Abstract
In plants, B3 transcription factors play important roles in a variety of aspects of their growth and development. While the B3 transcription factor has been extensively identified and studied in numerous species, there is limited knowledge regarding its B3 superfamily in pepper. Through the utilization of genome-wide sequence analysis, we identified a total of 106 B3 genes from pepper (Capsicum annuum), they are categorized into four subfamilies: RAV, ARF, LAV, and REM. Chromosome distribution, genetic structure, motif, and cis-acting element of the pepper B3 protein were analyzed. Conserved gene structure and motifs outside the B3 domain provided strong evidence for phylogenetic relationships, allowing potential functions to be deduced by comparison with homologous genes from Arabidopsis. According to the high-throughput transcriptome sequencing analysis, expression patterns differ during different phases of fruit development in the majority of the 106 B3 pepper genes. By using qRT-PCR analysis, similar expression patterns in fruits from various time periods were discovered. In addition, further analysis of the CaRAV4 gene showed that its expression level decreased with fruit ripening and located in the nucleus. B3 transcription factors have been genome-wide characterized in a variety of crops, but the present study is the first genome-wide analysis of the B3 superfamily in pepper. More importantly, although B3 transcription factors play key regulatory roles in fruit development, it is uncertain whether B3 transcription factors are involved in the regulation of the fruit development and ripening process in pepper and their specific regulatory mechanisms because the molecular mechanisms of the process have not been fully explained. The results of the study provide a foundation and new insights into the potential regulatory functions and molecular mechanisms of B3 genes in the development and ripening process of pepper fruits, and provide a solid theoretical foundation for the enhancement of the quality of peppers and their selection and breeding of high-yield varieties.
Similar content being viewed by others
Introduction
Members of the B3 transcription factors are the superfamily specific to plants, and they all have at least one region known as the B3-binding structural domain1,2. The third basic region of VIVIPAROUS 1 (VP1) in maize is where the structural domain is first identified3. Around 110 amino acid residues of recognizable DNA make up the conserved B3 structure region, which has two short α-helices and seven β-barrels1,2. As far as we know, the B3 superfamily is composed of four subfamilies, including RELATED TO ABI3/VP1 (RAV), AUXIN RESPONSE FACTOR (ARF), LEAFY COTYLEDON2-ABSCISIC ACID INSENSITIVE3-VAL (LAV), and REPRODUCTIVE MERISTEM (REM), analyzed genetic structure and systemic development1. The B3 superfamily has been found in numerous species including mustard, rice, grapes, cucumbers, maize, bryophytes, pineapple, oak, cocoa, soybeans, Brassica rapa, Arabidopsis thaliana, tobacco, hazelnuts, eagle beans, citrus, spinach, and red algae4,5,6,7,8,9,10,11,12. However, it has not been detected in significant vegetable crops such as peppers. Members of the B3 superfamily serve critical regulatory activities that are required for plant growth and stress response. RAV subfamily proteins have a B3 domain at the end that identifies the CACCTG motif. Additionally, certain RAV subfamily members include an AP2/EREBP domain that binds to the CAACA sequence at the beginning13. Members of the RAV subfamily control the formation of lateral roots, the timing of flowering, and seed growth. Additionally, they assist in signal transmission, biotic and abiotic stress response, and leaf senescence promotion14,15,16,17,18,19,20,21. LAV subfamily members each have a single B3-binding structural domain that recognizes the Sph / RY element of the CATGCA sequence. Each of them also possesses a motif called ethylene response element binding factor-associated amphipathic repressor (EAR), which inhibits the expression of genes linked to seed maturation by acting as a transcriptional repressor. Additionally, certain ones include the zf-CW domains, which change the dynamics and structure of chromosomes, interact with histone methylation, and regulate gene expression. The LAV subfamily mainly regulates seed development, induction of somatic embryogenesis, embryonic development, and adversity stresses6,22,23,24,25,26,27,28. The REM subfamily typically consists of several B3 binding domains, although there hasn't been much research done on the unique DNA-binding capacity of the REM subfamily's B3 domain. The genes REM1, VRN1, and VDD play a role in various processes including cell division, formation of floral tissues, development of embryos, and pollen development29,30,31,32,33. Furthermore, while the DNA binding specificity of ARF, RAV, LAV, and REM has been studied, few research have thoroughly examined the structure, expression, and function of the B3 superfamily in plants.
The ARF subfamily of the B3 gene may be essential for fruit growth and maturity. The ARF subfamily is a part of the auxin-responsive factors and contains a B3-binding domain at the beginning, which identifies the TGTCTC element (indole-3-acetic acid -responsive) in gene promoter that respond to growth hormone. Furthermore, it has a C-terminal AUX/IAA-interacting domain that promotes interactions between ARFs and AUX/IAA inhibitors, as well as an intermediate region to determine whether ARF activates or represses target genes34,35. The ARF subfamily members play a role in controlling various developmental processes influenced by auxin, including the establishment of adventitious roots, the growth of flowers and fruits, the production of seeds36,37,38,39,40. A few studies have been carried out on the ARF subfamily in fruit development and maturation, as mentioned in certain reports. For instance, in Arabidopsis thaliana, the inhibition of carpel development into fruits is observed due to ARF8, and mutations in ARF8 lead to the formation of seedless fruits without the need for pollination and fertilization41. Additionally, the closure of germ in infertile flowers is hindered in ARF8/ARF6 double mutants, and flower maturity is completely blocked in ARF6/ARF8 mutants42. Recent studies have revealed that ARF genes affect tomato fruit set, development, ripening, and quality43,44,45,46,47. Little is known about how ARF members govern fruit development in various plants since the regulatory mechanism of fruit development is frequently species-specific.
Pepper (Capsicum annuum) is a plant in the Solanaceae that is widely grown for both vegetables and spices48. Due to its considerable diversity in color and form49, it offers a commendable model for investigating the growth of non-climacteric fruits. Fruit development and ripening are physiological processes specific to plants, mainly involving expansion, sweetening, and increased pigmentation50,51. Significant physiological, biochemical, morphological, and molecular changes exist at this stage, with phytohormones and transcription factors being determinants of fruit growth, development, and ripening52,53. The transcription factors MADS-box, MYB (v-myb avian myeloblastosis viral oncogene homolog), NAC (NAM/ATAF1/CUC2), and SPL (squamosa promoter-binding protein-like) all contribute to the development of fruits54,55,56,57. Despite the B3 transcription factor's significance in regulating fruit growth, an exhaustive study of the B3 superfamily in pepper has not yet been carried out. More importantly, since the molecular mechanisms of pepper fruit development and maturity processes have not been fully explained, it is unclear whether the B3-transcript factor regulates pepper fruit development and maturity.
We examined the genome-wide analysis of the B3 superfamily in pepper for this study. The pepper genome contains 106 B3 genes, whose chromosomal locations, gene structures, promoter cis-acting elements, subcellular locations, and phylogenetic relationships have been examined. We also studied at the conserved motifs and biochemical characterization of CaB3 proteins. In addition, we used high-throughput transcriptome analysis to discover many CaB3 genes linked to pepper fruit maturity. Quantitative Real-time PCR (qRT-PCR), a technique that generates numerical data, was used to confirm the expression patterns of the selected genes. The main goal of this work was to increase knowledge of the possible role of the B3 superfamily gene in the maturation and development of pepper fruits.
