly beneficial in our study, where model simplicity and interpretability are paramount.
In evaluating the fitness of the WOA, we adopt a cross-validation with five folds. This partitions the data into five subsets, iteratively using one subset for validation and the rest for training. Such a technique strengthens the model’s validation on different data samples and mitigates the risk of overfitting, leading to a more reliable performance evaluation56.
Also, while L1 regularization naturally promotes feature sparsity, our approach further penalizes the fitness score based on the number of features the logistic regression model selects. This penalty is designed to encourage the selection of the most relevant features and enhance interpretability.
With its unique integration of logistic regression with L1 regularization, k-fold cross-validation, and feature selection penalty, our proposed fitness function is a robust tool in the whale optimization algorithm. It adheres to the principles of parsimony and generalizability and aligns to achieve high predictive accuracy while maintaining model simplicity. This approach has significant implications for applications in various domains, particularly those involving high-dimensional datasets where feature selection is critical in model performance.
Transfer function
In the research, a sigmoid transfer is defined for use in WOA. The sigmoid function is mathematically depicted as Eq. (9).
The sigmoid function can receive a numerical input in real numbers and subsequently act upon it by transforming input data into a finite range between 0 and 1. The sigmoid function converts the continuous output of the whale optimization algorithm into a binary format amenable to indicating the inclusion or exclusion of features. By employing a threshold value on the sigmoid, established at 0.5, we can ascertain the appropriateness of incorporating a feature, denoted as ‘1’, or removing it as ‘0’.
Optimal features selected
WOA is a metaheuristic algorithm with stochastic characteristics; hence, it generates marginally distinct feature sets for every dataset in ten (10) different runs. WOA was executed ten times for every dataset. The selected frequency for each feature was determined by tallying the occurrences across ten separate runs indicating the prominence of each feature. The features that exhibit the highest occurrence among the various iterations are considered potential optimal features due to their higher consistency and relevance. To determine the final set of optimal features, a threshold value of 80% was established. This threshold ensures that only those features that appear in at least 80% of the experimental runs (a minimum of 8 out of 10 runs) will be selected. The optimal features chosen are subjected to individual evaluation by executing the classifiers. The performance of these features is documented for further analysis. The findings offer valuable insight into the effects of feature selection on model performance. Using the methodology above, the experiment endeavours to alleviate the stochastic nature inherent in the whale optimization algorithm (WOA) and produce a resilient collection of optimal features for diagnosing heart disease risk. Table 2 outlines the various datasets and the optimal features identified and selected.
Visualization after feature selection
The DBSCAN Outlier Detection scatter plots across the datasets illustrate the DBSCAN algorithm’s ability to identify clusters and outliers. The clusters appear more distinct in the selected features compared to the broader spread seen in the full feature set. This suggests that feature selection has potentially removed noisy variables, allowing for more apparent patterns to emerge. The Correlation Heatmaps provide a detailed view of the interdependencies between features. After feature selection, the heatmaps are generally less cluttered, with fewer variables exhibiting strong correlations. This reduction in multicollinearity can benefit many machine learning models, as it tends to enhance model interpretability and performance. The boxplots also highlight the distribution and variance of each feature within the datasets. Post-feature selection, the plots are fewer but more focused, often showing a reduced number of extreme outliers. This indicates that feature selection has likely discarded features with extreme values that could skew the model’s learning process. The shapes of the target distributions are generally consistent before and after feature selection, signifying that the selected features maintain the original structure of the target variable. However, in some cases, the distribution appears more balanced post-feature selection, which may positively influence model performance, especially in classification tasks. In all, the visualization suggests that feature selection has streamlined the datasets, potentially improving the efficiency and efficacy of subsequent analyses. By focusing on the most informative features, we expect that the chosen subsets will provide clearer, more relevant insights and facilitate the development of more robust predictive models. Figures 9, 10, 11, 12, 13 and 14 present a visual representation of the various features after performing feature selection.
Training and testing models
After selecting the optimal features, the data is passed to the prediction model for training and testing. The models used are Support Vector Machine, Decision Tree, Random Forest, Multi-Layer Perceptron, Recurrent Neural Network, Adaptive Boosting, Long Short-Term Memory, Extreme Gradient Boosting, K-Nearest Neighbors, and Naïve Bayes classifiers. The choice of prediction models used ranges from classical ML classifiers, ensemble classifiers, and deep learning classifiers to get a broader perspective on performance.
