Abstract
Plant-derived smoke has been shown to promote plant growth and seed germination, but its roles and mechanisms in response to nutrient deficiency stress remain unclear. Plants respond to phosphorus (P) deficiency by undergoing morphological, physiological, and transcriptional changes in order to improve nutrient uptake efficiency. Here, we showed that rice straw-derived smoke water could promote root growth in rice (Oryza sativa cv. Nipponbare) grown under P-sufficient and P-deficient conditions. Transcriptome analysis of the root tissues identified 1309 genes up-regulated and 1311 genes down-regulated by smoke water under P-deficient conditions. The GO terms ‘glutathione transferase activity’ and ‘photosynthesis—light reaction’ were found to be significantly enriched among the genes that were up- and down-regulated by smoke water, respectively. Biochemical analysis showed that smoke water reduced P-deficient-induced accumulation of H2O2 and malondialdehyde (MDA), a lipid peroxidation marker, reduced sucrose contents, but increased Fe accumulation. Furthermore, smoke water suppressed the expression of strigolactone biosynthesis genes, which were strongly induced by P deficiency as an adaptive strategy to improve root P uptake. These results revealed a potential mechanism by which smoke water promotes root growth and interacts with P deficiency-induced transcriptional regulation to mitigate P deficiency stress in rice.
Similar content being viewed by others
Introduction
Biomass burning in open fields is a common agricultural practice in several areas, particularly in Asia1. This burning activity typically takes place before and after cultivation as a quick and easy way of eliminating previous crop residues and weeds. Although fire and smoke have been shown to promote germination, especially for fire-prone plant species, such practices not only cause air pollution but also kill beneficial soil microorganisms2. Recently, plant-derived smoke and aqueous extracts obtained from smoke have received a lot of attention in agriculture and horticulture due to their potent effect similar to that of plant growth regulators3.
Smoke contains a number of different compounds, including biologically active compounds with potential agricultural use such as karrikins (KARs) and butenolides4,5. Smoke water has been reported to promote seed germination and growth in several plant species, including Arabidopsis, cucumber, scotch marigold, and rice6,7. However, the mechanisms by which smoke water promotes plant growth and its interaction with nutrient deficiency stress are not fully understood.
Phosphorus (P) is an essential macronutrient that plays a crucial role in energy metabolism and plant growth and development. However, P is often bound to soil particles and is relatively immobile, making it unavailable to plants even when P is present in the soil8. Plants adapt to P availability by undergoing morphological, physiological, and transcriptional changes in order to improve nutrient uptake efficiency9. Modification of root system architecture in response to P deficiency is one of the strategies to improve P acquisition10. In rice (Oryza sativa L.), the ability of roots to elongate under P deficiency varied greatly among rice populations11,12. Previous studies have shown that a gene encoding the protein kinase Pstol1 (Phosphorus-starvation tolerance 1), which is located in a major quantitative trait locus for P-deficiency tolerance, Pup1, is responsible for enhanced root growth and large root system, contributing to increased nutrient uptake and P-deficiency tolerance13. Overexpression of the gene could promote root growth and improve P-deficiency tolerance in rice cv. Nipponbare, whose genome lacks PSTOL1 and does not exhibit P deficiency-induced root elongation traits11,13.
In this study, the effects of rice straw-derived smoke water treatment on root growth of Nipponbare rice seedlings under P-sufficient and P-deficient conditions were investigated in a hydroponic system. Transcriptomic analysis of root tissues treated with smoke water revealed that smoke water altered the expression of genes involved in oxidative stress, photosynthesis, Fe deficiency, and strigolactone biosynthesis. The accumulation of H2O2, an oxidative stress marker, sucrose, and P and Fe in root tissues were also determined to investigate the mechanism of smoke water in promoting rice root growth and mitigating P deficiency stress.
Results
Smoke water promotes rice root growth
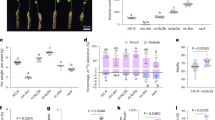
Rice seedlings (cv. Nipponbare) were grown hydroponically in P-sufficient (HP) or deficient (LP) conditions and treated with various concentrations of smoke water in the nutrient solutions. Nipponbare root growth was slightly inhibited under P deficiency, consistent with previous reports11 (Fig. 1). Smoke water treatments promoted shoot and root growth in a dose-dependent manner under both HP and LP conditions. When 0.2%, 1% or 2% smoke water was applied, root elongation could be promoted under LP condition at higher levels than under HP condition (Fig. 1). This suggests that smoke water could enhance P deficiency-induced root elongation in Nipponbare.
Smoke water enhanced P deficiency-induced root elongation in rice. (a) Rice seedlings (cv. Nipponbare) were grown hydroponically in P-sufficient (HP) or deficient (LP) conditions and treated with various concentrations of smoke water in the nutrient solutions. Scale bar, 10 cm. (b) Quantification of root lengths. Data are means ± s.d. (n = 4 biological replicates with 6 plants each). Different letters indicate significant differences (p < 0.05) according to Duncan’s multiple range test (DMRT).