Results
Identification of B3 genes in the pepper genome
The pepper (C. annuum) genome contained a total of 106 genes belonging to the B3 superfamily. The putative pepper B3 proteins exhibited significant variation in their amino acid lengths, with the 106 B3 superfamily genes encoding proteins ranging from 90 to 1186 amino acids. Additionally, these proteins range from 4.47 to 10.49 in isoelectric point and 10.34–133.98 KDa in molecular weight. Based on their homologous sequences in Arabidopsis thaliana, the four subfamilies RAV, ARF, LAV, and REM are used to group the 106 members of the pepper B3 superfamily1,6 (Supplementary Table S1). In pepper, REM has been found to be the largest B3 subfamily, with a total of 70 CaREM genes identified. ARF is the second largest subfamily, with 23 CaARF genes identified; The LAV and RAV subfamilies are much smaller, with eight CaLAV and five CaRAV genes respectively identified in pepper, accounting for 66%, 21.7%, 7.6%, and 4.7% of the total identified B3 family genes, respectively.
Phylogenetic analysis of the pepper B3 genes
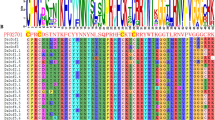
A phylogenetic tree was generated for the B3 genes in pepper in order to gain insight into the evolutionary relationships within the B3 superfamily (Fig. 1). The findings indicated that the pepper B3 superfamily genes were categorized into four primary lineages, exhibiting notable variations in the member count within each cluster. Excluding CaRAV5 and 10 REM members (CaREM23, CaREM10, CaREM24, CaREM57, CaREM43, CaREM61, CaREM56, CaREM8, CaREM59, and CaREM48), the B3 proteins of the same subfamily formed a cluster in the same branch during the analysis. Furthermore, 23 ARF subfamily genes were mostly distributed in nine sister pairs (CaARF12/9,CaARF8/23,CaARF14/17,CaARF3/6,CaARF2/1,CaARF5/21,CaARF4/20,CaARF18/16,CaARF19/10) with the remaining five CaARFs not matching from CaARF12 to CaARF 7 (from left to right). In the REM subfamily, there were only 20 paralogous pairs for 70 genes, and the remaining 30 genes were mismatched, whereas there were one (CaRAV1/2) and two paralogous pairs(CaLAV5/6, CaLAV4/7) for each of the five RAV subfamily genesand eight subfamily genes, respectively. The direct homology of the CaB3 genes in various subfamilies is illustrated by this. Upon comparing the evolutionary trends of B3 proteins in pepper and Arabidopsis thaliana (Fig. 2, Supplementary Table S2), RAV, ARF, and REM subfamilies were shown to cluster together with members of their respective subfamilies on a single branch. Notably, all the major branches and sub-branches included interspecific members. Nevertheless, REM subfamily members exhibit distinct distribution patterns across various branches of the phylogenetic tree, with the majority consisting of intraspecific members. These results illustrate that one of the fundamental features of the REM subfamily is its relative diversity6.
Chromosomal distribution of the pepper B3 genes
We performed a mapping study to find the distribution of the pepper B3 genes across chromosomes in order to better understand the chromosome location of the pepper B3 superfamily (Fig. 3). In addition to the 24 CaB3s found on various scaffolds (Supplementary Table S1), the rest of the CaB3s were unevenly distributed among the 12 linkage groups (LG) of pepper. The density of CaB3 genes per chromosome was variable, with only one gene (0.9%) on chromosome 5 (CaARF9) and as many as 16 genes on chromosome 1 (15.1%). At the extremities of the chromosomes, CaB3 genes showed relatively high concentrations, with the lower end of chromosome 1 having the highest concentration. It is important to note that there is no direct relation between the number of CaB3 genes and chromosomal length. For instance, chromosome 2, despite being shorter, contains 14 CaB3 genes, while chromosome 5, which is longer, only has one CaB3 gene.
Collinearity and duplication analysis of the B3 genes in pepper
In order to enhance our comprehension of the expansion patterns and evolutionary connections within the pepper B3 superfamily, we conducted analyses on duplication events and collinearity. The findings indicated that out of all the pepper B3 genes identified, there were a combined total of four instances of tandem duplication on chromosomes 1, 4, 6, and scaffold757 (Supplementary Tables S1, S2 and Fig. 3). Additionally, 13 occurrences of segmental duplication were observed on 10 chromosomes, excluding chromosomes 1 and 8 (Fig. 4). The expansion of the pepper B3 superfamily may be influenced by segmentation and the replication process occurring in tandem. Calculating the ratios of Ka (non-synchronous substitution rate) and Ks (symmetric substitution ratio) is beneficial for evolutionary study. The results showed that the Ka/Ks (non-synonymous/synonymous substitution rate) ratios in the pepper B3 superfamily ranged from 0.15 to 0.89, all of which were below 1. These ratios were caused by the duplication of gene pairs.
Collinearity studies were performed on the dicots (soybean, tomato, and A. thaliana), as well as the monocots (rice), so as to more thoroughly examine the evolutionary process of the pepper B3 superfamily (Fig. 5). According to the findings, AtB3, SlB3, and GmB3 exhibited collinearity with 8, 44, and 13 CaB3 genes, respectively, while no collinearity was observed with OsB3.The above crops formed 9, 44, and 23 pairs of orthologous genes (Supplementary Table S4). These differences may have been influenced by variations in the timing of evolutionary divergence of the B3 gene among species.
(A). Collinearity Analysis the correlation between pepper and soybean. (B). Analysis of collinearity between pepper and tomato plants. (C). Collinearity analysis was conducted between pepper and Arabidopsis plants. (D). Analysis of collinearity between pepper and rice. Duplicated CaB3 pairs are shown as red lines, while segmental duplications are shown as gray lines.
Structure and protein motif analyses of the pepper B3 genes
The emergence of structural polymorphism in genes during the evolution of the superfamily is common and can assist the co-selection of genes for additional functions in response to environmental changes58. Out of the 106 CaB3s that are categorized into four subfamilies, the majority of REM subfamily genes consist of 4–14 exons, while ARF subfamily genes possess more than three exons. RAV subfamilies, on the other hand, exclusively contain either one or four exons, whereas LAV subfamilies exhibit a range of 6–13 exons (Fig. 6). Within the REM subfamily, adjacent genes may exhibit variations in the number of exons within their respective phylogenetic trees. In addition, the examination of motifs revealed 15 distinct sequences within the CaB3 protein sequence, and the findings indicated that all CaB3 genes possess a designated B3 domain motif. The same family had similar patterns, which differed greatly among the different subfamilies. For instance, the RAV genes possess motif6 and motif9, while the ARF genes possess motif1 and motif3. On the other hand, motif5 is exclusively present in the REM genes, and the ARF family exclusively contains motif4, motif12, and motif10 (Fig. 6, Supplementary Figure S1). The various patterns of the subfamily genes within the population indicate that the CaB3 superfamily has multiple functions.