Model evaluation metrics
The models are evaluated using accuracy, precision, recall, AUC and F1 score. The evaluation metrics utilized and the reasons for the choice are discussed next.
Accuracy
Accuracy measures the frequency with which a model accurately predicts outcomes. This metric provides a simple and intuitive understanding of how well the model performs regarding correct classifications57,58 making it a valuable choice for this work. The formula for calculating accuracy is provided in Eq. (10).
Precision
Precision primarily focuses on minimizing false positives to avoid unnecessary stress and medical interventions for patients59. High precision indicates that the model accurately identifies heart disease cases, facilitating informed clinical decision-making60. This is crucial in building trust in the model’s diagnostic capabilities, hence its usage in our work. The precision formula is listed as Eq. (11).
Recall
Recall, also called sensitivity, is the ratio of correctly classified positive samples to all samples assigned to the positive class11. It shows the percentage of positive samples that are correctly classified. Given that a high recall is achieved by missing as few positive instances as possible, this metric is also thought to be among the most crucial for medical research. It is depicted by Eq. (12).
AUC
The Area Under the Curve (AUC) of the Receiver Operating Characteristic (ROC) is a metric that measures the model’s ability to distinguish between the negative and positive classes61. It is calculated based on the ROC curve, which plots the True Positive Rate (TPR) against the False Positive Rate (FPR) at various thresholds62. Computed using Eq. (13), a higher AUC value indicates overall model performance, signifying a high rate of correctly identified positive cases (sensitivity) and a low rate of false positives.
F1 score
Simple accuracy can be misleading as a model could inaccurately appear highly accurate by predominantly predicting the majority class63,64. To avoid this issue, F1 score is used. F1 score is the balance mean precision and recall65 and defined by the Eq. (14).
Model hyperparameters
Table 3 lists the optimal hyperparameters for each of the ten models utilized in this work.
Experimental setup
In this study, we conduct experiments on a Dell Latitude 5430 laptop with a 12th Gen Intel(R) Core (TM) i5 1245U CPU, 16 GB of RAM, and 12 logical processors running Windows 10.
Ethical and informed consent
All ethical and informed consent for data use has been taken care of by the data providers.
Results and discussions
This section discusses the results obtained from the baseline studies and the experiments conducted with feature selection (WOA).
Baseline experiments without feature selection
The baseline experiments in this subsection serve as a critical foundation, providing valuable insights and setting a benchmark for the subsequent evaluation of feature selection methods. The experimentation results are captured in Tables 4, 5 and 6.
Experiments with WOA feature selection
This subsection discusses the findings from experiments with WOA feature selection.
Evaluating completion time for WOA feature selection
In this subsection, the study delves into the completion time when WOA feature selection is employed. We find that the duration varies across different datasets. Factors such as the number of agents, iterations, and features in the data directly influence the completion time; more factors result in a longer duration. Moreover, the observations in each dataset also impact the model completion time. For instance, despite the Z-AlizadehSani and Cleveland UCI datasets containing 303 records, the former requires more time for feature selection. Taking the Framingham dataset as an example, the highest time consumption occurred with this dataset. The completion times for 10, 20, 30, 40, 50, and 100 agents/iterations were 7.56 s, 182.26 s, 383.06 s, 747.03 s, 1072.46 s, and 6656.57 s, respectively. The SA Heart dataset, with times of 0.46 s, 11.80 s, 30.05 s, 58.56 s, 91.20 s, and 547.01 s, had a longer completion time than the Cleveland dataset, even though the Cleveland dataset possesses three additional features. This discrepancy could be due to the marginally more significant number of records in the Cleveland UCI dataset. In summary, the time required for WOA to complete the feature selection process increases with the number of features and records in each dataset. Figure 15 presents the findings about feature selection time using Whale Optimization.
Average estimation of evaluation metrics
To obtain a general overview of the evaluation metrics, an average is computed for each metric after ten (10) runs for each of the five (5) datasets and all ten (10) models. The results show a general improvement across all metrics and datasets. The average metrics are elaborated in Figs. 16, 17, 18, 19, and 20, depicted per dataset.