Transcriptome profiling of smoke water-treated rice roots
To understand the mechanisms by which smoke water promotes root elongation and response to P deficiency, we performed RNA-seq analysis of root tissues derived from rice seedlings treated with or without 1% smoke water (+S or −S) under HP or LP conditions for 3 days. Each treatment included three biological replicates. mRNA libraries were sequenced using the Illumina NovaSeq platform. After pre-processing, filtering low-quality reads, and alignment to the RAP-DB reference genome (IRGSP 1.0.21), comparison of expression profiles was performed using DESeq214. Genes that were significantly differentially expressed by more than 2 folds (|log2fold change| > 1 and adjusted p-value < 0.05) were included in the differentially expressed gene (DEG) list for further analysis.
To identify smoke water-responsive genes in HP condition, the treatment +S/HP was compared with −S/HP; 841 and 609 genes were up- and down-regulated by smoke water treatment, respectively. In LP condition (+S/LP vs. −S/LP), smoke water up-regulated 1309 genes and down-regulated 1311 genes, with 608 and 395 genes also up- and down-regulated, respectively, by smoke water in HP condition. Among all smoke water-responsive genes, there were 35 genes that showed opposite regulation by smoke water under HP and LP conditions (up-regulated in one and down-regulated in the other) and were excluded from the Venn diagram (Fig. 2a, Supplementary Table S1).
Transcriptome analysis of roots treated with smoke water under P-sufficient and -deficient conditions. Root tissues were harvested from 10-day-old rice seedlings grown hydroponically in Pi-sufficient (HP) or deficient (LP) conditions and treated with or without 1% smoke water (+S or −S) for 3 days. (a) Venn diagram showing the overlap between the lists of smoke water up- and down-regulated genes (|log2fold change| > 1; adjusted p-value < 0.05) in HP and LP conditions. (b) Hierarchically clustered heatmap displaying the log2FC values of all smoke water-responsive or P-deficiency-responsive genes. (c) GO biological process (BP) and molecular function (MF) term enrichment analysis of the smoke water up- and down-regulated gene lists.
To identify P deficiency-responsive genes, the treatment −S/LP was compared with −S/HP; 969 and 700 genes were up- and down-regulated by P deficiency, respectively. Expression heatmap analysis of 4281 genes, which were significantly differentially expressed in at least one of the comparisons (+S/LP vs. −S/LP, +S/HP vs. −S/HP, and −S/LP vs. −S/HP), showed that the effect of smoke water on gene expression is quite distinct from the effect of P deficiency on root transcriptomes (Fig. 2b). In addition, the effects of smoke water treatment on root transcriptomes under both LP and HP conditions were similar, but the degrees of expression fold changes under LP condition were higher.
Gene ontology enrichment analysis
To investigate the effects of smoke water on biological processes and molecular functions, we performed gene ontology (GO) enrichment analysis of the lists of smoke water-responsive genes under LP condition (Fig. 2c). GO terms significantly enriched for the smoke water-up-regulated gene list included glutathione transferase activity (GO:0004364), defense response (GO:0006952), amine metabolic process (GO:0009308), and lipid metabolic process (GO:0006629). GO terms significantly enriched for the smoke water-down-regulated gene list included photosynthesis (GO:0015979), photosynthesis—light reaction (GO:0019684), and terpenoid metabolic process (GO:0006721).
Oxidative stress-related genes
The term ‘glutathione transferase activity’ was enriched among the smoke water-up-regulated genes with 10.35 fold enrichment. A number of genes encoding glutathione-s-transferases (GSTs) were upregulated by smoke water in both HP and LP conditions. Compared to the control (−S/HP), the expression levels were strongly upregulated by smoke in LP more than in HP (Fig. 3a, Supplementary Table S1). We identified 26 OsGSTUs, five OsGSTFs and one OsGSTZ up-regulated by smoke water, with the exception of OsGSTU36 and OsGSTU43 that were significantly down-regulated.
Smoke water promoted expression of genes encoding glutathione-S-transferases (GSTs) and reduced accumulation of H2O2 and oxidative stress markers in P-deficient roots. (a) Heatmap represents log2FC values of genes in the −S/LP, +S/HP or +S/LP compared with −S/HP conditions. Only genes that showed statistical significance in at least one of the comparisons were included in this figure. (b,c) Quantification of H2O2 (b) and malondialdehyde (MDA) (c) in P-deficient roots. (d,e) Total P contents in shoots (d) and roots (e) of rice seedlings. Data are means ± s.d (n = 3 biological replicates). Different letters indicate significant differences (p < 0.05) according to Duncan’s multiple range test (DMRT).
GSTs play important roles in the detoxifying mechanisms in response to abiotic stresses and stress tolerance in plants15. Previous studies have shown that P deficiency, as well as other abiotic stresses, induced the accumulation of H2O2 and malondialdehyde (MDA), a lipid peroxidation marker caused by oxidative stress16. Quantification of H2O2 and MDA levels in the roots showed that P-deficient roots accumulated higher levels of H2O2 and MDA and that smoke water treatment could remarkably reduce both H2O2 and MDA (Fig. 3b,c). This suggests that smoke water treatment is able to reduce P deficiency-induced oxidative stress, potentially by upregulating the expression of GSTs.
To test whether smoke water treatments mitigate P deficiency-induced stress by improving plant P status, total P contents in shoots and roots were determined. LP treatments dramatically reduced P contents in both shoots and roots, whereas smoke water treatments did not significantly increase P contents when compared to the control of the same P condition (Fig. 3d,e). This illustrated that both the −S/LP and +S/LP samples were similarly P-deficient. Phosphate (Pi) starvation-induced (PSI) genes, including Purple Acid Phosphatases (PAPs), Pi Transporters (PTs), OsSPX, and OsSQD2 genes, were also upregulated at similar levels in both −S/LP and +S/LP samples (Supplementary Fig. S1). Taken together, these results suggest that smoke water did not alter degrees of P deficiency as reflected by total P contents and transcript levels of most genes involved in Pi starvation responses but could mitigate P-deficiency stress.
