Abstract
Diffuse large B-cell lymphoma (DLBCL) is the most common subtype of lymphoma in dogs with a multicentric form. This study aimed to assemble 41 variants of the previously reported genes and to investigate these variants in canine DLBCL using the Agena MassARRAY platform. These variants were chosen based on the high prevalence observed in canine B- and T-cell lymphomas, their significance for target therapy, and compatibility for multiplex PCR amplification. Lymph node biopsy was performed from 60 dogs with B-cell lymphoma comprising 47 purebred and 13 crossbred dogs. All dogs presented single nucleotide polymorphisms (SNPs) at HYAL4 and SATB1 genes. The lesser mutual SNPs were observed at SEL1L, excluding a cocker spaniel, and c-Kit, with the exception of a pug and a French bulldog. Even though no statistical association was noted between each SNP and dog breed, purebreds were 3.88 times more likely to have a SNP at FLT3 rs852342480 (95%CI 0.50–45.03, p = 0.26), 3.64 times at TRAF3 F306X (95%CI 0.58–42.50, p = 0.43) and 2.66 times at TRAF3 E303EX (95%CI 0.56–13.12, p = 0.31). Also, DLBCL dogs (CHOP-based treatment) with c-Kit T425= had a poorer prognosis with shorter median overall survival times (OST) than dogs with the wild type. Dogs treated with COP chemotherapy and contained 3–5 variants at SEL1L were associated with decreased median OST. Therefore, this SNP’s lymphoma panel provides valuable information that we can use to outline a prognosis and develop a treatment plan for the targeted therapy of each dog.
Similar content being viewed by others
Introduction
Multicentric lymphoma is the most relevant anatomical form in dogs. Based on cellular morphology, cell lineage and topography, the most common subtypes of B-cell lymphoma in dogs are diffuse large B-cell lymphoma (DLBCL) and marginal zone lymphoma (MZL)1. Peripheral T-cell lymphoma (PTCL) and T-zone lymphoma (TZL) are frequently reported as T subtypes1. Both MZL and TZL are low-grade and have indolent behaviors that cause a low mortality rate2. In contrast, high-grade lymphomas such as DLBCL and PTCL are related to poorer prognoses and a shorter survival time3. The CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisolone) protocol is the standard-of-care in dogs with naïve intermediate- and high-grade lymphoma4; however, refractory and relapsed diseases present major barriers to successful treatment. Therefore, novel strategies such as immunotherapy and target drugs have been investigated to enhance the effectiveness of therapy in canine lymphomas5. An autologous heat shock protein peptide chaperone (HSPPC) vaccine was evaluated the efficacy of antitumor effect in dogs with B-cell lymphoma treated with CHOP-based protocol. Vaccinated dogs compared to placebo group showed promising results which prolonged time to progression and lymphoma-specific survival with no local or systemic adverse effects6,7. Anti-canine CD20 monoclonal antibody has also been developed for treating B-cell lymphomas. The generation of this chimeric antibody against CD20 showed good binding affinity to canine B cells and induced antibody-dependent cell-mediated cytotoxicity (ADCC) against canine neoplastic B cells in vitro and in vivo studies8,9,10. Small molecules such as NEMO-binding domain (NBD) peptide and NEDD-8 activating enzyme inhibitor (Pevonedistat) inhibited nuclear factor kappa-B (NF-kB) activity, commonly upregulated in B-cell lymphoma, and induced apoptosis of malignant B lymphocytes11,12. This might be useful for future clinical application in lymphoma dogs that have genetic mutation in the NF-kB pathway.
Gene expression analysis and next-generation sequencing technology have been used to investigate the potential risk genes and probable signaling pathways that may cause specific diseases and cancers. In humans, global gene expression profiling has indicated two subtypes of DLBCL: neoplastic cells that derive from germinal center B cells (GCB DLBCL) or those from post-germinal activated B cells (ABC DLBCL)13. The ABC subgroup is notably stimulated through B-cell receptors including many of the NF-kB target genes14, whereas genetic mutations in GCB DLBCL are frequently observed in chromatin modifiers and histone proteins such as KMT2D, MYD88, CARD11, EZH2 and CREBBP15,16. Interestingly, Richards et al.17 investigated gene expression profiling in canine B-cell lymphomas and found similarities to humans. Based on the immunohistochemistry and gene expression pattern, two distinct groups were classified as the ABC-like and the GCB-like DLBCL. The GCB-like group had higher expression of IRAK1BP1 and STAT4, while the ABC type had increased expression of NF-kB pathway genes. Furthermore, the canine ABC-like group had significantly poorer the progression-free and overall survival times compared to the GCB subgroup resembling with human DLBCL.