Evaluating metrics using optimal features
The KNN model performs best across all the metrics and for all the datasets. The results significantly improved over the same experiments on all datasets without WOA feature selection. In some instances, KNN with WOA records 100% across metrics (combined dataset), indicating the influence of WOA on a model’s performance. XGB models perform very well across all the metrics on all datasets except the Framingham dataset. Though XGB performs lower on the Framingham dataset than on all the other datasets, the performance across all the valuation metrics is significantly improved over values recorded without WOA feature selection on the Framingham dataset. In essence, the model has improved due to the feature selection just lower than recorded for other datasets when feature selection is employed. The RF classifier also performs well on all the datasets except the Framingham dataset, which performs well on all the metrics apart from the lowest precision (4.78%). The LSTM model records the highest, 99.80%, on all the metrics except the AUC, which recorded 99.81%. The best performance is recorded on the combined dataset for all the models. Similar performance improvement is seen across all the other datasets for the deep learning models. Generally, the ensemble models (RF, XGB and AdaBoost recorded consistent and improved performance across all the datasets on the evaluation metrics than all but KNN for the classical ML models. The deep learning models perform comparatively well, especially on larger datasets. It was also observed that though the SA heart dataset has a relatively small number of features (9 features), WOA selected seven, subsequently improving the model metrics across all the models. This shows that even if there are not many features, WOA can still select the optimal features if they exist. The experiments consistently improve the evaluation metrics when the results from the model with feature selection are compared to those without. The evaluation metrics are captured in Tables 7, 8 and 9.
Comparative studies
This section performs a comparative study with other related works, comparing the metrics used in the study with values from the metrics of the optimal features used in this work. Works used are the most recent studies in the domain that utilize the same datasets. Table 10 outlines recent studies with the same five datasets used in this work. We compared the features selected by the respective algorithms and their metrics reported and compared it with results from the optimal features in Tables 6, 7 and 8. The results when WOA is used show significant improvement.
The results show superior metrics recorded when WOA feature selection is used and the impact of WOA on the model performance. It is worth noting that this work measures five evaluation metrics over five different datasets using ten models, providing an exhaustive list of metrics capable of judging the performance reported for every model.
Table 10 lists the most recent works in the HD domain, with the last five being the results obtained from this study on the optimal features.
Limitations of the study
The study has data limitations as image heart disease datasets are out of the scope of this work. The experiments in this study are not performed for more than 100 agents and 100 iterations for the whale optimization algorithm due to computational constraints. Finally, the work is limited to only the heart disease domain.
Conclusion
The original WOA is designed for continuous optimization. In this work, however, we implement the solution space into a binary form, necessitating the transformation of the output from continuous to binary using a sigmoid transfer function. A thresholding process is then used to convert the values to a binary format. We specify a fitness function which involves training a logistic regression model using the selected features and evaluating its performance through cross-validation. Since the aim is feature selection, the logistic regression model is penalized using an L1 (Lasso) to produce more sparse models, where a subset of coefficients becomes exactly zero, technically performing feature selection. We then introduce a penalty term to discourage the selection of more features. The whale optimization algorithm for feature selection shows improved heart disease risk prediction evaluation metrics. With fewer yet relevant features, models significantly improved across all five metrics. This contributes to the ongoing efforts to enhance the effectiveness of cardiovascular health risk, providing valuable insights for future studies in this field.
Future works can consider using WOA on image datasets for feature extraction. How WOA compares to other swarm intelligence algorithms in the heart disease domain is an area worth researching. Extending the research to more than one dataset in domains other than Cardiovascular health is a useful area for future studies. In addition, future studies can consider more hyperparameter tuning of all the models to improve the results obtained.
Data availability
Combined dataset https://www.kaggle.com/datasets/johnsmith88/heart-disease-dataset, Z-AlizadehSani dataset https://www.kaggle.com/datasets/tanyachi99/zalizadeh-sani-dataset-2csv, Framingham dataset https://www.kaggle.com/datasets/captainozlem/framingham-chd-preprocessed-data, South African Heart dataset https://hastie.su.domains/ElemStatLearn/datasets/SAheart.data, Cleveland UCI dataset https://www.kaggle.com/datasets/cherngs/heart-disease-cleveland-uci.