Photosynthesis-related genes
The GO biological processes ‘photosynthesis’ and ‘photosynthesis—light reaction’ were significantly enriched in the smoke water-down-regulated gene list with 13.59 and 19.22 fold enrichment, respectively (Fig. 2c). Smoke water significantly down-regulated 5 genes encoding photosystem I (PSI) light-harvesting chlorophyll a/b-binding (LHCA) proteins, 8 genes encoding photosystem II (PSII) light-harvesting chlorophyll a/b-binding (LHCB) proteins, 9 genes encoding PSI reaction center subunits (PsaD, PsaE, PsaF, PsaG, PsaH, PsaK, PsaL, PsaN, and PsaO), and 8 genes encoding PSII components (Fig. 4a,b, Supplementary Table S1). In addition to the genes encoding PSI and PSII components, Ferredoxin 1 (Fd1), Plastocyanin (PC), and leaf-type ferredoxin-NADP+-oxidoreductase 1 (LFNR1) that are involved in electron transport, as well as OsPGR5 (Proton Gradient Regulation 5), which is required for PSI cyclic electron transport, were also down-regulated by smoke water. Moreover, smoke water treatment in the P-deficient condition also down-regulated the MYB transcription factor OsGLK1 (Golden 2-like 1), a major regulator of chloroplast development17. Measurement of sucrose contents in root tissues showed that P deficiency decreased sucrose contents by 42.57% in the absence of smoke water, and smoke water decreased sucrose contents by 67.89% and 65.69%, when compared to the control in HP and LP, respectively (Fig. 4c). Although root chloroplasts are capable of photosynthesis, their efficiency is much lower than that of shoots18. This finding suggests that, rather than having a negative impact directly on root carbon assimilation, smoke water may affect light-induced ROS production in the root chloroplasts, and shoot–root translocation or sink strength of the roots.
Smoke water repressed expression of genes involved in photosynthesis and reduced sucrose accumulation in roots. (a) Heatmap represents log2FC values of genes in the −S/LP, +S/HP or +S/LP compared with −S/HP conditions. Only genes that showed statistical significance in at least one of the comparisons were included in this figure. (b) MapMan overview of the genes functioning at the thylakoid membrane of the chloroplast. The individual genes are exhibited by small color squares, showing log2FC values of genes in the +S/LP vs. −S/LP comparison. (c) Sucrose contents in the roots. Data are means ± s.d. (n = 3 biological replicates). Different letters indicate significant differences (p < 0.05) according to Duncan’s multiple range test (DMRT).
Fe deficiency-related genes
The antagonistic interaction between P and Fe has been well established19,20,21. Under Fe deficiency, rice employs both reduction and chelation strategies to increase Fe acquisition and transports. In rice, Fe-deficiency-responsive genes include genes encoding ferric-chelate reductase (FRO), ferrous transporters (IRTs), deoxymugineic acid (DMA) biosynthesis enzymes, Fe (III)-DMA transporters (Yellow Stripe family transporters, YSLs), as well as transcription factors, OsIRO2 and OsIRO3, which are responsible for controlling the DMA synthesis and Fe transporter genes22,23.
In the absence of smoke water treatment, P deficiency down-regulated expression of genes involved in Fe homeostasis and increased Fe concentrations in shoots and roots (Fig. 5), consistent with previous reports20. This is partly due to increased availability of Fe in the Pi-deficient media19. Interestingly, smoke water treatment further enhanced suppression of the Fe-related genes under P deficiency. This expression data was correlated with the elevated Fe concentrations in the smoke water-treated Pi-deficient plants (Fig. 5). It is possible that smoke water could further increase Fe availability in the Pi-deficient media and/or increase Fe accumulation in plants due to the larger root system (Fig. 1).
Smoke water increased Fe contents and repressed Fe deficiency-related gene expression in P-deficient plants. (a) Heatmap represents log2FC values of genes in the −S/LP, +S/HP or +S/LP compared with −S/HP conditions. Only genes that showed statistical significance in at least one of the comparisons were included in this figure. (b,c) Fe contents in shoots (b) and roots (c) of rice seedlings. Data are means ± s.d. (n = 3 biological replicates). Different letters indicate significant differences (p < 0.05) according to Duncan’s multiple range test (DMRT).
Strigolactone-related genes
Strigolactone (SL) plays key roles in plant adaptation to nutrient deficiency by promoting arbuscular mycorrhiza (AM) colonization in order to increase nutrient foraging. In the absence of smoke water, P deficiency strongly induced expression of SL biosynthesis enzymes, but not SL signaling components, consistent with previous findings24. Smoke water treatment in HP condition did not alter expression of SL biosynthesis genes (Fig. 6). Interestingly, the P deficiency-induced SL biosynthesis gene expression was strongly suppressed in the presence of smoke water (Fig. 6). Moreover, smoke water also suppressed the upregulation of ZAS in response to P deficiency. The gene ZAS encodes a carotenoid cleavage dioxygenase (CCD) enzyme, zaxinone synthase, which produces zaxinone, an apocarotenoid metabolite that regulates growth and SL biosynthesis in rice25. These findings suggest that smoke water could alter the transcriptional control of these genes and affect the Pi starvation-induced mechanism. It also suggests that smoke water could alter the metabolic balance of these carotenoid-derived signaling molecules under P deficiency.