Several studies currently have used whole genome, whole exome and whole transcriptome (RNA-Seq) sequencing to explore the aberrant genes contributing to lymphomagenesis in specific dog breeds. Elvers et al.18 investigated the genetic risk factors in three lymphoma-predisposed breeds: boxers for T cells, cocker spaniels for B cells and golden retrievers for B and T cells. The authors found strong similarities between the mutations in both dog breeds with B-cell lymphoma, occurring in TRAF3-MAP3K14, FBXW7 and POT1. However, the boxer with T-cell lymphoma typically had mutations in the PTEN-mTOR pathway, which was dissimilar to the golden retriever with T-cell lymphoma that usually exhibited mutations in genes related to cellular metabolism. In addition to these findings, multiple somatic point mutations were identified in canine B-cell lymphoma including TRAF3, POT1, LMNB1 and MVB12A19,20,21. Recently, single nucleotide polymorphisms (SNPs) that affected SPAM1, HYAL4, HYLAP1, PTEN and SATB1 were noted in canine T-cell lymphomas22,23,24.
Even though fresh frozen tissue is more preferable for genetic mutation analysis due to conserving DNA quality, formalin-fixed paraffin embedded (FFPE) technique is a standard method in routine work in the pathology unit. The FFPE tissue could preserve the cellular morphology and keep at room temperature for several years. However, the fixation process usually causes DNA cross-linkage, degradation, and fragmentation which could interfere the accuracy of molecular studies especially RNA25. There are several studies have compared the DNA quality between fresh and FFPE specimens by using next generation sequencing. The results of mutation analysis from those two types of samples were comparable26. Moreover, FFPE tissues from dogs with call rate > 65% provided similar results of single nucleotide variation when compared to fresh whole blood samples27.
In the present study, we assembled a custom SNP panel of the previously reported genes that may drive lymphomagenesis in dogs. The criteria for variants selection were high prevalence in specific dog breeds, application for target therapy, and primer compatibility for multiplex polymerase chain reaction (PCR) with maximum ability of MassARRAY. This lymphoma SNP panel was then investigated in dogs with DLBCL, studying 47 purebred and 13 crossbred dogs, from archival FFPE samples. Beyond SNPs’ evaluation, the mutation genotyping panel was found to be different in each dog. This might be of relevance when it comes to outlining prognoses and selecting targeted therapies for affected dogs, by contributing toward increasing the treatment efficacy of personalized medicine.
Methods
Tissue samples and immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) tissues were obtained from the archive of the Department of Pathology, Faculty of Veterinary Science, Chulalongkorn University and SQ Reference Lab, China between 2011 and 2021. A statement to confirm that all methods were carried out in accordance with relevant guidelines, regulations and sampling procedures was approved by the Chulalongkorn University Animal Care and Use Committee.
All dogs had a history of generalized lymphadenopathy and were diagnosed as having nodal lymphomas based on the cytopathology and histopathology. Lymph node samples were collected at the time of presentation and before receiving chemotherapy. Immunostaining with CD3, CD20 and Pax5 and/or a clonality test were performed for lymphoma subtype classification following REAL/WHO (Revised European American Lymphoma/World Health Organization)1. The immunohistochemistry for CD20, CD3 and Pax5 was adapted from a previous study28. Briefly, tissue section was quenched endogenous peroxidase by 0.3% (v/v) H2O2 for 30 min and 5% bovine serum albumin was used for non-specific blocking for 20 min. Antigen was unmasked for CD3 and Pax5 by 10 mM citrate buffer (pH 6) in water bath at 95 °C for 20 min. Then, the slide was incubated with primary antibodies: CD20 (1:300, ab27093, Abcam, MA, USA), CD3 (1:10, PF. Moore, CA, USA), and Pax5 (1:100, 1EW, Leica, Newcastle Upon Tyne, UK) for 1.5 h. Secondary antibodies were applied for 30 min using EnVision system-HRP, mouse/rabbit (Dako, Glostrup, Denmark) for CD20 and Pax5 and ImmPRESS HRP, rat (Vector Laboratories Inc., CA, USA) for CD3. Visualization system for CD20 and Pax5 was DAB (Dako) and for CD3 was NovaRED (Vector Laboratories Inc.) as a substrate. Only DLBCL was included in this study. Demographic information was also recorded for each subject (Table 1). From 60 dogs, 26 dogs were treated with COP-based protocol (cyclophosphamide, vincristine, and prednisolone), 12 dogs received CHOP-based protocol, and the rest were loss of contact or death after diagnosis.