The exon–intron structure and motif organization of CaB3s. (A). Phylogenetic relationships of CaB3s. (B). Motif identification of CaB3 proteins using MEME. (C). The intron and exon structures of B3 genes. Exons are shown by the green boxes, while introns are symbolized by the lines through the boxes.
Cis-acting element analysis of the pepper B3 genes
The contact of the RNA polymerase with the promoter is a critical event in the initial phase of transcription, which is a critical step of gene expression. RNA polymerase binding affinity and gene expression level are influenced by the promoter's structure59. To further comprehend CaB3s' possible regulatory functions in plant development as well as growth, an analysis was conducted on a promoter sequence that is 2000 base pairs long preceding ATG. An overall number of 799 cis-acting elements were observed in the promoters of 106 CaB3s (Fig. 7, Supplementary Table S5). These components are divided into four categories: hormonal reactions, photoreactions, coercive responses, and growth and developmental regulation. Within the REM subfamilies, the most extensive category of cis-acting elements is comprised of hormone-responsive ones, making up 49.8% of all cis-acting elements. It responded more to abscisic acid, methyl jasmonate, and ethylene than the other components. Hormone-sensitive cis-acting elements comprised the majority (46.4%) of cis-acting elements in the ARF subfamily, exhibiting a higher response to methyl jasmonate, abscisic acid, and ethylene compared to other components. Hormone-sensitive cis-regulatory elements in the LAV subfamily constituted the majority, making up 52.6% of all cis-regulatory elements. Compared to other compounds, these elements responded more strongly to salicylic acid, ethylene, and methyl jasmonate. Hormone-responsive cis-acting elements made up 46.7% of all cis-acting elements in the RAV subfamily, making them the most prevalent kind. These elements exhibited a stronger response to methyl jasmonate, ethylene, and gibberellin compared to other components. Furthermore, a total of 270 (33.8%) cis-regulatory elements that are sensitive to light were discovered across all CaB3 genes. These elements consist of light-responsive components as well as GT binding sites that are linked to the light response. In addition, a total of 48 (6.0%) elements that respond to stress were found, which encompass stress-responsive elements, defense elements, and low-temperature responsive elements. Additionally, 68 (8.5%) elements linked to growth and developmental responses were found, such as elements regulating meristematic tissue development, transcription factors binding to CCAAT-box, elements related to fenestrated tissue cell differentiation, elements regulating circadian rhythm, elements regulating cell cycle, elements controlling zein metabolism, and elements specific to seed.
Prediction of cis- acting elements in the B3 genes of pepper. (A). Phylogenetic relationships of CaB3s. (B). Prediction of cis-elements in CaB3 promoters. The CaB3 gene is colored to signify various cis-acting elements and their corresponding positions. The promoter is 2 kb long. The promoter is 2 kb in length.
Expression patterns of B3 genes during pepper fruit ripening
In this study, we gathered pepper fruits at three distinct stages of development: mature green fruit (MG, 40 DPA), breaker fruit (BR, partially red fruits, 50 DPA), and red ripe fruit (RR, completely red fruits, 60 DPA). In order to find the expression of the B3 gene during fruit ripening in pepper, we next carried out high-throughput transcriptome sequencing analysis to look at the pattern of gene expression in the process of growth and ripening of the peppers. In the three stages of fruit development, it was found that 93 CaB3 genes had FPKM values greater than zero, while the expression of 13 CaB3 genes was undetectable throughout the three developmental phases (Supplementary Table S6) illustrates distinct expression patterns observed in the majority of pepper B3 genes across the three developmental stages. Within the LAV subfamily, the expression levels of CaLAV1/4/7/8 were higher throughout the three stages of fruit development, whereas CaLAV2/4/6 exhibited lower expression. The transcription level of CaLAV3 gene is zero. Genes related to the RAV subfamily have low expression levels in general. However, CaRAV1 exhibited the highest transcriptional abundance during red maturity, while CaRAV4 showed higher levels during green and broken maturity. The three phases of fruit development have shown high levels of expression for the ARF subfamily CaARF14/5/6, whereas the expression of CaARF2/10/18 has been observed to be very low in these three phases. The transcript abundance of REM subfamily was comparatively lower than other subfamilies, particularly CaREM1/3/6/7/19/25/32/36/40/44/46/57, which showed no expression throughout all three fruit developmental periods. However, CaREM21/22/49/53 had higher transcript abundance, suggesting the varied expression patterns within the REM subfamily. During the three developmental periods, there were duplicated gene pairs that exhibited diverse expression profiles. For instance, CaARF14 displayed high expression levels (FPKM ranging from 68.83 to 79.01), whereas its segmental duplicated gene, CaARF7, had low expression levels (FPKM between 2.63 and 3.73). These results suggest that functional divergence between the duplicated genes may have occurred throughout the process of evolution.
In order to further verify the dynamic expression pattern of CaB3 genes in different developmental stages of fruit, we selected five up-regulated genes (CaRAV1, CaREM61, CaREM62, CaARF8, CaARF10), and four down-regulated genes(CaRAV4, CaREM17, CaREM48, CaARF1) on the basis of their relatively high multiplicity of differences in transcript abundance at different stages (|log2FC|≥ 1, P ≤ 0.05), and the transcript levels of nine CaB3 genes were investigated by qRT-PCR to examine the relative expression levels of CaB3 genes at three developmental stages of pepper fruits. The transcriptome results agreed with the expression patterns of these genes during the three phases of development. During pepper fruit maturation (Fig. 9), the findings indicated significant expression of four CaB3 genes, namely CaRAV1, CaREM61, CaREM62, and CaARF8.Furthermore, the levels of CaREM62, CaREM61, CaARF8, and CaRAV1 were markedly increased in MG-BR-RR during the RR period, with increases of 2.18 times, 2.60 times, 3.19 times, and 9.17 times, in contrast to the BR period. Conversely, the expression of CaARF10 was initially decreased by 1.16 times in Mg-BR and subsequently increased by 2.63 times in BR-RR. Additionally, CaREM48, CaARF1, CaRAV4, and CaREM17 exhibited significant reductions in MG-BR-RR, with decreases of 4.68 times, 4.84 times, 6.76 times, and 12.55 times, respectively, during the RR period compared to the BR period.
Subcellular location of the pepper B3 genes
The Plant-mPLoc online tool served to explore the subcellular localization of CaB3 proteins. The nucleus was predicted to contain the majority of CaB3 proteins (Supplementary Table S1), where all CaARF proteins are found, consistent with previous research40. CaRAV proteins and CaLAV proteins are also located in the nucleus. 17 CaREM proteins were detected in both chloroplasts and the nucleus, whereas 47 CaREM proteins were only discovered in the nucleus. Chloroplasts held CaREM30, the cell membrane had CaREM25, the nucleus held CaREM1, and the cytoplasm held CaREM1. CaREM2 was in the Golgi, and CaREM11 and CaREM57 were in the mitochondrion and the nucleus. These findings highlight the relative diversity in characteristics among members of the REM subfamily. The homologous gene AtRAV1 of CaRAV4 is involved in primary root development, leaf senescence, tree bud length, flowering, and possible general growth regulation. Therefore, the downregulated gene CaRAV4 was selected to study its subcellular position, and a preliminary exploration was conducted on which cellular location RAV4 may play a regulatory role in the development of pepper fruits18,60. We selected one of the down regulated genes, CaRAV4, to verify the prediction of subcellular localization. In order to determine the subcellular localization of CaRAV4, CaRAV4-GFP vector was constructed and transferred into Agrobacterium, and the bacterial solution was injected into the lower epidermis of well-grown Nicotiana benthamiana leaves by transient expression, and the subcellular localization was observed by confocal microscopy after 48 h of incubation protected from light. The results showed that the CaRAV4-GFP fusion protein appeared green fluorescence in the nucleus, and the GFP fluorescence emitted by the fusion protein overlapped with the nuclear localization signal mCherry (Fig. 10), indicating that CaRAV4 localized in the nucleus. The location of CaRAV4 in the nucleus of cells was further confirmed through the transient expression of CaRAV4 in Nicotiana benthamiana leaves. So the evidence above suggests that most CaB3 proteins primarily operate within the nucleus.
