Abstract
Carbapenems are broad-spectrum antibiotics widely used for the treatment of human infections caused by multidrug-resistant (MDR) Gram-negative bacteria. However, emerging carbapenemase-producing Enterobacterales (CPE) are rising as a public threat to human and animal health. We screened clinical bacterial isolates from 241 dogs and 18 cats hospitalized at Veterinary Medical Teaching Hospital, Seoul National University, from 2018 to 2020 for carbapenemase production. In our study, 5 strains of metallo-β-lactamase NDM-5-producing Escherichia coli and Klebsiella pneumoniae were isolated from 4 different dogs. Multilocus sequence typing (MLST) results showed that all E. coli strains were ST410 and all K. pneumoniae strains were ST378. Whole genome analysis of the plasmid showed that blaNDM-5 is carried on a IncX3 plasmid, showing a high concordance rate with plasmids detected worldwide in human and animal isolates. The blaNDM gene was associated with the bleMBL gene and the ISAba125 element, truncated with the IS5 element. The results of this study show that CPE has already become as a threat to both animals and humans in our society, posing the necessity to solve it in terms of "One Health". Therefore, preventive strategies should be developed to prevent the spread of CPE in animal and human societies.
Similar content being viewed by others
Introduction
Carbapenem is considered the last resort antibiotic for multidrug-resistant (MDR) Gram-negative bacteria. Carbapenemases produced by bacteria can hydrolase antibiotics containing β-lactam rings, including carbapenems, with even higher potential than extended-spectrum β-lactamases (ESBLs)1. Among the β-lactamases categorized into four Ambler classes of A-D, there are three classes to which carbapenemases belong, namely, class A, class B and class D2. Among class B carbapenemases, New Delhi metallo-β-lactamase (NDM) is known to be more effective than other groups and can be inhibited by metal chelators such as EDTA and mercaptopropionic acid3.
The increasing global spread of CPE, including Klebsiella pneumoniae and Escherichia coli, is considered a public threat to human and animal society, and these bacteria are listed by the World Health Organization (WHO) as priority 1 critical pathogens4. E. coli is a commensal bacterium colonizing the mucosal layer of the mammalian colon, including that of human infants, within a few hours after birth that can occasionally cause disease in immunocompromised hosts or in those with breached barriers of the gastrointestinal tract, including patients with peritonitis5. K. pneumoniae is an opportunistic pathogen causing pneumonia, sepsis, UTIs, bacteremia, meningitis and pyogenic liver abscesses, often in immunosuppressed patients via hospital infection6. K. pneumoniae resides not only in the environment but also on medical devices such as urinary catheters and endotracheal tubes and is frequently disseminated between health care workers and patients in hospitals7. Both E. coli and K. pneumoniae live alongside humans and animals and are emerging as threats as they gain resistance against antimicrobial agents, including resistance against carbapenems.
Since the first report of the isolation of a K. pneumoniae NDM-bearing strain from a patient in Sweden in 2009, NDM has spread worldwide due to its location on mobile genetic elements such as plasmids, transposons and integrons8. Conjugative plasmids such as Incompatibility group (Inc) F, A/C, L/M, N and X are associated with dissemination of blaNDM via horizontal gene transfer (HGT)2. In 2011, NDM-5 was first detected in E. coli from the United Kingdom with mutations of two amino acid positions of 88 (Val → Leu) and 154 (Met → Leu) from NDM-1 that resulted in increased action against carbapenems9. NDM-5 is disseminated worldwide and has been detected in various countries including Australia10, Denmark11, Italy12,13,14, Switzerland15, the Netherlands16, China17, India18, South Korea19, and the USA20.
Although prescription of carbapenem in animal medicine is prohibited in any part of the world by the World Organisation for Animal Health (OIE), the blaNDM-5 gene has been detected worldwide not only in companion animals, including dogs and cats15, 21, but also in domestic animals17, 22, 23. In South Korea, CPE were first reported among E. coli of companion animals in 2018 and reported as New Delhi metallo-β-lactamase-5 (NDM-5)21. Regarding the increasing threat of CPE, continuous surveillance and genetic characterization of CPE isolates have been required to develop the control measures against their spread in human and animal society.