DNA extraction
Each FFPE block was shaved to 75–100 µm thickness in a sterile microcentrifuge tube. The samples were then deparaffinized with xylene and absolute ethanol, respectively, before genomic DNA extraction using a DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) following manufacturer’s instructions. The DNA concentration from each sample was quantified using a Nanodrop Lite Spectrophotometer (Thermo Scientific, MA, USA) and kept at -20 °C until use. The DNA samples were required to have a level of quality marked by at least a 260/280 absorbance ratio between 1.5 and 2.0 and concentration ≥ 5 ng/µl, when examined using a MassARRAY analyzer (Agena Bioscience, CA, USA).
Gene polymorphisms’ detection
Forty-one SNPs, in the mutated genes of dogs with lymphoma, were selected from previous studies (Table 2) based on the incidence in various dog breeds such as golden retriever, cocker spaniel, and mixed breed. The nucleotide locations were defined according to the sequence provided in the CanFam 3.1 reference genome (www.ensembl.org). The primer designed process was followed the manufacturer's online specific software for MassARRAY system, AgenaCx (Agena Bioscience). The compatibility of multiple primer sets were selected to avoid primer dimer and their concentrations with efficient amplification were used for the multiplex PCR. Each primer pair had repeatability from 85 to 100%. The multiplex PCR cocktail comprised 0.5 μL of PCR buffer (10X), 0.4 μL MgCl2 (25 mM), 0.1 μL dNTPs (25 mM), 0.2 μL PCR enzyme (5 U/μL), 1 μL amplification primer mix, 2 μL DNA (20 ng/μL) and HPLC-grade H2O to a total volume of 5 μL. The thermocycling conditions were 95 °C for 2 min followed by 45 cycles at 95 °C for 30 s, 56 °C for 30 s and 72 °C for 1 min, with a final incubation at 72 °C for 5 min.
To eliminate excess dNTPs from the previous step, 0.17 μL of 10X SAP buffer, 0.3 μL of SAP enzyme and 1.53 μL of HPLC-grade H2O were added to the step-one PCR products, to a total volume of 7 μL, and incubated in the thermal cycles of 37 °C for 40 min following by 85 °C for 5 min. The final step of the single-base extension reaction was performed with an IPLEX® Pro Reagent Kit (Agena Bioscience) to hybridize and elongate the extension primers at the nucleotide position of interest. For the single-base extension (ddNTPs), 0.2 μL IPLEX® Buffer (10X), 0.2 μL IPLEX® Terminator Mix (10X), 0.04 μL IPLEX® Pro Enzyme (33 U/μL) and 0.94 μL extension primers (0.58–1.21 μM) were mixed with the step-two products, and H2O was added to a total volume of 9 μL. The reaction was performed at 95 °C for 30 s, followed by five cycles of 95 °C for 5 s, 52 °C for 5 s and 80 °C for 5 s, for a total of 40 cycles, with a final extension at 72 °C for 3 min.
For desalination, 29 μL HPLC-grade H2O and 13 μL clean resin (96-well microplates) were added to the step-three extension products. Afterward, the supernatant was spotted onto a matrix-precoated SpectroCHIP® through a MassARRAY Nanodispenser and scanned using a MassARRAY Analyzer. The results were analyzed using MassARRAY Typer Software (v.4.1.8.3). The mutation was distinguished with TOF (time-of-flight) mass spectrometry on the basis of different molecular weights. The peaks in the mass spectrum were identified as mutations. Only those samples with a success rate greater than 80% were included in the analysis. Genotyping calls were viewed in call cluster plots, and peak intensities were reviewed in each respective sample spectrum. A SIFT (Sorting Intolerant From Tolerant) score from variant effector predictor in dog genome database was used to predict whether an amino acid substitution affects protein function (https://asia.ensembl.org/Multi/Tools/VEP).