Abbreviations
- WOA:
-
Whale optimization algorithm
- FS:
-
Feature selection
- HD:
-
Heart disease
- ML:
-
Machine learning
- DL:
-
Deep learning
- KNN:
-
K-nearest neighbor
- RF:
-
Random forest
- DT:
-
Decision tree
- RNN:
-
Recurrent neural network
- LSTM:
-
Long short-term memory
- XGBoost:
-
Extreme gradient boosting
- NB:
-
Naïve Bayes
- SVM:
-
Support vector machine
- MLP:
-
Multilayer perceptron
- CSRS:
-
Cuckoo search with the rough set
References
World Health Organization. Cardiovascular Diseases 2020. [Online] (Accessed 10 March 2022); https://www.who.int/health-topics/cardiovascular-diseases#tab=tab_1
Ghwanmeh, S., Mohammad, A. & Al-Ibrahim, A. Innovative artificial neural networks-based decision support system for heart diseases diagnosis. J. Intell. Learn. Syst. Appl. 5(3), 176–183 (2013).
Staffini, A. et al. Heart rate modeling and prediction using autoregressive models and deep learning. Sensors 22(1), 1–13 (2022).
Anshori, M. & Haris, M. S. Predicting heart disease using logistic regression. Knowl. Eng. Data Sci. 5(2), 188–196 (2023).
Shah, D., Patel, S. & Bharti, S. K. Heart disease prediction using machine learning techniques. SN Comput. Sci. 1, 1–6 (2020).
Wang, Y., Pan, Z. & Dong, J. A new two-layer nearest neighbor selection method for kNN classifier. Knowl.-Based Syst. 235, 107604 (2022).
Verma, E. P. & Singh, E. P. Human heart disease prediction system using enhanced decision tree algorithm in data mining. Int. J. Innov. Sci. Eng. Technol. 8(6), 1–7 (2021).
Bharti, R. et al. Prediction of heart disease using a combination of machine learning and deep learning. Comput. Intell. Neurosci. 2021, 11 (2021).
Amin, S. M., Kia, Y. & Dewi, K. Identification of significant features and data mining techniques in predicting heart disease. Telematics Inform. 36, 82–93 (2019).
Haq, A. U., Li, J., Memon, M. H., Memon, M. H., Khan, J. & Marium, S. M. Heart disease prediction system using model of machine learning and sequential backward selection algorithm for features selection, in IEEE 5th International Conference for Convergence in Technology (I2CT) (2019).
Hicks, S. A. et al. On evaluation metrics for medical applications of artificial intelligence. Sci. Rep. 12, 5979 (2022).
Benítez-Caballero, M. J., Medina, J., Ramírez-Poussa, E. & Ślȩzak, D. Bireducts with tolerance relations. Inf. Sci. 435, 26–39 (2018).
Zeniarja, J., Ukhifahdhina, A. & Salam, A. Diagnosis of heart disease using K-nearest neighbor method based on forward selection. J. Appl. Intell. Syst. 4(2), 39–47 (2019).
Farahat, A. K., Ghodsi, A. & Kamel, M. S. Efficient greedy feature selection for unsupervised learning. Knowl. Inf. Syst. 35(2), 285–310 (2013).
Wang, S., Chen, J., Guo, W. & Liu, G. Structured learning for unsupervised feature selection with high-order matrix factorization. Expert Syst. Appl. 140, 112878 (2020).
Pathan, M. S., Nag, A., Pathan, M. M. & Dev, S. Analyzing the impact of feature selection on the accuracy of heart disease. Healthc. Anal. 1, 100060 (2022).
Bommert, A., Sun, X., Bischl, B., Rahnenführer, J. & Lang, M. Benchmark for filter methods for feature selection in high-dimensional classification data. Comput. Stat. Data Anal. 143, 106839 (2020).
Ghosh, P., Azam, S., Karim, A., Jonkman, M., & Hasan, M. Z. Use of efficient machine learning techniques in the identification of patients with heart diseases, in 5th International Conference on Information System and Data Mining (ICISDM 2021) (2021).
Narsimhulu, K., Ramchander, N. S., & Swathi, A. An AI enabled framework with feature selection for efficient heart disease prediction, in 2022 5th International Conference on Contemporary Computing and Informatics (2022).