Based on the current situation of CPE, Korean isolates of CPE from companion animals were characterized phenotypically and genotypically. Also, the genetic characteristics of the isolates were revealed by comparison with those from other countries.
Results
Profiles and antimicrobial susceptibility of the isolates
A total of 5 carbapenemase-producing strains were isolated from 520 isolates and identified as 3 E. coli strains and 2 K. pneumoniae strains. Four strains were isolated in 2019 and one (DMCPEC3) in 2020. Three strains were isolated from urine samples, and the others were isolated from ear swab samples (Fig. 1). According to hospital records, the likelihood of physical contact between patients was low. Notably, one E. coli strain (DMCPEC2) and one K. pneumoniae strain (DMCPKP4) were isolated from different urine samples from the same dog. Although the dog was suffering from cystitis and receiving antibiotic treatment with enrofloxacin and amoxicillin/clavulanic acid, it did not show any improvement of clinical signs. The other strain of K. pneumoniae (DMCPKP1) was isolated from urine specimen of a dog patient with increased urine volume and odor. The patient was treated with amoxicillin/clavulanic acid and enrofloxacin but showed no improvement. The other 2 E. coli isolates (DMCPEC3, DMCPEC7) were isolated from ear swab samples of 2 different dogs. Interestingly, the two dogs had previously been treated in the same facility in Seoul, but the timing of each individual's visit was unknown. DMCPEC3 was isolated from a dog with ear pruritus in 2020, but has never been applied with antibiotics in our facility. DMCPEC7 was isolated from ear purulent exudate of other dog patient, which was treated with enrofloxacin.
General information of the NDM-5-producing Enterobacterales strains based on patient history, PFGE and MLST data. PFGE patterns were generated and analyzed in Dice similarity coefficient with Unweighted Pair-Group Method with Arithmetic Mean (UPGMA) dendrogram via BioNumerics, version 6.6 (Applied Maths NV, Belgium).
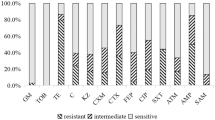
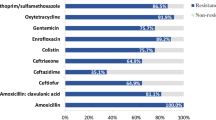
All 5 isolates showed resistance not only to carbapenems, including imipenem (> 256 µg/ml), meropenem (32–128 µg/ml), and ertapenem (32–64 µg/ml) (Table 1), but also to various other antibiotics. Resistant phenotypes included resistance to cefotaxime, ceftazidime, ceftriaxone, gentamicin, ampicillin, levofloxacin, norfloxacin, ofloxacin, nalidicic acid and tetracycline. However, all isolates were susceptible to polymyxin B and colistin. As for aztreonam and tobramycin, E. coli strains appeared susceptible to intermediate, while K. pneumoniae strains showed resistance. The resistant phenotypes were determined referring to the CLSI clinical interpretation breakpoints.
PCR amplification followed by sequencing revealed that all 5 strains of carbapenemase-harboring strains produced NDM-5, with two amino acid mutations at positions 88 (Val to Leu) and 154 (Met to Leu), regardless of bacterial species or source of isolation. The other four major carbapenemase (blaKPC, blaVIM, blaIMP, and blaOXA-48) markers were not detected in all isolates.
Genotypic relatedness according to MLST and PFGE
The genotypic relationships between isolated strains determined with MLST revealed that all E. coli strains belonged to ST410 and all K. pneumoniae strains belonged to ST378. The epidemiological similarity results between PFGE indicated high relatedness of the E. coli strains (> 93%) and the K. pneumoniae strains (99%).
Conjugation transferability and plasmid analysis
Conjugation assays confirmed the transferability of the blaNDM-5 gene in broth mating at frequencies of 1.0 × 10−4 to 1.0 × 10−5 (Table 2). All transconjugants were confirmed by PCR identification. In conjugation assay, transferability was proven not only in E. coli but also in the case where K. pneumoniae strains contributed as donors.