Data analysis
Kaplan–Meier survival analysis was used to analyze overall survival times (OST) from sex, breed, and chemotherapeutic protocol using GraphPad Prism version 9.2.0 (GraphPad Software, CA, USA). The SNP variants were selected to predict the OST and separately compared between the mutant and wild-type dogs received COP and CHOP. The p-value of survival analysis was calculated by medians of a log-rank test. The odds ratio of SNP location and dog breeds was calculated by Fisher’s exact test. P < 0.05 was understood to represent statistical significance.
Results
Study dogs
There were 63 dogs diagnosed as having nodal DLBCL, but three cases were excluded as they had call rates of less than 80%. Of the 60 remaining dogs, there were 47 purebreds and 13 crossbreed dogs (Table 1). The majority of the purebreds were golden retrievers (GRs), Shih Tzus and Labrador retrievers (LRs). The mean age of the dogs was 9.45 years old. Twenty-six dogs were males, sixteen were females, nine dogs were castrated males and nine dogs were spayed females.
Gene polymorphisms’ detection
Most of the primer variants had an efficiency of more than 80%, except for FLT3 rs23257447 T > C (63.3%), MET c.3804 C > G (66.7%), PTEN rs397513087 C > T (68.3%), SEL1L 8:53818371 G > A (43.3%) and ZC3H7A c.2792 A > G (16.67%). All dog breeds with DLBCL had mutations at SATB1Q420P (c.1259 A > C, Gln420Pro) and HYAL4 (rs8499846, 14:11791385, 14:11794735 and 14:11807161) (Table 2 and Fig. 1). The lesser variants were observed at c-Kit T425 = (rs22299980 A > G, Thr425 =) and SEL1L (8:53778185, 8:53785948, rs24507594, 8:53818371 and rs24560262). The c-Kit wild type was only noted in two French bulldogs and two pugs, while wild-type SEL1L presented in one cocker spaniel. For the POT1 and TRAF3 mutations-i.e., the primary SNPs in canine DLBCL-four variants of POT1R284C, F308X, R583*, F643S (c.850 C > T, c.927 del, c.1747 C > T and c. 1928 C > T) were accounted for 3–13% of all dogs and five variants of TRAF3K284*, F306X, E303EX, ID323–324X, -316–317PKX (rs851689319, c.906del, c.908dup, c.968–971del and c.942-949dup) varied from 6–30% of all DLBCL cases (Table 2 and Fig. 2). In addition, when we compared the FLT3 and TRAF3 variants between dog breeds, purebreds tended to have these variants more than crossbreeds; the odds ratio for FLT3 rs852342480 was 3.88 (95%CI: 0.50–45.03, p = 0.26), for TRAF3F306X was 3.64 (95%CI 0.58–42.50, p = 0.43) and for TRAF3E303EX was 2.66 (95%CI 0.56–13.12, p = 0.31). No particular breeds were overrepresented in each SNP with any statistical significance.
Survival analysis
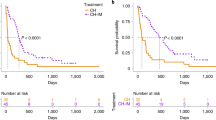
Among 38 dogs receiving chemotherapy, eight dogs were loss of contact after treatment and only 30 patients were included in our survival analysis. One dog was deceased from heat stroke and others were deceased from lymphoma. The median OST of sex (male vs. female) and breed (purebred vs. crossbred) were not statistical difference. However, the median OST of 20 dogs treating with COP was lower than 10 dogs treating with CHOP (79 days vs. 243 days, p = 0.006). The median OST was calculated and compared between the mutant (c-Kit, FLT3, POT1, TRAF3, HYAL4, SEL1L and SPAM1) and wild-type dogs in two different protocols as shown in Table 3. For CHOP group, dogs with the c-Kit mutation had decreased median OST (173 days) when compared to those with the wild type (461 days), with a statistically significant difference (p = 0.01, Fig. 3b). For COP, dogs accumulated with three to five SNPs on SEL11 had shorter median OST than dogs with ≤ 2 SNPs on SEL11 (37 days vs. 119 days, p = 0.03, Fig. 3a).
Kaplan–Meier survival analysis of overall survival time (OST) in DLBCL dogs treated with COP and CHOP protocols. (a) The median OST of COP-treated dogs with 3–5 variants on SEL1L was 37 days comparing to dogs with ≤ 2 variants on SEL1L (119 days, p = 0.03). (b) CHOP-treated dogs with mutant c-Kit had lower median OST than wild-type c-Kit. (173 and 461 days, respectively; p = 0.01).