Ditzler, G., Polikar, R. & Rosen, G. A sequential learning approach for scaling up filter-based feature subset selection. IEEE Trans. Neural Netw. Learn. Syst. 29(6), 2530–2544 (2017).
Taha, A., Hadi, A. S. & Bernard Cosgrave, S. M. A multiple association-based unsupervised feature selection algorithm for mixed data sets. Expert Syst. Appl. 212, 118718 (2023).
Mostafa, S. A. et al. Examining multiple feature evaluation and classification methods for improving the diagnosis of Parkinson’s disease. Cogn. Syst. Res. 54, 90–99 (2019).
Zhang, D. et al. Heart disease prediction based on the embedded feature selection method and deep neural network. Hindawi 2021, 1–9 (2021).
Hutamaputra, W., Mawarni, M., Krisnabayu, R. Y., & Mahmudy, W. F. Detection of coronary heart disease using modified K-NN method with recursive feature elimination, in 6th International Conference on Sustainable Information Engineering (2021).
Ang, J. C., Mirzal, A., Haron, H. & Hamed, H. N. A. Supervised, unsupervised, and semi-supervised feature selection: A review on gene selection. IEEE/ACM Trans. Comput. Biol. Bioinf. 13(5), 971–989 (2015).
Khaire, U. M. & Dhanalakshmi, R. Stability of feature selection algorithm: A review. J King Saud Univ. Comput. Inf. Sci. 34(4), 1060–1073 (2022).
Firdaus, F. F., Nugroho, H. A. & Soesanti, I. A review of feature selection and classification approaches for heart disease prediction. Int. J. Inf. Technol. Electric. Eng. 4(3), 75–82 (2020).
Ghosh, P. et al. Efficient prediction of cardiovascular disease using machine learning algorithms with relief and lasso feature selection techniques. IEEE Access 9, 19304–19326 (2021).
Pavya, K. & Srinivasan, B. Feature selection techniques in data mining: A study. Int. J. Sci. Dev. Res. 2(6), 594–598 (2017).
Acharjya, D. P. A hybrid scheme for heart disease diagnosis using rough set and cuckoo search technique. J. Med. Syst. 44(1), 1–16 (2020).
Mandal, M., Singh, P. K., Ijaz, M. F., Shafi, J. & Sarkar, R. A tri-stage wrapper-filter feature selection framework for disease classification. Sensors 21, 5571 (2021).
Arroyo, J. C. T. & Delima, A. J. P. An optimized neural network using genetic algorithm for cardiovascular disease prediction. J. Adv. Inf. Technol. 13(1), 95–99 (2022).
Khourdifi, Y. & Bahaj, M. Heart disease prediction and classification using machine learning algorithms optimized by particle swarm optimization and ant colony optimization. Int. J. Intell. Eng. Syst. 12(1), 242–252 (2019).
Prayogo, R. D. & Karimah, S. A. Hybrid feature selection with K-nearest neighbors for optimal heart failure detection, in 2022 12th International Conference on System Engineering and Technology (ICSET), Bandung, Indonesia (2022).
Rostami, M., Berahmand, K., Nasiri, E. & Forouzande, S. Review of swarm intelligence-based feature selection methods. Eng. Appl. Artif. Intell. 100, 104210 (2021).
Usman, A. M., Yusof, U. K. & Naim, S. Cuckoo inspired algorithms for feature selection in heart. Int. J. Adv. Intell. Inf. 4(2), 95–106 (2018).
Al-Tashi, Q., Rais, H., & Jadid, S. Feature selection method based on grey wolf optimization for coronary artery disease classification, in International Conference of Reliable Information and Communication Technology (2018).
Bakrawy, L. M. E. Grey Wolf optimization And Naive Bayes classifier incorporation for heart disease diagnosis. Aust. J. Basic Appl. Sci. 11(7), 64–70 (2017).
Chakraborty, C., Kishor, A. & Rodrigues, J. J. Novel enhanced-Grey Wolf optimization hybrid machine learning technique for biomedical data computation. Comput. Electric. Eng. 99, 107778 (2022).
David, V. K. Feature selection using Whale swarm algorithm and a comparison of classifiers for prediction of cardiovascular diseases. Int. J. Res. Anal. Rev. (IJRAR) 6(2), 123–130 (2019).