Complete sequences of all five NDM-5 harboring plasmids were obtained using the Illumina MiSeq Sequencing System. Plasmid sequencing and PCR replicon typing results identified all isolated plasmids as IncX3, with lengths varying from 45–46 kb, GC contents varying from 46.5 to 46.91% and 60–61 CDSs (Table 2). Downstream of blaNDM-5, the bleMBL gene encoding the bleomycin-resistant protein, the trpF gene encoding phosphoribosylanthranilate isomerase, the dsbC gene for oxidoreductase, the cutA gene fragment encoding the periplasmic divalent cation tolerance protein inserted by the IS26 element and the truncated umuD gene encoding a mutagenesis protein were identified.
Comparative plasmid genome analysis
The genetic load regions and the whole circular sequences of the five plasmids were compared with three reference IncX3 plasmids from three different countries (pNDM-5_A0917122 from South Korea, pNDM_MGR194 from India and pNDM5-SCNJ1 from China) (Figs. 2, 3). pEC2-NDM5 showed high identity and similar gene features with two reference plasmids, pNDM-5_A0917122 and pNDM_MGR194, isolated from our country and India, respectively. pNDM-5_A0917122 was the plasmid isolated from companion animal of South Korea, while pNDM_MGR194 was isolated from human patient of India. On the other hand, the middle parts of transposon IS26 in pEC3-NDM5 and pEC7-NDM5 were lost over a length of 567 base pairs. Plasmids from K. pneumoniae showed higher identity with pNDM5-SCNJ1 from human sputum of China, which was also detected from isolates of K. pneumoniae. The ISAba125 region was previously found to be truncated (166 bp upstream from the blaNDM-5 start codon) by the IS5 element, with 67 base pairs of remnants between the blaNDM-5 and IS5 elements26. While the ISAba125 region between the IS5 element and IS3000 element was found to be shortened by 112 base pairs in pNDM5-SCNJ1 compared with other IncX3 plasmids, the same gene area of K. pneumoniae in our study was completely lost. Two plasmids from K. pneumoniae strains, pKP1-NDM5 and pKP4-NDM5, showed 100% identity to each other.
Schematic map of comparative circular genome structure analysis of 8 IncX3 plasmids. Circular maps were used to illustrate and compare the backbone and the location of the genetic load region of plasmids. GC skew was featured based on data of pKP4-NDM5. Genome alignments were performed by Mauve24, and the circular map was generated with CIRCOS (http://circos.ca/).
Genetic load sequence context of IncX3 blaNDM–5 plasmids. Genes are denoted by arrowheads and colored based on class of gene function, sorting by replication, transposon, antimicrobial resistance genes and plasmid backbone elements. Gray shades denote shared regions with a high degree of homology. Easyfig 2.2.3 (https://github.com/mjsull/Easyfig/wiki) was used for this pairwise BLASTn alignment comparing analysis25. The accession numbers were: pNDM_MGR194 (KF220657.1); pNDM-5_A0917122 (MH094148); pEC2-NDM5 (MW415440); pEC3-NDM5 (MW415441); pEC7-NDM5 (MW415442); pNDM5-SCNJ1 (MK715437.1); pKP1-NDM5 (MW415443); pKP4-NDM5 (MW415444).
Discussion
This investigation identified Enterobacterales bearing NDM-5-producing IncX3 from companion animals. This was the first report in our country of K. pneumoniae as a carbapenemase producer from companion animals, whereas the presence of carbapenemase-producing E. coli was reported previously in our country21. Of note, two different isolates, one E. coli (DMCPEC2) and one K. pneumoniae (DMCPKP4) came from different urine samples collected from a single dog at different times. Their identical carbapenemase and plasmid features (99.85% identity) likely indicate the possibility of bacterial interspecies horizontal transfer of genetic elements inside the host, or on medical devices such as urinary catheter. Sporadic dissemination among companion animals or in animal hospitals is suspected, regarding the identical carbapenemase gene profiles, the identical genetic environment structures, the same MLST types and PFGE patterns showing the same pulsotype (> 93% along E. coli strains and > 99% along K. pneumoniae strains), and the genetic environment structure (ISAba125-blaNDM-5-bleMBL-trpF-dsbC-cutA-umuD).