Discussion
Our study developed a SNP panel of 41 locations and investigated in canine nodal DLBCL using a MassARRAY system. The MassARRAY panel could evaluate up to 40 variants from 192 samples in a single run and it is affordable in relation to other technologies available. The prevailing variants in DLBCL were SATB1 Q420P (96.67%), c-Kit rs22299980 (75.86%) and SEL1L 8:53778185 (74.58%). Among three locations, SATB1Q420P had a moderate impact on the protein function, with a SIFT score equal to 0.0 (deleterious). In contrast, a nonsense mutation of c-Kit had a low impact and an intron variant of SEL1L was a modifier.
Special AT-rich sequence-binding protein 1 (SATB1) is a global transcription regulator and chromatin organizer. SATB1 encodes a binding nuclear matrix protein that recruits the chromatin remodeling factor, to regulate the chromatin structure and gene expression. It regulates the gene expression in thymocytes and pre-B cells29. As mentioned in a previous report, this gene was mutated in 12–25% of T-cell lymphoma cases in boxers and golden retrievers18. Harris et al.22 also found that one non-boxer dog with PTCL had SNP at c. 1259 T > G in SATB1L420R. The author selected this SNP location followed aforementioned studies; however, different mutation (c. 1259 A > C) was used in this SNP panel. The polymorphisms of SATB1Q420P, Q420PX were observed in all purebred and mixed-breed dogs with DLBCL. In human cancers, this gene promotes tumor progression and metastasis in breast cancer, colorectal cancer and cutaneous T-cell lymphoma30,31,32. As the authors could not indicate the significance of mutated SATB1 and its expression in high-grade nodal lymphomas in dogs, further study is required to confirm the significant pathogenesis, and a target drug that could restore SATB1 function might be helpful for the treatment of canine lymphomas.
The c-Kit proto-oncogene is encoded as the transmembrane type-III receptor tyrosine kinase KIT, which is expressed in the hematopoietic progenitor of both myeloid and lymphoid cells. It plays a role in proliferation, cell survival and differentiation of hematopoietic precursors33,34. As c-Kit mutations are mainly reported in canine mast-cell tumors, few studies have investigated its mutation in canine leukemia and lymphoma. A nonsense mutation at codon 425 of exon 8 (rs22299980 A > G, Thr425 =) was observed in 73% (30/41) of dogs with acute myeloid leukemia35, and a similar polymorphism of c-Kit T425 = was described in mixed-breed dogs with lymphoma36. Although the c-Kit mutation was rarely reported in canine nodal lymphomas, we decided to choose the SNP location of the c-Kit gene and investigate in canine DLBCL with an increased sample size. Surprisingly, 44 dogs (75.86%) with DLBCL, from most breeds except French bulldogs and pugs, had mutated c-Kit. Synonymous variants of c-Kit T425 = were enriched in DLBCL, followed by frameshift variant of c-Kit T425TX (rs22299980 A > AG). However, the significance of the frameshift mutation in canine lymphomagenesis was undetermined in this study. Yamazaki et al.37 investigated the response rate of toceranib phosphate, one of the tyrosine kinase inhibitor (TKI) drugs used to treat unresectable high-grade mast-cell tumor with c-Kit mutation, in dogs with refractory T-cell lymphoma. The overall response rate of this monotherapy was only 40% in three dogs with partial remission and two dogs with stable disease. Therefore, the effectiveness and efficacy of TKI to treat canine lymphoma with c-Kit T425 = /T425TX require further investigation. In addition, c-Kit mutation has a significant prognosis in canine cutaneous mast cell tumors. Internal tandem duplication in exon 11 were approximately observed in 30–50% of higher grade mast cell tumors38. It has been associated with decreased survival times and progression-free survivals39,40. No evidence has reported on c-Kit mutation and its prognostic value in canine lymphoma. As such, its significance on prognosis needs further investigation in canine DLBCL.