Discussion
B3 controls the growth of fruits, seeds, flowers, and the aging of leaves in reaction to various biological and non-biological pressures and the transmission of hormonal signals20,21,33,46,61. B3 has been characterized genetically, and its associated functions have been studied in several species and have yielded valuable information5,7,8,9,10,11,12. We argue that this study is the first comprehensive exploration and analysis of the B3 superfamily and its expression in pepper plants on a genome-wide scale. This study identified a total of 106 CaB3s using a comprehensive investigation of the complete genome. These proteins have been classified into four subfamilies by phylogenetic analysis (Fig. 1), aligning with the Arabidopsis B3 protein grouping1, namely RAV, LAV, REM, and ARF subfamilies (Fig. 2). Pepper contains a lower amount of B3 protein compared to mustard (118) and a higher amount compared to rice (91)1, despite having four subfamilies. In relation to the quantity of ARF and REM subfamily genes in pepper, we noticed a pattern comparable to that of other organisms—a predominantly inverse relationship between the number of REM and ARF genes present within a species4. Like Arabidopsis and rice, the REM subfamily contains numerous clusters of B3 structural domain genes1. In peppers, it has been reported that there are nineteen ARF genes40. However, in this study, twenty-three ARF genes were identified, possibly due to the utilization of distinct parameters and additional selection criteria. The quantity of REM subgroups differs among different species, the role of the REM genes is seldom understood, and there could be notable duplications.
Exon–intron organization and motif analyses within gene families can further support and inform evolutionary relationships. The examination of various gene families from distinct species has revealed that closely related individuals typically possess conserved patterns and comparable arrangements of exons and introns within the same category. This finding supports the idea that these individuals have conserved functions and have evolved from a shared ancestor62,63. The current investigation involved the clustering and distribution of 106 B3 proteins from pepper, which were categorized into four branches. It was observed that genes within the same branch exhibited a tendency to possess an equal number of introns and share similar conserved motifs, although there were a few exceptions (Fig. 6). When analyzing the intron–exon structure of the pepper B3 genes, significant differences in the number and length of introns were found. This suggests that introns have either been lost or gained by these genes over the process of evolution. Pepper has been found to contain a total of eight B3 genes without introns, possibly indicating the preference for intron removal to enhance the efficiency of replication, transcription, and processing, as suggested by previous research64. Additionally, variations in the number of exons in genes from the same subcategory have a substantial influence on the evolution of supergene families65. A motif, which is a brief and conserved sequence of amino acids, is regarded as the fundamental functional component necessary for the processing and folding of proteins66. The phylogenetic analysis's validity was confirmed by examination of the conserved pattern in the pepper’s B3 gene since it revealed that genes belonging to the same branch of the phylogeny shared analogous motifs and had comparable motif organization. Furthermore, the presence of recurring motif patterns within individuals belonging to the identical subcategory of genes indicates duplication in both structure and function.
Studies on chromosomal localization in the past have indicated that genes belonging to the REM subfamily have a tendency to group together in the genome. Numerous species, including citrus, grapes, rice, tobacco, Arabidopsis, and others have shown this clustering phenomenon. In pepper, a comparable scenario was witnessed, wherein the most extensive gene cluster was situated on chromosome 1, close to the 300 Mb genomic area containing four REM genes (Fig. 3). Furthermore, it was discovered that numerous CaB3 genes were grouped together in various sections of the remaining chromosomes. The large number of duplicate genes may have promoted the clustering of REM genes.
The expansion of gene families is mostly a result of tandem and segmental duplications, which play a vital role in the development of genes and gene families. In this process, segmental duplication plays a more significant function than tandem duplication67. In our current study, we discovered 11 instances of segmental duplication and four instances of tandem duplication in relation to the pepper B3 gene. The results showed that the Ka/Ks ratios in the pepper B3 superfamily were below 1. This shows that the evolution of all gene in the pepper B3 superfamily has been impacted by purifying selection. In contrast to tandemly duplicated gene pairs (mean Ka/Ks = 0.54), segmentally duplicated gene pairs (mean Ka/Ks = 0.38) showed a variety of powerful purifying selects. Purification options are designed to maintain long-term stability by eliminating deleterious amino acid substitutions68. Therefore, it is essential to maintain and protect the B3 gene family's important role in the development of pepper. In conclusion, our results are consistent with past studies in that the evolution of pepper B3 genes predominantly by segmental duplication. Specifically, certain replicated genes were observed to be dispersed in sibling pairs within the phylogenetic tree, indicating their proximity to the shared progenitor. In addition, the high similarity of conserved motifs and exon–intron arrangements among duplicate genes suggests that they have functional redundancy in some biological processes. However, functional divergence between duplicate genes may have also occurred during long-term evolutionary processes.
The examination of collinearity can aid in the analysis of how the same gene family has evolved across various species. The collinearity of B3 proteins among pepper, tomato, Arabidopsis, soybean, and rice was examined in this investigation (Fig. 5). The results showed that no colinearity was found between B3 in pepper and rice because although extensive homology and colinearity were identified in grasses and dicotyledons plants, there was much less homology and colinearity between the two populations due to longer evolutionary distances and more genomic rearrangements. Whereas pepper belongs to dicotyledons plants and rice belongs to grasses plants, no covariance of the B3 gene was found in pepper and rice, suggesting that it may be the long evolutionary distances and different genome rearrangements experienced by the two groups that have led to this phenomenon69. The findings also indicated that the quantity of direct similarity occurrences of CaB3-SlB3 greatly surpassed that of CaB3-OsB3, implying that the separation of pepper and tomato, both belonging to the Solanaceae family, happened subsequent to the separation of the shared precursor of rice and dicotyledonous plants. Additionally, it suggested that pepper and tomato have a stronger genetic proximity to each other compared to soybean and Arabidopsis thaliana. The significant degree of collinear preservation between pepper and tomato implies that the B3 transcription factor in pepper potentially possesses a comparable structure and function to the immediate homologous gene in tomato.