Discussion
P deficiency has been shown to alter root system architecture to enhance P foraging and uptake in several species9,26. In rice, the effect of P deficiency on root system development depends on underlying genetic control11,27. Here, we demonstrated the growth-promoting effects of rice straw-derived smoke water on Nipponbare rice root under P-deficient condition, in which root growth was inhibited in the absence of smoke water treatment (Fig. 1). This suggests that smoke water could promote root elongation under P deficiency through mechanisms independent of PSTOL1, which functions to enhance root system but is absent from the Nipponbare genome13. Our transcriptome analysis revealed that smoke water modulated the expression of genes involved in oxidative stress, photosynthesis, and SL biosynthesis (Fig. 2). Based on the results, we proposed a schematic model of smoke water regulating rice root elongation and P deficiency responses (Fig. 7).
The antioxidative role of glutathione in reducing oxidative stress and preventing lipid peroxidation has been well documented28. In this study, smoke water strongly induced expression of GSTs, and the degree of induction was increased by P deficiency (Fig. 3a). Smoke water treatment also reduced H2O2 and MDA levels in P-deficient roots (Fig. 3b,c). These results suggest that smoke water may alleviate oxidative stress on roots induced by P deficiency29, consequently contributing to root elongation under P deficiency. A recent proteomic analysis has demonstrated that smoke water induced glutathione-related proteins and improved shoot growth in maize seedlings30. Glutathione also modulates auxin homeostasis to regulate root system adaptation to P deficiency in Arabidopsis31. Furthermore, it has been shown in wheat that combining P and Fe deficiency could promote expression of GSTs and recover root elongation, which was otherwise inhibited under Fe deficiency32. Wang et al. reported consistently that catechol, a major component of smoke, upregulated a number of genes related to redox homeostasis, leading to smoke-induced root cell elongation through spatial changes in redox status in Nicotiana attenuata roots33. Previous research has demonstrated that ROS controls the balance between cell proliferation and differentiation in roots, where O2·− accumulates in the meristem and H2O2 accumulates in the elongation zone, thereby determining meristem size and root length34. Further research into tissue-specific ROS distribution in root tips using histochemical assays would provide a better understanding of the effects of smoke water on root cell proliferation and differentiation.
Our transcriptome analysis showed that smoke water treatment repressed expression of photosynthetic genes, particularly those involved in the light reaction at the thylakoid membrane, and the repression was stronger under P deficiency (Fig. 4). It should be noted that, in our study, roots were grown under in vitro conditions with some exposure to light, which may contribute to the responses of the photosynthetic genes. P deficiency has been shown to reduce photosynthesis in shoots as well as the expression of photosynthetic genes in roots of Arabidopsis, soybean, and wheat35,36,37,38. Loss-of-function mutations of a gene encoding tyrosylprotein sulfotransferase (TPST) and overexpression of GLK transcription factors in Arabidopsis resulted in ectopically activated photosynthesis in the roots, increased light-induced ROS production in root cells, and hypersensitive root growth inhibition under P deficiency37. Although the biological significance of photosynthesis in roots remains unclear, this evidence demonstrates that suppression of photosynthetic gene expression in roots is required for sustained root growth in vitro under P deficiency37. Hence, smoke water application may promote rice root growth by suppressing the expression of photosynthetic genes, which may contribute to light-induced ROS production. Expression of photosynthetic genes is controlled by several transcription factors, including HY5, PIFs, GLKs, GNC, and GNL39,40,41. Only OsGLK1 was found to be significantly differentially expressed by smoke water in our transcriptome analysis. Hydroquinone, a bioactive compound in plant-derived smoke42, has been shown to inhibit transcription of photosynthetic genes in marine diatoms43. The bioactive compounds in smoke water that are responsible for the transcriptional regulation of photosynthetic genes will need to be identified through additional research.
Sucrose not only functions as a source of energy for cellular metabolism, but also acts as a signaling molecule in multiple pathways to coordinate the growth of above-ground shoots with the underground roots44. P deficiency has been reported to increase sugar accumulation in bean, soybean, and rice roots, decrease sugar accumulation in cucumber roots, and remain unchanged in Arabidopsis roots44,45,46. In our observation, sucrose contents were reduced in P-deficient and smoke water-treated roots (Fig. 4c). Previous studies have shown that the effects of P deficiency on root growth vary depending on plant genotype, growth conditions (soil, hydroponic, or agar), light conditions, and root illumination11,47,48,49. These factors may explain the variation in sugar contents observed across studies. Interestingly, despite the reduced sucrose content, our results showed that smoke water promoted root growth, suggesting that smoke water may promote metabolic efficiency. P deficiency has been shown to induce lysigenous aerenchyma formation in some genotypes of rice, maize, and common bean. This adaptive trait contributes to decreased respiration and increased P utilization efficiency, thus increasing metabolic efficiency and allowing plants to develop additional root structures, such as longer roots, lateral roots, and root hairs, to explore the soil50,51. Further study is needed to determine whether smoke water can promote aerenchyma formation and improve metabolic efficiency in P-deficient conditions.