Shahid, A. H. & Singh, M. A novel approach for coronary artery disease diagnosis using hybrid particle Swarm optimization based emotional neural network. Biocybern. Biomed. Eng. 40(4), 1568–1585 (2020).
Asadi, S., Roshan, S. & Kattan, M. W. Random forest swarm optimization-based for heart diseases diagnosis. J. Biomed. Inform. 115, 103690 (2021).
Wankhede, J., Kumar, M. & Sambandam, P. Efficient heart disease prediction-based on optimal feature selection using DFCSS and classification by improved Elman-SFO. IET Syst. Biol. 14(6), 380–390 (2020).
Sureja, N., Chawda, B. V. & Vasant, A. A novel salp swarm clustering algorithm for prediction of the heart diseases. Indones. J. Electric. Eng. Comput. Sci. 25(1), 265–272 (2022).
Lee, C.-Y. & Zhuo, G.-L. A hybrid Whale optimization algorithm for global optimization. Mathematics 9, 1477 (2021).
Mirjalili, S. & Lewis, A. The Whale optimization algorithm. Adv. Eng. Softw. 95, 51–67 (2016).
Pham, Q.-V., Mirjalili, S., Kumar, N., Alazab, M. & Hwang, W.-J. Whale optimization algorithm with applications to resource allocation in wireless networks. IEEE Trans. Veh. Technol. 69(4), 4285–4297 (2020).
Alameer, Z., Elaziz, M. A., Ewees, A. A., Ye, H. & Jianhua, Z. Forecasting gold price fluctuations using improved multilayer perceptron neural network and whale optimization algorithm. Resour. Policy 61, 250–260 (2019).
Ay, Ş, Ekinci, E. & Garip, Z. A comparative analysis of meta-heuristic optimization algorithms for feature selection on ML-based classification of heart-related diseases. J. Supercomput. 79, 11797–11826 (2023).
Mezher, M. A. Genetic folding (GF) algorithm with minimal kernel operators to predict stroke patients. Appl. Artif. Intell. 1, 2022 (2022).
Nguyen, H. T., Cao, A. H., & Bui, P. H. D. Electrocardiogram-based heart disease classification with machine learning techniques, in International Conference on Computational Collective Intelligence (2023).
Deza, A. & Atamturk, A. Safe screening for logistic regression with ℓ0–ℓ2 regularization, in 14th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2022) (2022).
Qin, J. & Lou, Y. L1–2 regularized logistic regression, in 53rd Asilomar Conference on Signals, Systems, and Computers (2019).
Emmert-Streib, F. & Dehmer, M. High-dimensional LASSO-based computational regression models: Regularization, shrinkage, and selection. Mach. Learn. Knowl. Extr. 1(1), 359–383 (2019).
Patil, A. R. & Kim, S. Combination of ensembles of regularized regression models with resampling-based lasso feature selection in high dimensional data. Mathematics 8(1), 110 (2020).
Wong, T.-T. & Yeh, P.-Y. Reliable accuracy estimates from k-fold cross validation. IEEE Trans. Knowl. Data Eng. 32(8), 1586–1594 (2020).
Chicco, D. & Jurman, G. The advantages of the matthews correlation coefficient (mcc) over f1 score and accuracy in binary classification evaluation. BMC Genom. 21(1), 1–13 (2020).
Chicco, D., Tötsch, N. & Jurman, G. The matthews correlation coefficient (mcc) is more reliable than balanced accuracy, bookmaker informedness, and markedness in two-class confusion matrix evaluation. BioData Min. 1(14), 1–22 (2021).
Sukegawa, S. et al. Multi-task deep learning model for classification of dental implant brand and treatment stage using dental panoramic radiograph images. Biomolecules 11(6), 815 (2021).
Seliya, N., Khoshgoftaar, T. M., & Hulse, J. V. A study on the relationships of classifier performance metric, in 2009 21st IEEE International Conference on Tools with Artificial Intelligence (2009).
Ma, W. & Lejeune, M. A. A distributionally robust area under curve maximization model. Oper. Res. Lett. 48(4), 460–466 (2020).