The level to which genes are expressed at different developmental stages is closely related to the function of those genes. We learned more about the potential functions of these genes by examining how the B3 genes in pepper expressed at different phases of fruit development. In the MG stage, ARF, REM genes, and a few LAV genes were highly expressed, except for RAV genes; specifically, the top three expression peaks were all ARF genes (CaARF6, CaARF5, CaARF14). During the BR stage, the majority of genes with high expression belonged to the ARF and REM subfamilies, with the exception of CaLAV7, CaLAV4, and CaLAV8. The three genes with the highest expression values were CaARF5, CaREM53, and CaARF14.During the RR phase, the ARF and REM subfamilies consisted of all highly expressed genes, excluding CaLAV7, CaLAV4, CaRAV1, and CaLAV8. Notably, the three highest expression peaks were attributed to ARF genes, namely CaARF5, CaARF8, and CaARF14.Based on this observation, it can be inferred that the elevated levels of auxin in the maturing pepper fruit stimulate the transcription of these ARF genes. Research observed a limited number of genes that were constantly expressed whereas a big number of genes were found to be selectively transcribed during particular developmental phases after analyzing transcriptome data during the three stages of pepper fruit development. CaARF21, CaARF12, and CaARF17 exhibited an initial increase followed by a subsequent decrease throughout fruit development, reaching their highest point at the BR stage. This suggests that these three genes have the ability to control the entire process of fruit development. In tomato, the SlARF8 gene represses the formation of fruit initiation70, whereas in our study CaARF8 expression gradually increased during pepper fruit development and was higher at the red ripening (RR) stage, suggesting that CaARF8 is also involved in the regulation of fruit initiation in pepper; down-regulation of SlARF4 expression affects pectin synthesis and improves ripening fruit firmness71, however, CaARF4 seems to be a positive regulator, and the expression of CaARF4 gradually decreases with the decrease of fruit firmness in pepper; the repression of SlARF7 transcription leads to the initiation of fruit set in tomato before pollination and fertilization72, and the expression of CaARF7 decreases and then increases in the process of fruit development, which may play different roles at different periods of fruit development in pepper. SlARF9 negatively regulates cell division in the early fruit development73, and it is possible that SlARF9 negatively regulates cell division at the early stage of fruit development in pepper. CaARF9 expression gradually decreases during the development of chili fruits, and cell division is not inhibited and the fruits are enlarged. In Arabidopsis, ARF6 and ARF8 coordinate the development of petals and reproductive organs during the transition from closed buds to mature fertile flowers, which contributes to efficient fertilization and subsequent fruit development74. CaARF6 expression gradually decreased during pepper fruit development, while CaARF8 expression gradually increased during pepper fruit development, which was different from the expression pattern in Arabidopsis. It is possible that there is no redundancy effect during pepper fruit development. Above of all, ARF is vital for regulating tomato fruit growth43,44,45,46,47. The majority of genes showed distinct reactions to the various stages of pepper fruit growth, based on the transcriptome data (Fig. 8). Meanwhile, some genes were highly expressed at all three developmental stages. CaARF5 and CaARF14 exhibited significant expression levels during the MG, BR, and RR phases, indicating their crucial involvement in the advancement of various developmental stages. CaREM58, CaREM41, and CaREM56 were detected exclusively during a single developmental stage, indicating their specific involvement in a particular phase of fruit development. It was found that CaREM36 and CaREM32 in the tandem duplicate gene pair CaREM35/CaREM36 and the segmental duplicate pair CaREM29/CaREM32 were not expressed in all three fruit developmental periods, whereas CaREM35 and CaREM29 were expressed in all three fruit developmental periods, suggesting that the absence of expression of some B3 genes may be due to gene duplication. The expression pattern of pepper REM genes is supported by earlier research on the expression and transcriptional characterization of NtREM as well as other REM genes75,76.
Heat map of 106 CaB3 genes showing that they are highly expressed during pepper fruit ripening. (A). Pepper undergoes three distinct phases of fruit ripening: mature green fruit (MG, 40 DPA), breaker fruit (BR, which is partially red and occurs at 50 DPA), and red ripe fruit (RR, fully red and observed at 60 DPA). (B). Hierarchical clustering of CaB3 genes in different fruit developmental periods. The scale bar indicates log2 normalized fragments per kilobase per million reads (FPKM) values. Red denotes high expression, while green denotes low expression, as shown by the color gradient.
Furthermore, the findings were additionally confirmed through quantitative PCR (Fig. 9), suggesting that the ARF gene could potentially have a significant impact on fruit development, aligning with prior investigations41,42. During fruit development, the RR and MG phases exhibited a strong expression of the two REM genes, indicating their importance in fruit maturity. From the above, we can make a hypothesis that up-regulated genes are positively regulated and down-regulated genes are negatively regulated during the development of pepper fruits, and the inconsistency in the expression trend of some genes suggests that they play different roles at different times, and may be suppressed or facilitated by endogenous hormones within the stage that lead to this result. Future studies on the processes governing the growth and maturity of pepper fruit will benefit greatly from the insights provided by the B3 gene. To learn more about the precise regulation mechanism of B3 in the development of pepper fruit, a more comprehensive study is needed (Fig. 10).
Subcellular location of the CaRAV4. The transgenic tobacco plants display images depicting the GFP signals of both formations. Confocal microscopy revealed the presence of green fluorescence. The protein mCherry is responsible for nuclear localization, while the protein GFP is responsible for green fluorescence. Visible light is used for bright field imaging, and the merged image combines the bright field with mCherry and GFP. Bar = 50 μm.
Conclusion
In this study, we conducted a systematic analysis of the B3 gene family in Capsicum annuum. We identified the B3 genes and analyzed their phylogenetic relationships, chromosomal distribution, gene structure, colinearity and its replication events, promoter elements, subcellular localization, and tissue-specific expression. The results of this study revealed the dynamic transcriptional patterns of CaB3s during fruit development, especially the ARFs genes may directly regulate the fruit development of pepper, suggesting that CaB3s may be involved in the formation and development of pepper fruits. The present study provides an important foundation and new insights for further research on the mechanism of B3 genes regulating the development and maturation of pepper fruits, and provides a solid theoretical basis for the enhancement of the quality of peppers and the selection and breeding of their high-yield varieties. However, the specific molecular mechanisms of B3 genes regulation in the development of pepper fruits should be studied more comprehensively and deeply in the future.
Materials and methods
Identification of B3 genes in pepper
In order to accurately and comprehensively identify the B3 superfamily in pepper, the genomic protein data of pepper was downloaded from the Ensemble Plants database (https://plants.ensembl.org Genomic version: Capsicum annuum cv.CM334 release 1.55) obtained11. We use Arabidopsis B3 protein as the query sequence (https://www.arabidopsis.org/) Conducted BLAST search and then retrieved from Pfam database (http://pfam.xfam.org/) Download the HMM (Hidden Markov Model) map of the B3 domain (PF02362), and use Hmmer 3.0 to re identify whether it contains the B3 domain (with an e-value cutoff of 1e-5). After removing redundant sequences, all nonredundant proteins with conserved B3 domains were assigned as members of the pepper B3 family. Utilizing ExPASy (https://web.expasy.org/protparam/) Calculate the biochemical characteristics of the B3 gene family in chili peppers. The subcellular localization of each CaB3 protein was predicted using the Plant-mPLoc online tool (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/)77.