Our findings demonstrate P deficiency-induced transcriptomic changes, including upregulation of the PSI genes, downregulation of Fe homeostasis genes, and upregulation of SL biosynthesis genes, which are consistent with previous reports19,32. Under P deficiency, smoke water treatment did not alter the expression of the PSI genes but enhanced the suppression of Fe homeostasis genes. It is possible that smoke water may affect P acquisition efficiency in soil-grown plants, potentially by promoting root growth and improving foraging capacity. However, in this study, the smoke water-treated P-deficient plants could not benefit from the larger root system because the amount of available Pi in hydroponic conditions was limited and finite. Further studies are needed to study the effects of smoke water on root growth and P uptake in the soil. In contrast, Fe uptake was enhanced by smoke water under P deficiency, which could be attributed to the increase of Fe availability in the Pi-deficient nutrient solution19, as well as the larger root system of the smoke water-treated plants.
Pi and nitrate deficiencies have been shown to induce strigolactone biosynthesis, which plays crucial roles in regulating root adaptation by modulating auxin transport24. Moreover, SLs in root exudates not only promote symbiosis with arbuscular mycorrhiza (AM) to increase nutrient uptake, but also trigger seed germination of parasitic weeds52,53. Our results revealed that smoke water treatment inhibit the expression of SL biosynthesis genes, which were strongly induced in P-deficient plants. This suggests that smoke water interacts with the Pi starvation-induced transcriptional control and may reduce levels of SLs exuded by the roots, which could consequently affect mycorrhization. On the other hand, it is also possible that smoke water may contain compounds that have similar properties as SLs, and thus the presence of such compounds could feedback regulate endogenous SL biosynthesis under P deficiency54. In this case, the chemicals may have the potential to induce germination of root parasitic weeds before farming, an agricultural technique known as suicidal germination55. Indeed, karrikins, which are plant growth regulators derived from plant material smoke, have structural similarities to strigolactones and function via a signaling pathway closely related to that of SL5. Further experiments are needed to determine whether smoke water reduces SL levels in plants and root exudates, how that might affect SL function in plant growth and development, including Pi starvation responses and AM colonization, and whether smoke water could affect germination of the parasitic weeds.
Conclusion
Smoke water has been used in agriculture in various ways, including as a seed treatment, foliar spray, or soil amendment, and several studies have shown positive effects of smoke water on seed germination and plant growth. However, the effects of smoke water on plant growth and development could vary depending on the type of plant and the concentration and composition of smoke water3,7,56. In this study, we showed the promoting effects of rice straw-derived smoke water on root growth of P-deficient rice and provided further understandings of potential mechanisms through transcriptional modulation of genes involved in oxidative stress, photosynthesis, and SL biosynthesis. Further research is needed to fully understand its potential as a plant growth promoter across different plant species and under different environmental conditions.
Materials and methods
Production of plant-derived smoke water solution
Rice straw-derived smoke water was prepared using the method adapted from a previous study7. In brief, one kilogram of dried rice straws was burned in a closed burning system connected to a vacuum to draw the smoke through 1-L water in the bottle, in which the water-soluble compounds in the smoke could dissolve. The smoke–water solution was then used in the subsequent experiments.
Plant materials and growth condition
Seeds of rice (Oryza sativa L.) cv. Nipponbare were obtained from the Center of Excellence in Environment and Plant Physiology, Chulalongkorn University, as used in the previous study57. Seeds were surface-sterilized and pre-cultured in water for 2 days, followed by half-strength Yoshida’s solution58 for 2 days under dark conditions. Then, the seedlings were grown in full-strength Yoshida’s solution under light conditions for 4 days before removing the seed endosperm and being transferred to P-sufficient (320 µM NaH2PO4, HP) and P-deficient (0.8 µM NaH2PO4, LP) conditions in the presence of various concentrations of smoke water. The seedlings were treated for 8 days for growth and biochemical analyses or 3 days for RNA extraction. The nutrient solutions were adjusted to pH 5.8 and renewed every 2 days. The seedlings were grown in a growth room at 30 °C with a 12 h/12 h light–dark cycle.
RNA extraction, cDNA library construction and RNA-Seq
Whole root tissues were collected from treated plants and immediately frozen in liquid nitrogen. Three biological replicates, each consisting of 6 plants, were used for each treatment. Total RNA was extracted from the root tissues using PureLink RNA Mini Kit based on the manufacturer’s instructions (Invitrogen, USA) and genomic DNA was removed using DNase I (Thermo Fisher Scientific, USA). The quality of the total RNA was assessed by the Agilent 2100 Bioanalyzer (Agilent Technologies, USA), and all samples had an RNA integrity number (RIN) greater than 8.9. Purification of mRNA, library construction, and sequencing were carried out by Apical Scientific Sdn. Bhd. (Malaysia) using the Illumina NovaSeq 6000 sequencer (Illumina, USA) to obtain 2 × 150 bp paired-end reads according to the manufacturer’s instructions. The RNA-seq raw data were deposited in the Sequence Read Archive of the National Center for Biotechnology Information under accession number PRJNA924970.