The common SNP locations identified in canine DLBCL were POT1 and TRAF318. The telomere-binding protein protection of telomeres 1 (POT1), encoded by POT1, serves as providing telomere maintenance, and its dysfunction can lead to defective telomere replication, in turn, leading to genomic instability and enhanced carcinogenesis41. The adaptor protein TNF receptor associated factor 3 (TRAF3) is a tumor-suppressor gene that plays a critical role in B lymphocyte survival. A TRAF3 mutation leads to upregulation of the NF-kB pathway19. Both POT1 and TRAF3 mutations have been reported in human and canine B-cell lymphoma18,19,21. Elvers et al.18 found POT1 and TRAF3 mutations in 17–20% of B-cell lymphoma cases in both golden retrievers and cocker spaniels. Therefore, our study selected SNP mutations of the POT1 and TRAF3 genes, following Smith et al.21. Each dog with a POT1 mutation tended to have more than one enriched variant, with a similar observation in TRAF3. Therefore, the novel target drugs against POT1 and TRAF3 might have therapeutic potential for treating DLBCL in dogs.
As lymphoma-risked genes depend on hereditary or somatic mutations, the best way to determine the significant genes is in one population, such as in a predisposing breed like boxers or retriever dogs. Our study focused on the application of the MassARRAY technique to detect nominated lymphoma variants from a valuable database in various dog breeds with the DLBCL subtype. Thus, it was possible that the candidate SNPs were not discovered in some dogs or were discovered at lower rates than usual. Moreover, few primers of each SNP location in our study had an amplification efficiency lower than 60%, especially in ZC3H7AH931R. Although the true prevalence could not be determined in our study, nine out of ten dogs (90%) had a mutation in ZC3H7AH931R. The missense mutation of ZC3H7A was also reported in canine DLBCL, with predictable functional consequences20.
In summary, the SNPs panel of one variant/gene (C-kit, SATB1, TP53, MVB12A, ZC3H7A, ZNHIT6, MET, PTEN, HYALP1, MYC, and LMNB1), three variants/gene (FLT3 and SPAM1), four variants/gene (POT1 and HYAL4), five variants/gene (SEL1L), and 11 variants/gene (TRAF3) were determined in dogs with diffuse large B-cell lymphoma. Most of the variants could be detected in both purebred and mixed-breed dogs, excluding MYCS75F, TRAF3A531VX, L399X, D551X, T477X, VIDSQA322–327VX, MRGEY478–482I and LMNB1S307L. The application of the MassARRAY technique to discover SNPs provides informative data on each subject and could be used to identify a prognosis and develop a treatment strategy for each lymphoma dog.
References
Valli, V. E. et al. Classification of canine malignant lymphomas according to the World Health Organization criteria. Vet. Pathol. 48, 198–211. https://doi.org/10.1177/0300985810379428 (2011).
Valli, V. E., Vernau, W., de Lorimier, L. P., Graham, P. S. & Moore, P. F. Canine indolent nodular lymphoma. Vet. Pathol. 43, 241–256. https://doi.org/10.1354/vp.43-3-241 (2006).
Valli, V. E., Kass, P. H., San Myint, M. & Scott, F. Canine lymphomas: association of classification type, disease stage, tumor subtype, mitotic rate, and treatment with survival. Vet. Pathol. 50, 738–748. https://doi.org/10.1177/0300985813478210 (2013).
Vail, D. M., Thamm, D. H. & Liptak, J. M. in Withrow and MacEwen's Small Animal Clinical Oncology 688–772 (2019).
Thamm, D. H. Novel Treatments for Lymphoma. Vet. Clin. North Am. Small Anim. Pract. 49, 903–915. https://doi.org/10.1016/j.cvsm.2019.04.004 (2019).
Marconato, L. et al. Opportunities and challenges of active immunotherapy in dogs with B-cell lymphoma: a 5-year experience in two veterinary oncology centers. J. Immunother. Cancer 7, 146. https://doi.org/10.1186/s40425-019-0624-y (2019).
Marconato, L. et al. Randomized, placebo-controlled, double-blinded chemoimmunotherapy clinical trial in a pet dog model of diffuse large B-cell lymphoma. Clin. Cancer Res. 20, 668–677. https://doi.org/10.1158/1078-0432.CCR-13-2283 (2014).
Mizuno, T. et al. Generation of a canine anti-canine CD20 antibody for canine lymphoma treatment. Sci. Rep. 10, 11476. https://doi.org/10.1038/s41598-020-68470-9 (2020).
Rue, S. M. et al. Identification of a candidate therapeutic antibody for treatment of canine B-cell lymphoma. Vet. Immunol. Immunopathol. 164, 148–159. https://doi.org/10.1016/j.vetimm.2015.02.004 (2015).