Phylogenetic analysis of the pepper B3 genes
The B3 protein from pepper was compared to the downloaded amino acid sequences of Arabidopsis B3 using ClustalW with the default parameters78. The maximum likelihood (ML) technique was used to build an important phylogenetic tree with 1000 bootstrap replicates using IQ-TREE (multicore version 2.2.0)79.
Chromosomal distribution and conserved motif analyses of B3 in pepper
Data regarding the placement of the B3 gene in pepper chromosomes was obtained from the Genome General Characterization File. The chromosomal locations of Pepper B3 were determined using the internet resource MG2C (http://mg2c.iask.in/mg2c_v2.1/). MEME (http://meme-suite.org/) was used to examine the conserved motifs of the pepper B3 gene, with a maximum of 15 conserved motifs set80. Visualization of the genetic structure and conservative sequence of the pepper B3 family was done using TBtools81.
Cis-acting element, collinearity, and duplication analysis of pepper B3 genes
We were able to obtain a 2000 bp section of the pepper B3 gene sequence before the start codon using TBtools. We next used the PlantCare website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to submit this extracted sequence for analysis of its cis-acting elements82. To assess the collinearity of B3 genes within and between species and to map collinearity, we utilized MCScanX83 and Circos84. Furthermore, the Ka/Ks values of the B3 gene pairs were calculated using the (natural gradient descent, NG) method by TBtools in order to understand the selection pressure on the B3 gene in pepper during the process of evolution.
Plant materials
The tested pepper (C. annuum) is a self-selected ’chili pepper inbred line gd-75–3-10 by the research group. It was cultivated in a light-culture room with alternating periods of 16 h of light and 8 h of darkness at a temperature of 22–25 °C. At the time of fruiting, fruit samples were gathered from MG fruits (40 DPA), BR fruits (partially red fruits, 50 DPA), and RR fruits (fully red fruits, 60 DPA). Three sets of biological duplicates were enclosed in aluminum foil, rapidly frozen in liquid nitrogen, and preserved at a temperature of − 80 °C. Afterward, the specimens were preserved at a temperature of − 80 °C. The samples were stored simultaneously.
Transcriptome sequencing to analyze CaB3 gene expression levels
For transcriptome analysis of the mentioned samples of MG fruit (40 DPA), BR fruit (partially red fruits, 50 DPA), and RR fruit (completely red fruits, 60 DPA) of pepper, three biological replicates were collected. Novogene (Beijing, China)85 performed the extraction of total RNA, creation and testing of libraries for quality, as well as the sequencing, analysis, and annotation of the transcriptome. The R package was used to generate heatmaps and to standardize expression levels using FPKM values. The "full" clustering approach was used to conduct a hierarchical cluster analysis. FPKM (fragments per kilobase per million mapped reads) was used to represent these transcripts.
qRT-PCR analysis
B3 genes with significantly different expression levels were screened among the three maturation stages. These genes were chosen from the RNA-seq data due to their significant variations in transcript abundance across the stages, with a relatively high multiplicity (|log2FC|≥ 1, P ≤ 0.05). To ascertain the expression patterns, nine genes were chosen for qRT-PCR analysis. For qRT-PCR analysis of the mentioned samples of MG fruit (40 DPA), BR fruit (partially red fruits, 50 DPA), and RR fruit (completely red fruits, 60 DPA) of pepper, three biological replicates were collected, the RNA-prep Pure Total RNA Extraction Kit for Polysaccharide-Polyphenol Plants (Tiangen, Beijing, China) was utilized for extraction. Sangyo Bioengineering Corporation (Shanghai, China) designed and synthesized the Total RNA primers, as listed in Supplementary Table S7. The RNA that was extracted underwent reverse transcription to cDNA with the FastKing gDNA Dispelling RT SuperMix (Tiangen, Beijing, China). Fluorescence quantitative PCR was conducted utilizing an ABIVi-iA7 instrument (Hangzhou, China). CaUBI-3 was reported to have the most stable expression level in pepper under abiotic stress and different tissue types; therefore, this gene can be recommended as the best reference gene for qRT-PCR analysis in pepper86. Gene expression levels were determined by employing the 2−ΔΔCt technique87. The data were examined for statistical significance and visualized using GraphPad Prism 8.0. The significance level of 0.05 was used to conduct multivariate comparisons through one-way ANOVA and Duncan's multiple range test.
Subcellular localization analysis
To confirm the anticipated outcomes, Nicotiana benthamiana plants were chosen for transient expression using CaRAV4.To generate a vector of CaRAV4-GFP under the control of the P35S promoter, the coding sequence of CaRAV4 was combined with the N-terminus of GFP in pEGOEP35S-H-GFP. Agrobacterium strain GV3101 carrying the CaRAV4-GFP vector was mixed in equal proportions with the Agrobacterium strain GV3101 with the mCherry Nucleus marker before infiltration, and the infiltrated leaves were directly observed with an FV1000 laser confocal microscope after 48 h of infiltration. The emission filters for GFP and mCherry were set at 488 nm and 561 nm, correspondingly.
Data availability
The self-tested RNA-Seq data of pepper fruits at different fruit stages in this study can be obtained at the Short Read Archive (SRA) of NCBI with BioProject PRJNA1017123.
Material availability
The pepper material used for the experiment is a self-selected and purified strain by our research group, and has not yet been registered as a variety. We have the right to collect plants, conduct experimental and field research on peppers, and comply with relevant institutional, national, and international standards and legislation.
References
Swaminathan, K., Peterson, K. & Jack, T. The plant B3 superfamily. Trends Plant Sci. 13, 647–655 (2008).
Yamasaki, K., Kigawa, T., Seki, M., Shinozaki, K. & Yokoyama, S. DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci. 18, 267–276 (2013).
Suzuki, M., Kao, C. Y. & McCarty, D. R. The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell. 9, 799–807 (1997).
Wang, Y. et al. Systematic analysis of plant-specific B3 domain-containing proteins based on the genome resources of 11 sequenced species. Mol Biol Rep. 39, 6267–6282 (2012).
Peng, F. Y. & Weselake, R. J. Genome-wide identification and analysis of the B3 superfamily of transcription factors in Brassicaceae and major crop plants. Theor. Appl. Genet. 126, 1305–1319 (2013).
Romanel, E. A., Schrago, C. G., Couñago, R. M., Russo, C. A. & Alves-Ferreira, M. Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLOS One. 4, e5791 (2009).
Wang, W. B. et al. Genome-wide analysis of the B3 transcription factors reveals that RcABI3/VP1 subfamily plays important roles in seed development and oil storage in castor bean (Ricinus communis). Plant Divers. 44, 201–212 (2022).
Ahmad, B. et al. Genomic organization of the B3-domain transcription factor family in grapevine (Vitis vinifera) and expression during seed development in seedless and seeded cultivars. Int. J. Mol. Sci. 20, 4553 (2019).
Liu, Z. et al. Genome-wide analysis of the citrus B3 superfamily and their association with somatic embryogenesis. BMC Genomics. 21, 305 (2020).
Ruan, C. C. et al. Genome-wide characterization and expression profiling of B3 superfamily during ethylene-induced flowering in pineapple (Ananas comosus). BMC Genomics. 22, 561 (2021).