Plasmids can transport multiple antibiotic resistance genes (ARGs) through conjugation between heterogeneous species as well as the identical species, which makes plasmids crucial for bacterial colonization and virulence potential27. Incompatibility group IncX3 plays a major role in dissemination of antibiotic resistance genes and is known to harbor blaNDM-1 and blaNDM-5 rather than other blaNDM variants2. IncX3 plasmids carrying various carbapenemase genes seem to be disseminated worldwide, mainly in China28. A recent investigation performed in China revealed that the IncX3 plasmid harboring blaNDM-5 is disseminated among Enterobacterales in both humans and animals22, 29. In animals, IncX3 plasmids carrying blaNDM have been detected in E. coli from swine and chickens in various regions in China17, 30. The identification of IncX3 from China and South Korea is not a surprise, considering the global dissemination of these plasmids. Among carbapenemase-harboring plasmids, the IncX3 plasmid is also dominant in South Korea, showing dissemination within diverse bacterial species, followed by IncFII and IncA/C19. blaNDM has been suggested to be disseminated from Acinetobacter baumannii to Enterobacterales via the mobile gene element ISAba12531. All five isolates in our study support this idea, with association of transposon elements of the upstream ISAba125 element. As described and predicted previously, the ISAba125 element was lost in K. pneumoniae strains examined in our study via horizontal transfer or host response mechanisms29.
E. coli ST410 is known for interspecies transmission along people, environment, wildlife and companion animals32, 33. Additional isolation of blaNDM-5 from E. coli ST410 in our country is posing threat of human infection from companion animal or vice versa. A recent study from Denmark suggested E. coli ST410 as a new high-risk clone, causing recurrent infections such as bloodstream infections and carrying carbapenemases such as NDM-5 or OXA-18134. E. coli ST410 is reported as NDM-5 producer in various countries worldwide, including China35, the United Kingdom36 and South Korea21. In South Korea, ST410 is the 3rd most dominant NDM-producing clinical E. coli strain, which includes reports from companion animals19, 21, 37. Therefore, detailed investigation to discover the role and dissemination of E. coli ST410 in our country is necessary. To prevent public outbreak, infection control across people, environment, wildlife and companion animal based on One Health Approach is needed.
K. pneumoniae ST378 has never been reported as CPE, even though NDM-5-producing K. pneumoniae has been reported worldwide from various strains16, 23, 38,39,40,41,42. While the predominant NDM-producing clinical K. pneumoniae strain is ST1061 in South Korea19, ST378 was reported as a common sequence type among ESBL- and/or AmpC β-lactamase-producing clinical K. pneumoniae isolates in Taiwan43. However, information on these clones of ST378 is still scarce, and further monitoring investigations are needed to avoid additional dissemination in our country. Considering that the plasmid of this new strain is similar to the previously described ones, it should be considered as additional evidence of horizontal spread of carbapenemase harboring IncX3 plasmids between Enterobacterales, discovered from companion dog hosts.
While the transmission route of these CPE isolates is still unclear, there are several possible hypotheses. First is circulation inside veterinary teaching hospital environment, considering genetic similarity between CPE strains and their urinary tract infection (possibly urinary catheter infection). Transmission via physical contact of companion animals is also considerable, regarding that MLST types of carbapenemase-harboring E. coli isolates discovered in companion animals in our country are identical, as ST41021. Also considering that these isolates were from a tertiary referral hospital in South Korea, it could be indicating unauthorized usage of carbapenems in local veterinary hospitals in our country. Interestingly, two of the host dogs had previously visited the same animal hospital, but it was not known whether they had been in contact, and this information alone could not reach a definite conclusion. Lastly, it could be acquired from contaminated feed or drinking water, or from contact with their human owners.