Data processing and bioinformatics analysis
Following pre-processing and filtering of low-quality reads, paired-end clean reads were aligned to the RAP-DB reference genome (IRGSP 1.0.21) using HISAT2 (version 2.1.0)59. Reads were assembled into transcripts using StringTie (version 2.0.6)60. Differential gene expression was performed using DESeq2 (version 1.31.6)14 and genes with an adjusted p-value < 0.05 and |fold change| > 2 (equivalent to |shrunken log2fold change| > 1) were considered to be significantly differentially expressed. Hierarchical clustering and heatmap analysis were performed using ‘hclust’ and ‘heatmap.2’ packages in R (version 3.5.3). Heatmaps showing expression levels of selected genes were plotted using ClustVis web tools61.
To investigate the effects of smoke water on biological processes and molecular functions, we performed gene ontology (GO) enrichment analysis of the list of genes responsive to smoke water under P-deficient conditions using agriGo v2.0 with Fisher’s exact test, Yekutieli (FDR under dependency < 0.05)62. Lists of the enriched GO terms were then summarized by REViGO63. Terms with FDR < 0.005 and fold enrichment > 2 were plotted using R script64. The log2FC values of genes in the +S/LP vs. −S/LP comparison were also imported into MapMan software65 to visualize genes functioning at the thylakoid membrane of the chloroplast.
Measurement of P and Fe contents
Root and shoot samples were dried at 70 °C until a constant weight was recorded. To determine total P and Fe contents, dry root and shoot samples were digested with concentrated HNO3 using microwave digestion (UltraWAVE, Milestone, CT, United States). P contents were determined by the ammonium molybdate colorimetric assay66. In brief, diluted supernatants were mixed with molybdate blue reagent (0.4% w/v ammonium molybdate in 0.5 M H2SO4 and 10% w/v ascorbic acid; 6:1 ratio) and then incubated at 40 °C for 20 min. The absorbance was read at 820 nm, and the P content was calculated by comparing with a standard curve. Fe contents in the supernatant were determined using atomic absorption spectroscopy (Analytik Jena ZEEnit 700P, Germany).
Measurement of H2O2 and MDA
Frozen tissue was ground and extracted with 0.1% trichloroacetic acid (TCA) on ice, and the supernatant was collected for H2O2 and MDA measurements. H2O2 was detected by mixing 0.5 ml of the supernatant with 0.5 ml of a 10 mM potassium phosphate buffer (pH 7.0) and 1 ml of 1 M potassium iodide, and measuring the absorbance at 390 nm. The H2O2 content was calculated using a standard curve67. MDA was detected by mixing 0.5 ml of the supernatant with 1 ml of a 0.5% thiobarbituric acid (TBA) solution, incubating the reaction mixture in boiling water for 30 min, and measuring the absorbance at 532 nm for the TBA-MDA complex and at 600 nm for non-specific turbidity correction. The MDA content was calculated using A532, A600 and the molar extinction coefficient of the MDA (115 mM−1 cm−1)68.
Measurement of sucrose content
Sucrose content was extracted and measured following the method used in the previous study69. In brief, sucrose in ground frozen tissue was extracted with 80% ethanol by incubating at 80 °C for 40 min. The supernatant was collected, and the extraction process was repeated twice. The supernatant was then mixed with 0.2 ml of 2 M NaOH and heated at 100 °C for 5 min. The reaction mixture was added to 2.8 ml 30% HCl and 0.8 ml 0.1% resorcinol in 95% ethanol, and then incubated at 80 °C for 10 min. After the reaction was stopped, the absorbance was read at 480 nm, and the sucrose content was calculated using a standard curve.
Statistical analysis
For quantitation of root length, the experiments included 4 biological replicates, each consisting of 6 plants. For measurement of P, Fe, H2O2, MDA, and sucrose contents, the experiments were performed with 3 biological replicates, each consisting of 8 plants. Means and standard deviations (SD) were calculated and analyzed by one-way analysis of variance (ANOVA). Mean comparison was calculated according to Duncan’s multiple range test (DMRT) using IBM SPSS Statistics 20.
Ethical approval
Experimental research on plants in this study, including the collection of plant material, complies with relevant institutional, national, and international guidelines and legislation.
Data availability
The RNA-seq data for all samples are available at the Sequence Read Archive of the National Center for Biotechnology Information under accession number PRJNA924970. All other data generated or analysed during this study are included in this published article.
References
Chang, D. & Song, Y. Estimates of biomass burning emissions in tropical Asia based on satellite-derived data. Atmos. Chem. Phys. 10, 2335–2351 (2010).
Mandal, K. G. et al. Rice residue-management options and effects on soil properties and crop productivity. J. Food Agric. Environ. 2, 224–231 (2004).
Kulkarni, M. G., Light, M. E. & Van Staden, J. Plant-derived smoke: Old technology with possibilities for economic applications in agriculture and horticulture. S. Afr. J. Bot. 77, 972–979 (2011).
Light, M. E., Burger, B. V., Staerk, D., Kohout, L. & Van Staden, J. Butenolides from plant-derived smoke: Natural plant-growth regulators with antagonistic actions on seed germination. J. Nat. Prod. 73, 267–269 (2010).
Nelson, D. C., Flematti, G. R., Ghisalberti, E. L., Dixon, K. W. & Smith, S. M. Regulation of seed germination and seedling growth by chemical signals from burning vegetation. Annu. Rev. Plant Biol. 63, 107–130 (2012).
Khan, M. H. U. et al. Bacillus safensis with plant-derived smoke stimulates rice growth under saline conditions. Environ. Sci. Pollut. Res. 24, 23850–23863 (2017).