Weiskopf, K. et al. Eradication of canine diffuse large B-cell lymphoma in a murine xenograft model with CD47 blockade and anti-CD20. Cancer Immunol. Res. 4, 1072–1087. https://doi.org/10.1158/2326-6066.CIR-16-0105 (2016).
Assumpcao, A., Lu, Z., Marlowe, K. W., Shaffer, K. S. & Pan, X. Targeting NEDD8-activating enzyme is a new approach to treat canine diffuse large B-cell lymphoma. Vet. Comp. Oncol. 16, 606–615. https://doi.org/10.1111/vco.12428 (2018).
Gaurnier-Hausser, A. & Mason, N. J. Assessment of canonical NF-kappaB activity in canine diffuse large B-cell lymphoma. Methods Mol. Biol. 1280, 469–504. https://doi.org/10.1007/978-1-4939-2422-6_29 (2015).
Alizadeh, A. A. et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511. https://doi.org/10.1038/35000501 (2000).
Wright, G. et al. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. U S A 100, 9991–9996. https://doi.org/10.1073/pnas.1732008100 (2003).
Schmitz, R. et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl. J. Med. 378, 1396–1407. https://doi.org/10.1056/NEJMoa1801445 (2018).
Zhang, J. et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat. Med. 21, 1190–1198. https://doi.org/10.1038/nm.3940 (2015).
Richards, K. L. et al. Gene profiling of canine B-cell lymphoma reveals germinal center and postgerminal center subtypes with different survival times, modeling human DLBCL. Cancer Res. 73, 5029–5039. https://doi.org/10.1158/0008-5472.CAN-12-3546 (2013).
Elvers, I. et al. Exome sequencing of lymphomas from three dog breeds reveals somatic mutation patterns reflecting genetic background. Genome Res. 25, 1634–1645. https://doi.org/10.1101/gr.194449.115 (2015).
Bushell, K. R. et al. Genetic inactivation of TRAF3 in canine and human B-cell lymphoma. Blood 125, 999–1005. https://doi.org/10.1182/blood-2014-10-602714 (2015).
Giannuzzi, D. et al. Mutational landscape of canine B-cell lymphoma profiled at single nucleotide resolution by RNA-seq. PLoS ONE 14, e0215154. https://doi.org/10.1371/journal.pone.0215154 (2019).
Smith, P. A. D., Waugh, E. M., Crichton, C., Jarrett, R. F. & Morris, J. S. The prevalence and characterisation of TRAF3 and POT1 mutations in canine B-cell lymphoma. Vet. J. 266, 105575. https://doi.org/10.1016/j.tvjl.2020.105575 (2020).
Harris, L. J. et al. Canine CD4+ T-cell lymphoma identified by flow cytometry exhibits a consistent histomorphology and gene expression profile. Vet. Comp. Oncol. 17, 253–264. https://doi.org/10.1111/vco.12460 (2019).
Labadie, J. D. et al. Genome-wide association analysis of canine T zone lymphoma identifies link to hypothyroidism and a shared association with mast-cell tumors. BMC Genomics 21, 464. https://doi.org/10.1186/s12864-020-06872-9 (2020).
McDonald, J. T. et al. Comparative oncology DNA sequencing of canine T cell lymphoma via human hotspot panel. Oncotarget 9, 22693–22702. https://doi.org/10.18632/oncotarget.25209 (2018).
Esteve-Codina, A. et al. A comparison of RNA-Seq results from paired formalin-fixed paraffin-embedded and fresh-frozen glioblastoma tissue samples. PLoS ONE 12, e0170632. https://doi.org/10.1371/journal.pone.0170632 (2017).
Gao, X. H. et al. Comparison of fresh frozen tissue with formalin-fixed paraffin-embedded tissue for mutation analysis using a multi-gene panel in patients with colorectal cancer. Front. Oncol. 10, 310. https://doi.org/10.3389/fonc.2020.00310 (2020).
Dear, J. D., Sykes, J. E. & Bannasch, D. L. Quality of DNA extracted from formalin-fixed, paraffin-embedded canine tissues. J. Vet. Diagn. Invest. 32, 556–559. https://doi.org/10.1177/1040638720929637 (2020).
Sirivisoot, S., Techangamsuwan, S., Tangkawattana, S. & Rungsipipat, A. Pax5 as a potential candidate marker for canine B-cell lymphoma. Vet. Med-Czech 62, 74–80. https://doi.org/10.17221/100/2016-Vetmed (2017).