In this study, the CPE strains were isolated from clinical lesions, and each strain caused ear and urinary tract clinical signs in the host dog. Therefore, the possibility of these strains to be infected and become human clinical pathogens should be considered. Also, bacterial screening of human owner is required to figure out the possibility of human-to-animal transmission, or vice versa.
In conclusion, additional emergence of CPE in this study shows the dissemination of carbapenemase in our society, which is already a public concern considering the forbidden usage of carbapenems in animals. Therefore, further investigation is necessary to unveil the role of IncX3 plasmids carrying blaNDM-5 and the evidence of transmission between human owners and companion animals.
Methods
Bacterial strain identification and carbapenemase gene detection
Specimens, urine and swabs of skin and ears, were collected from suspected of being infected companion dogs and cats hospitalized at Veterinary Medical Teaching Hospital, Seoul National University, from 2018 to 2020. The specimens were collected by professional veterinarians in accordance with the Guide for the Care and Use of Laboratory Animals and the Animal Welfare Act. From the collected specimens, a total of 520 clinical isolates from 241 dogs and 18 cats were isolated using sheep blood agar (Hangang, Gyeonggi, Korea) for the diagnostic isolation and stored in Tryptic Soy Broth (Thermo Fisher Scientific Oxoid Ltd., Basingstoke, UK) with 50% glycerol at − 70 °C for further epidemiological studies. The stored clinical specimens were subject to screening for carbapenem resistance on CHROMagar mSuperCARBA agar (CHROMagar). Total DNA of surviving isolates was purified by a Wizard Genomic DNA Purification Kit (Promega, Madison, WI) and amplified using PCR specific primers detecting 5 widespread carbapenemase genes (blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA-48) using previously designed primers44,45,46,47. Sequences were identified by the Sanger sequencing method and compared with GenBank data (www.ncbi.nlm.nih.gov/GenBank) with the Basic Local Alignment Search Tool (BLAST) network service. Microbial species were confirmed with matrix-assisted laser desorption ionization–time of flight-mass spectrometry (MALDI–TOF–MS; Bruker Daltonik GmbH, Bremen, Germany) and further confirmed using 16S rRNA sequencing with the primer pair 27F/1492R.
In vitro antibiotic susceptibility testing
Minimum inhibitory concentrations (MIC) of the isolates were determined for 35 antimicrobial agents: imipenem, meropenem, ertapenem, cephradine, cefotaxime, ceftazidime, ceftriaxone, cefepime, amikacin, gentamicin, kanamycin, kanamycin B, neomycin, streptomycin, tobramycin, ampicillin, amoxicillin, chloramphenicol, florfenicol, trimethoprim, enrofloxacin, levofloxacin, norfloxacin, ofloxacin, polymyxin B, colistin, erythromycin, aztreonam, penicillin G, oxacillin, nalidixic acid, sulfamethoxazole, tetracycline, tigecycline and azlocillin. Either broth microdilution method or Etest (bioMérieux, Marcy L'Etoile, France) strip method on Mueller–Hinton agar (Difco Laboratories, Detroit, MI) were applied depending on availability of antibiotics. E-test methods were used for 8 antimicrobial agents: ceftazidime, ceftriaxone, cefepime, gentamicin, tobramycin, colistin, aztreonam, tigecycline. The others were tested via broth microdilution method. Both methods were performed and interpreted following the recommendation of the Clinical and Laboratory Standards Institute (CLSI) interpretation criteria48. E. coli strain ATCC 25922 was used as a quality control strain.