Elsadek, M. A. & Yousef, E. A. Smoke-water enhances germination and seedling growth of four horticultural crops. Plants 8, 104 (2019).
Richardson, A. E., Hocking, P. J., Simpson, R. J. & George, T. S. Plant mechanisms to optimise access to soil phosphorus. Crop Pasture Sci. 60, 124 (2009).
Zhang, Z., Liao, H. & Lucas, W. J. Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J. Integr. Plant Biol. 56, 29 (2014).
Lynch, J. P. Root phenotypes for improved nutrient capture: An underexploited opportunity for global agriculture. New Phytol. 223, 548–564 (2019).
Shimizu, A., Yanagihara, S., Kawasaki, S. & Ikehashi, H. Phosphorus deficiency-induced root elongation and its QTL in rice (Oryza sativa L.). Theor. Appl. Genet. 109, 1361–1368 (2004).
Pinit, S. et al. Hyperspectral and genome-wide association analyses of leaf phosphorus status in local Thai indica rice. PLoS One 17, e0267304 (2022).
Gamuyao, R. et al. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488, 535–539 (2012).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Nianiou-Obeidat, I. et al. Plant glutathione transferase-mediated stress tolerance: Functions and biotechnological applications. Plant Cell Rep. 36, 791–805 (2017).
Zhang, K., Liu, H., Tao, P. & Chen, H. Comparative proteomic analyses provide new insights into low phosphorus stress responses in maize leaves. PLoS One 9, e98215 (2014).
Chen, M. et al. GOLDEN 2-LIKE transcription factors of plants. Front. Plant Sci. 7, 1509 (2016).
Kobayashi, K. et al. Photosynthesis of root chloroplasts developed in Arabidopsis lines overexpressing GOLDEN2-LIKE transcription factors. Plant Cell Physiol. 54, 1365–1377 (2013).
Misson, J. et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. U.S.A. 102, 11934–11939 (2005).
Zheng, L. et al. Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol. 151, 262–274 (2009).
Xie, X., Hu, W., Fan, X., Chen, H. & Tang, M. Interactions between phosphorus, zinc, and iron homeostasis in nonmycorrhizal and mycorrhizal plants. Front. Plant Sci. 10, 1172 (2019).
Ogo, Y. et al. OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol. Biol. 75, 593–605 (2011).
Kobayashi, T. et al. Jasmonate signaling is activated in the very early stages of iron deficiency responses in rice roots. Plant Mol. Biol. 91, 533–547 (2016).
Sun, H. et al. Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J. Exp. Bot. 65, 6735–6746 (2014).
Wang, J. Y. et al. The apocarotenoid metabolite zaxinone regulates growth and strigolactone biosynthesis in rice. Nat. Commun. 10, 810 (2019).
Saengwilai, P. J., Bootti, P. & Klinnawee, L. Responses of rubber tree seedlings (Hevea brasiliensis) to phosphorus deficient soils. Soil Sci. Plant Nutr. 69, 78–87. (2023).
Liu, D. Root developmental responses to phosphorus nutrition. J. Integr. Plant Biol. 63, 1065–1090 (2021).
Hasanuzzaman, M., Nahar, K., Anee, T. I. & Fujita, M. Glutathione in plants: Biosynthesis and physiological role in environmental stress tolerance. Physiol. Mol. Biol. Plants 23, 249–268 (2017).
Juszczuk, I., Malusà, E. & Rychter, A. M. Oxidative stress during phosphate deficiency in roots of bean plants (Phaseolus vulgaris L.). J. Plant Physiol. 158, 1299–1305 (2001).
Aslam, M. et al. Molecular responses of maize shoot to a plant derived smoke solution. IJMS 20, 1319 (2019).
Trujillo-Hernandez, J. A. et al. A glutathione-dependent control of the indole butyric acid pathway supports Arabidopsis root system adaptation to phosphate deprivation. J. Exp. Bot. 71, 4843–4857 (2020).
Kaur, G. et al. Physiological and molecular responses to combinatorial iron and phosphate deficiencies in hexaploid wheat seedlings. Genomics 113, 3935–3950 (2021).
Wang, M. et al. Catechol, a major component of smoke, influences primary root growth and root hair elongation through reactive oxygen species-mediated redox signaling. New Phytol. 213, 1755–1770 (2017).
Eljebbawi, A., Guerrero, Y. D. C. R., Dunand, C. & Estevez, J. M. Highlighting reactive oxygen species as multitaskers in root development. iScience 24, 101978 (2021).
Wu, P. et al. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 132, 1260–1271 (2003).
Zeng, H. et al. Genome-wide identification of phosphate-deficiency-responsive genes in soybean roots by high-throughput sequencing. Plant Soil 398, 207–227 (2016).
Kang, J. et al. Suppression of photosynthetic gene expression in roots is required for sustained root growth under phosphate deficiency. Plant Physiol. 165, 1156–1170 (2014).
Wang, J. et al. Transcriptome analysis in roots and leaves of wheat seedlings in response to low-phosphorus stress. Sci. Rep. 9, 19802 (2019).
Zubo, Y. O. et al. Coordination of chloroplast development through the action of the GNC and GLK transcription factor families. Plant Physiol. 178, 130–147 (2018).
Toledo-Ortiz, G. et al. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 10, e1004416 (2014).
Bi, Y.-M. et al. Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity: A GATA gene for chlorophyll and glucose sensitivity. Plant J. 44, 680–692 (2005).