Sofaer, H. R., Hoeting, J. A. & Jarnevich, C. S. The area under the precision-recall curve as a performance metric for rare binary events. Methods Ecol. Evol. 10(4), 565–577 (2018).
He, H. & Garcia, E. A. Learning from imbalanced data. Trans. Knowl. Data Eng. 21(9), 1263–1284 (2009).
Ribeiro, M. T., Singh, S., & Guestrin, C. Why should I trust you? Explaining the predictions of any classifier, in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and data Mining (2016).
Chicco, D. & Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 and accuracy in binary classification evaluation. BMC Genom. 21(1), 1–13 (2020).
Wadhawan, S. & Maini, R. ETCD: An effective machine learning based technique for cardiac disease prediction with optimal feature subset selection. Knowl. Based Syst. 255, 109709 (2022).
Kolukisa, B. & Bakir-Gungor, B. Ensemble feature selection and classification methods for machine learning-based coronary artery disease diagnosis. Comput. Stand. Interfaces 84, 103706 (2023).
Fajri, Y. A. Z. A., Wiharto, W. & Suryani, E. Hybrid model feature selection with the bee swarm optimization method and Q-learning on the diagnosis of coronary heart disease. Information 14(15), 1–15 (2023).
El-Shafiey, M. G., Hagag, A., El-Dahshan, E. S. A. & Ismail, M. A. A hybrid GA and PSO optimized approach for heart-disease prediction based on random forest. Multimed. Tools Appl. 81, 18155–18179 (2022).
Budholiya, K., Shrivastava, S. K. & Sharma, V. An optimized XGBoost based diagnostic system for effective prediction of heart disease. J. King Saud Univ. Comput. Inf. Sci. 34(7), 4514–4523 (2022).
Owusu, E., Boakye-Sekyerehene, P. & Appati, J. K. Computer-aided diagnostics of heart disease risk prediction. Comput. Intell. Neurosci. 2021, 3152618 (2021).
Mienye, I. D. & Sun, Y. An improved ensemble learning approach for the prediction of heart disease risk. Inform. Med. Unlocked 20, 100402 (2020).
Rahim, A. et al. An integrated machine learning framework for effective prediction of cardiovascular diseases. IEEE Access 9, 106575–106588 (2021).
Krishnani, D., Kumari, A., Dewangan, A., Singh, A., & Naik, N. S. Supervised machine learning algorithms prediction of coronary heart disease using, in TENCON 2019 - 2019 IEEE Region 10 Conference (TENCON) (2019).
Mahmoud, W. A., Aborizka, M. & Amer, F. A. E. Heart disease prediction using machine learning and data mining techniques: Application of framingham dataset. Turk. J. Comput. Math0 Educ. (TURCOMAT) 12(14), 4864–4870 (2021).
Nalluri, S., Saraswathi, R. V., Ramasubbareddy, S., Govinda, K., Swetha, E. Chronic heart disease prediction using data mining techniques, in Engineering and Communication Technology, Advances in Intelligent Systems and Computing, 903–912 (2020).
Anuradha, P. & David, V. K. Feature selection and prediction of heart diseases using gradient boosting algorithms, in Proceedings of the International Conference on Artificial Intelligence and Smart Systems (ICAIS-2021) (2021).
Gonsalves, A. H., Thabtah, F., Mohammad, R. M. A. & Singh, G. Prediction of coronary heart disease using machine learning: An experimental analysis. ACM 12(5), 28–36 (2019).
Gokulnath, C. B. & Shantharajah, S. P. An optimized feature selection based on genetic approach and support vector machine for heart disease. Cluster Comput. 22, 14777–14787 (2019).
Cenitta, D., Arjunan, R. V. & Prema, K. V. Ischemic heart disease prediction using optimized squirrel search feature selection algorithm. IEEE Access 10, 122995–123006 (2022).
Author information
Authors and Affiliations
Contributions
S.A.A.—Initial Draft, Experimentation, Analysis; J.K.A.—Idea Initialization, Supervision, Analysis, Final Draft; E.O.—Data Mugging, Planning, Supervision, Final Draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Atimbire, S.A., Appati, J.K. & Owusu, E. Empirical exploration of whale optimisation algorithm for heart disease prediction. Sci Rep 14, 4530 (2024). https://doi.org/10.1038/s41598-024-54990-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-54990-1
Keywords
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.