Pulse-field gel electrophoresis and multilocus sequence typing
The genetic relationships between isolated strains were analyzed via pulse-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). As recommended in the CDC’s protocol for E. coli, agarose plugs containing genomic DNA of the three E. coli and two K. pneumoniae isolates were digested with XbaI restriction enzyme and separated for 18 h at 14 °C using a CHEF-Mapper PFGE system at 6 V/cm (Bio-Rad, Hercules, CA)49. A lambda ladder (Bio-Rad) was used as a DNA size marker. PFGE patterns were then analyzed, and a Dice similarity coefficient with the unweighted pair-group method with arithmetic mean (UPGMA) dendrogram was generated using BioNumerics, version 6.6 (Applied Maths NV, Belgium). MLST of E. coli was performed using seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) following a standardized protocol as previously described and assigned using an online database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli)50. MLST of K. pneumoniae was also performed using seven housekeeping genes (rpoB, gapA, mdh, pgi, phoE, infB, and tonB), and assignment was based on an online database for K. pneumoniae (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html)51.
Conjugation assay
A conjugation assay was performed for all of the NDM-harboring isolates with sodium azide-resistant E. coli J53 as the recipient strain in a 1:1 ratio in broth. Each 2 mL culture of donor and recipient cells in the logarithmic phase was resuspended in fresh trypticase soy broth (TSB) and mixed before overnight incubation at 30 °C without agitation. Transconjugants were selected on trypticase soy agar (TSA) plates containing both sodium azide (100 μg/ml; Sigma Chemical Co., St. Louis, Mo.) and meropenem (1 μg/ml: Sigma-Aldrich, St. Louis, MO). Species identifications were confirmed using a MALDI-TOF MS Biotyper and 16S rRNA sequencing. The presence of blaNDM genes was confirmed by PCR analysis. The conjugation transfer frequency of carbapenemase-producing genes was expressed as transconjugants per donor cell (T/D) following methods previously described52.
Plasmid sequencing and mapping
Plasmid DNA was isolated using a QIAGEN Plasmid Mini Kit (QIAGEN, Germany) and sequenced with the Illumina MiSeq Sequencing System (Illumina, San Diego, CA, USA), generating 300 bp paired-end reads (1 Gbp per sample). FastQC (v.0.11.5) was used for sequence quality analysis, after which the sequences were filtered and trimmed using the program Trimmomatic (v.0.36). SPAdes (v3.13.0) software (https://github.com/ablab/spades)53 was utilized for de novo assembly, and Prokka (v.1.10) was used for annotation. To identify antibiotic resistance genes, annotation of coding sequences (CDSs) was performed with bioinformatic tools including the ARG-ANNOT (Antibiotic Resistance Gene-ANNOTation) database, the NCBI Prokaryotic Genome Annotation Pipeline (www.ncbi.nlm.nih.gov/books/NBK174280), and ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/). For comparison, three other related plasmids, namely, pNDM-5_A0917122 (companion animal isolate from South Korea, accession number MH094148), pNDM_MGR194 (human isolate from India, accession number KF220657.1), and pNDM5-SCNJ1 (human isolate from China, accession number MK715437.1), were aligned and interpreted with the BLAST network service. Multiple plasmid alignments were performed by Mauve24, and the circular maps of plasmid were generated using CIRCOS (http://circos.ca). Plasmid mapping for genetic load sequence was performed and visualized using Easyfig version 2.2.3 (https://github.com/mjsull/Easyfig/wiki) software25.
Incompatibility typing of the blaNDM-5 plasmid was additionally confirmed from plasmid sequencing results in silico by PCR-based replicon typing54, 55. Plasmid nucleotide sequences have been deposited in GenBank with the following accession nos.: MW415440 (pEC2-NDM5), MW415441 (pEC3-NDM5), MW415442 (pEC7-NDM5), MW415443 (pKP1-NDM5), and MW415444 (pKP4-NDM5).
Data availability
Publicly available datasets were analyzed in this study. This data can be found in Table 2 for all accession numbers. All data generated or analyzed during this study have been submitted with this manuscript. All genetic information of the plasmids was deposited in GenBank. Therefore, all data from this study are available publically.