Kamran, M. et al. Hydroquinone; A novel bioactive compound from plant-derived smoke can cue seed germination of lettuce. Front. Chem. 5, 30 (2017).
Yang, C. et al. Allelochemical induces growth and photosynthesis inhibition, oxidative damage in marine diatom Phaeodactylum tricornutum. J. Exp. Mar. Biol. Ecol. 444, 16–23 (2013).
Hammond, J. P. & White, P. J. Sucrose transport in the phloem: Integrating root responses to phosphorus starvation. J. Exp. Bot. 59, 93–109 (2007).
Naureen, Z. et al. Effect of phosphate nutrition on growth, physiology and phosphate transporter expression of cucumber seedlings. Plant Physiol. Biochem. 127, 211–222 (2018).
Luquet, D., Zhang, B. G., Dingkuhn, M., Dexet, A. & Clément-Vidal, A. Phenotypic plasticity of rice seedlings: Case of phosphorus deficiency. Plant Prod. Sci. 8, 145–151 (2005).
Cabrera, J., Conesa, C. M. & Pozo, J. C. May the dark be with roots: A perspective on how root illumination may bias in vitro research on plant–environment interactions. New Phytol. 233, 1988–1997 (2022).
Zheng, Z., Wang, Z., Wang, X. & Liu, D. Blue light-triggered chemical reactions underlie phosphate deficiency-induced inhibition of root elongation of Arabidopsis seedlings grown in petri dishes. Mol. Plant 12, 1515–1523 (2019).
Wissuwa, M., Gamat, G. & Ismail, A. M. Is root growth under phosphorus deficiency affected by source or sink limitations?. J. Exp. Bot. 56, 1943–1950 (2005).
Fan, M., Zhu, J., Richards, C., Brown, K. M. & Lynch, J. P. Physiological roles for aerenchyma in phosphorus-stressed roots. Funct. Plant Biol. 30, 493 (2003).
Pujol, V. & Wissuwa, M. Contrasting development of lysigenous aerenchyma in two rice genotypes under phosphorus deficiency. BMC Res. Notes 11, 60 (2018).
Yoneyama, K. et al. How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation?. Planta 235, 1197–1207 (2012).
Nopphakat, K., Runsaeng, P. & Klinnawee, L. Acaulospora as the dominant arbuscular mycorrhizal fungi in organic lowland rice paddies improves phosphorus availability in soils. Sustainability 14, 31 (2021).
Wang, J. Y. et al. Efficient mimics for elucidating zaxinone biology and promoting agricultural applications. Mol. Plant 13, 1654–1661 (2020).
Kountche, B. A. et al. Suicidal germination as a control strategy for Striga hermonthica (Benth.) in smallholder farms of sub-Saharan Africa. Plants People Planet 1, 107–118 (2019).
Calabrese, E. J. & Agathokleous, E. Smoke-water commonly induces hormetic dose responses in plants. Sci. Total Environ. 765, 142776 (2021).
Sripinyowanich, S. et al. Overexpression of a partial fragment of the salt-responsive gene OsNUC1 enhances salt adaptation in transgenic Arabidopsis thaliana and rice (Oryza sativa L.) during salt stress. Plant Sci. 213, 67–78 (2013).
Yoshida, S. Routine procedure for growing rice plants in culture solution. In Laboratory Manual for Physiological Studies of Rice 61–66 (International Rice Research Institute, 1976).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Metsalu, T. & Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 43, W566–W570 (2015).
Tian, T. et al. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129 (2017).
Supek, F., Bošnjak, M., Škunca, N. & Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800 (2011).
Bonnot, T., Gillard, M. & Nagel, D. A simple protocol for informative visualization of enriched gene ontology terms. Bio-Protoc. 9, e3429 (2019).
Thimm, O. et al. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939 (2004).
Pinit, S., Chadchawan, S. & Chaiwanon, J. A simple high-throughput protocol for the extraction and quantification of inorganic phosphate in rice leaves. Appl. Plant Sci. 8, e11395 (2020).
Alexieva, V., Sergiev, I., Mapelli, S. & Karanov, E. The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat: Effect of drought and UV on pea and wheat. Plant Cell Environ. 24, 1337–1344 (2001).
Hodges, D. M., DeLong, J. M., Forney, C. F. & Prange, R. K. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207, 604–611 (1999).
Zeng, Y. et al. Detection of sugar accumulation and expression levels of correlative key enzymes in winter wheat (Triticum aestivum) at low temperatures. Biosci. Biotechnol. Biochem. 75, 681–687 (2011).
Acknowledgements
The authors would like to thank Supachitra Chadchawan for advice on the project. This research was funded by Thailand Science Research and Innovation Fund Chulalongkorn University (BCG66230006) and National Research Council of Thailand (NRCT) (N42A640329) to J.C. and Science Achievement Scholarship of Thailand (SAST) scholarship to L.A.
Author information
Authors and Affiliations
Contributions
S.P. and J.C. designed experiments, analysed data, prepared figures and wrote the manuscript. S.P. and L.A. performed experiments. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pinit, S., Ariyakulkiat, L. & Chaiwanon, J. Rice straw-derived smoke water promotes rice root growth under phosphorus deficiency by modulating oxidative stress and photosynthetic gene expression. Sci Rep 13, 14802 (2023). https://doi.org/10.1038/s41598-023-41987-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-41987-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.