Xia, F. et al. Insight into the B3 transcription factor superfamily and expression profiling of B3 genes in axillary buds after topping in tobacco (Nicotiana tabacum). Genes. 10, 164 (2019).
Verma, S. & Bhatia, S. A comprehensive analysis of the B3 superfamily identifies tissue-specific and stress-responsive genes in chickpea (Cicer arietinum) 3. Biotech. 9, 346 (2019).
Kagaya, Y., Ohmiya, K. & Hattori, T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucl. Acids Res. 27, 470–478 (1999).
Liu, C. & Zhang, T. Expansion and stress responses of the AP2/EREBP superfamily in cotton. BMC Genomics. 18, 118 (2017).
Lee, S. C., Choi, D. S., Hwang, I. S. & Hwang, B. K. The pepper oxidoreductase CaOXR1 interacts with the transcription factor CaRAV1 and is required for salt and osmotic stress tolerance. Plant Mol. Biol. 73, 409–424 (2010).
Gao, Y. et al. Molecular characterization and systematic analysis of NtAP2/ERF in tobacco and functional determination of NtRAV-4 under drought stress. Plant Physiol. Bioch. 156, 420–435 (2020).
Zhao, L. et al. CmRAV1 shows differential expression in two melon (Cucumis melo L.) cultivars and enhances salt tolerance in transgenic Arabidopsis plants. Acta Bioch. Bioph. Sin. 51, 1123–1133 (2019).
Matías-Hernández, L., Aguilar-Jaramillo, A. E., Marín-González, E., Suárez-López, P. & Pelaz, S. RAV genes: regulation of floral induction and beyond. Ann. Bot-London. 114, 1459–1470 (2014).
Hung, K. T. & Kao, C. H. Hydrogen peroxide is necessary for abscisic acid-induced senescence of rice leaves. J. Plant. Physiol. 161, 1347–1357 (2004).
Woo, H. R. et al. The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J. Exp. Bot. 61, 3947–3957 (2010).
Breitel, D. A. et al. Auxin response factor 2 intersects hormonal signals in the regulation of tomato fruit ripening. PLOS Genet. 12, e1005903 (2016).
Carbonero, P., Iglesias-Fernández, R. & Vicente-Carbajosa, J. The AFL subfamily of B3 transcription factors: evolution and function in angiosperm seeds. J. Exp. Bot. 68, 871–880 (2017).
Wójcikowska, B. et al. LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis, via YUCCA-mediated auxin biosynthesis. Planta. 238, 425–440 (2013).
Stone, S. L. et al. Leafy cotyledon2 encodes a B3 domain transcription factor that induces embryo development. P. Natl. Acad. Sci. USA. 98, 11806–11811 (2001).
Freitas, N. C. et al. In silico and in vivo analysis of ABI3 and VAL2 genes during somatic embryogenesis of Coffea arabica: competence acquisition and developmental marker genes. Plant Cell. Tissue Organ. Cult. Pctoc. 137, 599–611 (2019).
Khandelwal, A. et al. Role of ABA and ABI3 in desiccation tolerance. Science. 327, 546 (2010).
Suzuki, M., Wang, H. H. & McCarty, D. R. Repression of the leafy cotyledon 1/B3 regulatory network in plant embryo development by VP1/abscisic acid insensitive 3-LIKE B3 genes. Plant Physiol. 143, 902–911 (2007).
Veerappan, V. et al. A novel HSI2 mutation in Arabidopsis affects the PHD-like domain and leads to derepression of seed-specific gene expression. Planta. 236, 1–17 (2012).
Mantegazza, O. et al. Analysis of the Arabidopsis REM gene family predicts functions during flower development. Ann. Bot-London. 114, 1507–1515 (2014).
Franco-Zorrilla, J. M. et al. Identification of genes specifically expressed in cauliflower reproductive meristems. Molecular characterization of BoREM1. Plant Mol. Biol. 39, 427–436 (1999).
Levy, Y. Y., Mesnage, S., Mylne, J. S., Gendall, A. R. & Dean, C. Multiple roles of arabidopsis VRN1 in vernalization and flowering time control. Science. 297, 243–246 (2002).
Mendes, M. A. et al. MADS domain transcription factors mediate short-range DNA looping that is essential for target gene expression in Arabidopsis. Plant Cell. 25, 2560–2572 (2013).
Caselli, F. et al. REM34 and REM35 control female and male gametophyte development in Arabidopsis thaliana. Front Plant Sci. 10, 1351 (2019).
Guilfoyle, T. J. & Hagen, G. Getting a grasp on domain III/IV responsible for auxin response factor–IAA protein interactions. Plant Sci. 190, 82–88 (2012).
Boer, D. R. et al. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell. 156, 577–589 (2014).
Pekker, I., Alvarez, J. P. & Eshed, Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of Kanadi activity. Plant Cell. 17, 2899–2910 (2005).
Kumar, R., Tyagi, A. K. & Sharma, A. K. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol. Genet. Genomics. 285, 245–260 (2011).
Ge, J. et al. Tobacco TTG2 regulates vegetative growth and seed production via the predominant role of ARF8 in cooperation with ARF17 and ARF19. BMC Plant Biol. 16, 126 (2016).
Ren, Z., Liu, R., Gu, W. & Dong, X. The Solanum lycopersicum auxin response factor SlARF2 participates in regulating lateral root formation and flower organ senescence. Plant Sci. 256, 103–111 (2017).
Zhang, H., Cao, N., Dong, C. & Shang, Q. Genome-wide identification and expression of ARF gene family during adventitious root development in hot pepper (Capsicum annuum). Hortic Plant J. 3, 151–164 (2017).
Goetz, M., Vivian-Smith, A., Johnson, S. D. & Koltunow, A. M. Auxin response factor8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell. 18, 1873–1886 (2006).
Nagpal, P. et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 132, 4107–4118 (2005).
Sagar, M. et al. SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 161, 1362–1374 (2013).
Lupi, A. C. D. et al. Solanum lycopersicum GOLDEN 2-LIKE 2 transcription factor affects fruit quality in a light- and auxin-dependent manner. PLOS One. 14, e212224 (2019).
Liu, S. et al. Tomato AUXIN RESPONSE FACTOR 5 regulates fruit set and development via the mediation of auxin and gibberellin signaling. Sci. Rep. 8, 2971 (2018).
Yuan, Y. et al. SlARF10, an auxin response factor, is involved in chlorophyll and sugar accumulation during tomato fruit development. J. Exp. Bot. 69, 5507–5518 (2018).
Yuan, Y. et al. Auxin response factor 6A regulates photosynthesis, sugar accumulation, and fruit development in tomato. Hortic. Res. Eng. 6, 85 (2019).
Liu, F. et al. Pepper hub, an informatics hub for the chili pepper research community. Mol. Plant. 10, 1129–1132 (2017).
Rivera, A. et al. Assessing genetic and phenotypic diversity in pepper (Capsicum annuum) landraces from North-West Spain. Sci. Hortic. Amst. 203, 1–11 (2016).
Giovannoni, J. J. Genetic regulation of fruit development and ripening. Plant Cell. 16, S170–S180 (2004).
Yu, K. et al. Transcriptome changes during fruit development and ripening of sweet orange (Citrus sinensis). BMC Genomics. 13, 10 (2012).