References
Nordmann, P., Naas, T. & Poirel, L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17, 1791 (2011).
Kopotsa, K., Osei Sekyere, J. & Mbelle, N. M. Plasmid evolution in carbapenemase-producing Enterobacteriaceae: A review. Ann. N Y Acad. Sci. 1457, 61–91. https://doi.org/10.1111/nyas.14223 (2019).
Somboro, A. M., Sekyere, J. O., Amoako, D. G., Essack, S. Y. & Bester, L. A. Diversity and proliferation of metallo-β-lactamases: a clarion call for clinically effective metallo-β-lactamase inhibitors. Appl. Environ. Microbiol 84, e00698-e1618 (2018).
Tacconelli, E. et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327 (2018).
Kaper, J. B., Nataro, J. P. & Mobley, H. L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140 (2004).
Paczosa, M. K. & Mecsas, J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661 (2016).
Rock, C. et al. Frequency of Klebsiella pneumoniae carbapenemase (KPC) and non-KPC-producing Klebsiella contamination of Healthcare workers and the environment. Infect. Control Hosp. Epidemiol. 35, 426 (2014).
Yong, D. et al. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother 53, 5046–5054 (2009).
Hornsey, M., Phee, L. & Wareham, D. W. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother 55, 5952–5954 (2011).
Wailan, A. M., Paterson, D. L., Caffery, M., Sowden, D. & Sidjabat, H. E. Draft genome sequence of NDM-5-producing Escherichia coli sequence type 648 and genetic context of blaNDM-5 in Australia. Genome Announc. 3 (2015).
Hammerum, A. M., Littauer, P. & Hansen, F. Detection of Klebsiella pneumoniae co-producing NDM-7 and OXA-181, Escherichia coli producing NDM-5 and Acinetobacter baumannii producing OXA-23 in a single patient. Int. J. Antimicrob. Agents 5, 597–598 (2015).
Piazza, A. et al. Detection of ST1702 Escherichia coli blaNDM-5 and blaCMY-42 genes positive isolates from a Northern Italian hospital. New Microbiol. 41, 230–231 (2018).
Giufrè, M. et al. Emergence of NDM-5-producing Escherichia coli sequence type 167 clone in Italy. Int. J. Antimicrob. Agents 52, 76–81 (2018).
Alba, P. et al. Carbapenemase IncF-borne blaNDM-5 gene in the E. coli ST167 high-risk clone from canine clinical infection Italy. Vet. Microbiol. 256, 9045 (2021).
Peterhans, S. et al. First report of a blaNDM-5-harbouring Escherichia coli ST167 isolated from a wound infection in a dog in Switzerland. J. Glob. Antimicrob. Resist. 15, 226–227 (2018).
Bathoorn, E., Rossen, J. W., Lokate, M., Friedrich, A. W. & Hammerum, A. M. Isolation of an NDM-5-producing ST16 Klebsiella pneumoniae from a Dutch patient without travel history abroad, August 2015. Euro Surveill. 20, 30040 (2015).
Ho, P.-L. et al. IncX3 epidemic plasmid carrying blaNDM-5 in Escherichia coli from swine in multiple geographic areas in China. Antimicrob. Agents Chemother. 62, 1 (2018).
Krishnaraju, M. et al. Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J. Med. Microbiol. 33, 30 (2015).
Yoon, E.-J. et al. New Delhi metallo-beta-lactamase-producing Enterobacteriaceae in South Korea between 2010 and 2015. Front. Microbiol. 9, 571 (2018).
de Man, T. J., Perry, K. A., Avillan, J. J., Rasheed, J. K. & Limbago, B. M. Draft genome sequence of a New Delhi metallo-β-lactamase-5 (NDM-5)-producing multidrug-resistant Escherichia coli isolate. Genome Announc. 3 (2015).
Hong, J. S. et al. First detection of New Delhi metallo-β-Lactamase-5-producing Escherichia coli from companion animals in Korea. Microb. Drug Resist. 25, 344–349 (2019).