Cai, S., Han, H. J. & Kohwi-Shigematsu, T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 34, 42–51. https://doi.org/10.1038/ng1146 (2003).
Han, H. J., Russo, J., Kohwi, Y. & Kohwi-Shigematsu, T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature 452, 187–193. https://doi.org/10.1038/nature06781 (2008).
Fredholm, S. et al. SATB1 in malignant T cells. J. Invest. Dermatol. 138, 1805–1815. https://doi.org/10.1016/j.jid.2018.03.1526 (2018).
Zhang, J. et al. SATB1 expression is associated with biologic behavior in colorectal carcinoma in vitro and in vivo. PLoS ONE 8, e47902. https://doi.org/10.1371/journal.pone.0047902 (2013).
Arber, D. A., Tamayo, R. & Weiss, L. M. Paraffin section detection of the c-kit gene product (CD117) in human tissues: value in the diagnosis of mast cell disorders. Hum. Pathol. 29, 498–504. https://doi.org/10.1016/s0046-8177(98)90066-1 (1998).
Nocka, K. et al. Expression of c-kit gene products in known cellular targets of W mutations in normal and W mutant mice–evidence for an impaired c-kit kinase in mutant mice. Genes Dev. 3, 816–826. https://doi.org/10.1101/gad.3.6.816 (1989).
Usher, S. G., Radford, A. D., Villiers, E. J. & Blackwood, L. RAS, FLT3, and C-KIT mutations in immunophenotyped canine leukemias. Exp. Hematol. 37, 65–77. https://doi.org/10.1016/j.exphem.2008.09.005 (2009).
Bóna, G. & Šiviková, K. Detection of mutations in selected proto-oncogenes of canine lymphoma. Folia Veterinaria 61, 34–39. https://doi.org/10.1515/fv-2017-0036 (2017).
Yamazaki, H. et al. Effects of toceranib phosphate (Palladia) monotherapy on multidrug resistant lymphoma in dogs. J. Vet. Med. Sci. 79, 1225–1229. https://doi.org/10.1292/jvms.16-0457 (2017).
Downing, S., Chien, M. B., Kass, P. H., Moore, P. E. & London, C. A. Prevalence and importance of internal tandem duplications in exons 11 and 12 of c-kit in mast cell tumors of dogs. Am. J. Vet. Res. 63, 1718–1723. https://doi.org/10.2460/ajvr.2002.63.1718 (2002).
Takeuchi, Y. et al. Validation of the prognostic value of histopathological grading or c-kit mutation in canine cutaneous mast cell tumours: a retrospective cohort study. Vet. J. 196, 492–498. https://doi.org/10.1016/j.tvjl.2012.11.018 (2013).
Webster, J. D. et al. The role of c-KIT in tumorigenesis: evaluation in canine cutaneous mast cell tumors. Neoplasia 8, 104–111. https://doi.org/10.1593/neo.05622 (2006).
Pinzaru, A. M. et al. Telomere Replication Stress Induced by POT1 Inactivation Accelerates Tumorigenesis. Cell Rep. 15, 2170–2184. https://doi.org/10.1016/j.celrep.2016.05.008 (2016).
Acknowledgements
S.S. is supported by the Ratchadapisek Somphot Fund for Postdoctoral Fellowships, Chulalongkorn University.
Funding
This research was funded by the Thailand Science Research and Innovation Fund, Chulalongkorn University (CU_FRB65_food(21)_185_31_04) and the Center of Excellence for Companion Animal Cancer, Faculty of Veterinary Science, Chulalongkorn University.
Author information
Authors and Affiliations
Contributions
S.S. contributed to the investigation, methodology, validation, formal analysis and writing, including review and editing the original draft. T.K., S.T. and Ay.R. contributed to the methodology, supervision, review and editing. K.C. and T.L. contributed to the methodology and resources. A.R. contributed to the conceptualization, funding acquisition, methodology, resources, supervision, review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sirivisoot, S., Kasantikul, T., Techangamsuwan, S. et al. Evaluation of 41 single nucleotide polymorphisms in canine diffuse large B-cell lymphomas using MassARRAY. Sci Rep 12, 5120 (2022). https://doi.org/10.1038/s41598-022-09112-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-09112-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.