Adams-Phillips, L., Barry, C. & Giovannoni, J. Signal transduction systems regulating fruit ripening. Trends Plant Sci. 9, 331–338 (2004).
Tang, D., Gallusci, P. & Lang, Z. Fruit development and epigenetic modifications. New Phytol. 228, 839–844 (2020).
Sharma, D., Koul, A., Kaul, S. & Dhar, M. K. Tissue-specific transcriptional regulation and metabolite accumulation in tomato (Solanum lycopersicum). Protoplasma. 257, 1093–1108 (2020).
Cao, H. et al. Tomato transcriptional repressor MYB70 directly regulates ethylene-dependent fruit ripening. Plant J. 104, 1568–1581 (2020).
Gao, J., Zhang, Y., Li, Z. & Liu, M. Role of ethylene response factors (ERFs) in fruit ripening. Food Qual. Saf. 4, 15–20 (2020).
Forlani, S., Mizzotti, C. & Masiero, S. The NAC side of the fruit: tuning of fruit development and maturation. BMC Plant Biol. 21, 238 (2021).
Liu, Z., Fan, M., Li, C. & Xu, J. H. Dynamic gene amplification and function diversification of grass-specific O-methyltransferase gene family. Genomics. 111, 687–695 (2019).
Shang, H., Li, W., Zou, C. & Yuan, Y. Analyses of the NAC transcription factor gene family in gossypium raimondii Ulbr.: Chromosomal location, structure, phylogeny, and expression patterns. J. Integr. Plant. Biol. 55, 663–676 (2013).
Mandal, D., Datta, S., Raveendar, G., Mondal, P. K. & Nag, C. R. RAV1 mediates cytokinin signaling for regulating primary root growth in Arabidopsis. Plant J. 113(1), 106–126 (2023).
Tsukagoshi, H., Morikami, A. & Nakamura, K. Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. P. Natl. Acad. Sci. USA. 104, 2543–2547 (2007).
Tang, R. et al. Genome-wide identification and function analyses of heat shock transcription factors in potato. Front. Plant Sci. 7, 490 (2016).
Li, D. et al. Genome-wide analysis of WRKY gene family in the sesame genome and identification of the WRKY genes involved in responses to abiotic stresses. BMC Plant Biol. 17, 152 (2017).
Deutsch, M. & Long, M. Intron—exon structures of eukaryotic model organisms. Nucl. Acids Res. 27, 3219–3228 (1999).
Xu, G., Guo, C., Shan, H. & Kong, H. Divergence of duplicate genes in exon-intron structure. P Natl. Acad. Sci. USA. 109, 1187–1192 (2012).
Bailey, T. L., Johnson, J., Grant, C. E. & Noble, W. The MEME suite. Nucl. Acids Res. 43, W39–W49 (2015).
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D. & May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10 (2004).
Sironi, M., Cagliani, R., Forni, D. & Clerici, M. Evolutionary insights into host-pathogen interactions from mammalian sequence data. Nat. Rev. Genet. 16, 224–236 (2015).
Villarino, G. H. et al. Transcriptomic signature of the SHATTERPROOF2 expression domain reveals the meristematic nature of arabidopsis gynoecial medial domain. Plant Physiol. 171, 42–61 (2016).
Tang, H. et al. Synteny and collinearity in plant genomes. Science. 320, 486–488 (2008).
Goetz, M. et al. Expression of aberrant forms of auxin response FACTOR8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol. 145, 351–366 (2007).
Kim, S. et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278 (2014).
Guillon, F. et al. Down-regulation of an auxin response factor in the tomato induces modification of fine pectin structure and tissue architecture. J. Exp. Bot. 59, 273–288 (2008).
De Jong, M., Wolters-Arts, M., Feron, R., Mariani, C. & Vriezen, W. H. The solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 57, 160–170 (2009).
De Jong, M. et al. Solanum lycopersicum AUXIN response factor 9 regulates cell division activity during early tomato fruit development. J. Exp. Bot. 66, 3405–3416 (2015).
Liu, N. et al. Down-regulation of auxin response factors 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J. Exp. Bot. 65, 2507–2520 (2014).
Chou, K. C. & Shen, H. B. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLOS One. 5, e11335 (2010).
Larkin, M. A. Clustal W and clustal X version 2.0.. Bioinformatics. 23(21), 2947–2948 (2007).
Minh, B. Q. et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic Era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Bailey, T. L. et al. MEME suite: Tools for motif discovery and searching. Nucl. Acids Res. 37, W202–W208 (2009).
Chen, C. et al. TBtools—an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202 (2020).
Lescot, M. et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucl. Acids Res. 30, 325–327 (2002).
Wang, Y. et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucl. Acids Res. 40, e49 (2012).
Krzywinski, M. et al. Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Li, Y. et al. The genome of Dioscorea zingiberensis sheds light on the biosynthesis, origin and evolution of the medicinally important diosgenin saponins. Hortic. Res. 9, 165 (2022).
Wan, H. et al. Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum). Biochem. Bioph. Res. Co. 416, 24–30 (2011).
Cui, J. et al. LncRNA33732-respiratory burst oxidase module associated with WRKY1 in tomato- Phytophthora infestans interactions. Plant J. 97, 933–946 (2019).
Acknowledgements
The authors thank the College of Agriculture, Guizhou University and Technology for the equipment used in this study.
Funding
This research was supported by Platform construction project of Engineering Research Center for Protected Vegetable Crops in Higher Learning Institutions of Guizhou Province (Qian Jiao Ji [2022] No. 040); Guizhou Provincial Science and Technology Support Program Targeted Key Projects (Qian Ke He Zhi Cheng [2022] Key No. 010); National Natural Science Foundation of China (31760576), Guizhou Provincial Scientific and Technological Achievements Application and Industrialization Plan Key Projects (Qian Ke He Chengguo [2020] No.1Z005); Advantageous industrial cluster construction project of Guizhou pod pepper supported by Ministry of Agriculture and Rural Affairs/Department of Agriculture and Rural Affairs of Guizhou Province (Nong Can Fa [2020] No. 2/Qian Nong Fa [2020] No. 67).
Author information
Authors and Affiliations
Contributions
T.W. and C.L. developed the experimental design and methodology. T.W., Y.W., and M.C. performed experimental work. S.S. and J.W. analysed the data. S.J., X.W. and J.H. carried out the plant material treatment. The manuscript was authored by T.W., D.X., Y.H., Y.R., and W.L. Each author participated in the creation of the experimental plan and the data analysis, offered suggestions for the paper, and gave the work their final approval before publishing. All authors have read and given their approval to the final, published version of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, T., Long, C., Chang, M. et al. Genome-wide identification of the B3 transcription factor family in pepper (Capsicum annuum) and expression patterns during fruit ripening. Sci Rep 14, 2226 (2024). https://doi.org/10.1038/s41598-023-51080-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-51080-6
This article is cited by
-
Identification of ARF genes in Juglans Sigillata Dode and analysis of their expression patterns under drought stress
Molecular Biology Reports (2024)
-
Characterization of leucine aminopeptidase (LAP) activity in sweet pepper fruits during ripening and its inhibition by nitration and reducing events
Plant Cell Reports (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.