Liu, Z. et al. Emergence of IncX3 plasmid-harbouring blaNDM-5 dominated by Escherichia coli ST48 in a goose farm in Jiangsu, China. Front. Microbiol. 10, 2002 (2019).
He, T. et al. Occurrence and characterization of blaNDM-5-positive Klebsiella pneumoniae isolates from dairy cows in Jiangsu China. J. Antimicrob. Chemother. 72, 90–94 (2016).
Darling, A. E., Mau, B. & Perna, N. T. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS one 5, e11147 (2010).
Sullivan, M. J., Petty, N. K. & Beatson, S. A. Easyfig: A genome comparison visualizer. Bioinformatics 27, 1009–1010 (2011).
Yuan, Y. et al. blaNDM-5 carried by a hypervirulent Klebsiella pneumoniae with sequence type 29. Antimicrob. Resist. Infect. Control 8, 1–9 (2019).
Cottell, J. L. et al. Complete sequence and molecular epidemiology of IncK epidemic plasmid encoding blaCTX-M-14. Emerg. Infect. Dis. 17, 645 (2011).
Mouftah, S. F. et al. Epidemic IncX3 plasmids spreading carbapenemase genes in the United Arab Emirates and worldwide. Infect. Drug Resist. 12, 1729 (2019).
Tian, D. et al. Dissemination of the blaNDM-5 Gene via IncX3-Type Plasmid among Enterobacteriaceae in Children. Msphere 5, 1 (2020).
Liu, Z. et al. Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a sequence type 48 Escherichia coli strain. Antimicrob. Agents Chemother. 61, 1 (2017).
Poirel, L. et al. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 1087–1089 (2012).
Schaufler, K. et al. Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410—another successful pandemic clone?. FEMS Microbiol. Ecol. 92, 1 (2016).
Falgenhauer, L. et al. Circulation of clonal populations of fluoroquinolone-resistant CTX-M-15-producing Escherichia coli ST410 in humans and animals in Germany. Int. J. Antimicrob. Agents 47, 457–465 (2016).
Roer, L. et al. Escherichia coli sequence type 410 is causing new international high-risk clones. Msphere 3, e00337-e1318 (2018).
Li, X. et al. Dissemination of blaNDM-5 gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob. Resist. Infect. Control 7, 59 (2018).
Reynolds, M. et al. Occurrence and characterization of Escherichia coli ST410 co-harbouring blaNDM-5, blaCMY-42 and blaTEM-190 in a dog from the UK. J. Antimicrob. Chemother. 74, 1207–1211 (2019).
Baek, J. Y. et al. Plasmid analysis of Escherichia coli isolates from South Korea co-producing NDM-5 and OXA-181 carbapenemases. Plasmid 104, 102417 (2019).
Balm, M. et al. Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clin. Microbiol. Infect. 19, E421–E423 (2013).
Shin, J., Baek, J. Y., Chung, D. R. & Ko, K. S. Instability of the IncFII-type plasmid carrying blaNDM-5 in a Klebsiella pneumoniae isolate. J. Microbiol. Biotechnol. 27, 1711–1715 (2017).
Liu, P.-p., Liu, Y., Wang, L.-h., Wei, D.-d. & Wan, L.-G. Draft genome sequence of an NDM-5-producing Klebsiella pneumoniae sequence type 14 strain of serotype K2. Genome Announc. 4 (2016).
Khalifa, H. O., Soliman, A. M., Ahmed, A. M., Shimamoto, T. & Shimamoto, T. NDM-4-and NDM-5-producing Klebsiella pneumoniae coinfection in a 6-month-old infant. Antimicrob. Agents Chemother. 60, 4416–4417 (2016).
Wang, Z. et al. Outbreak of blaNDM-5-Harboring Klebsiella pneumoniae ST290 in a Tertiary Hospital in China. Microb. Drug Resist. 25, 1443–1448 (2019).
Lin, W.-P. et al. The antimicrobial susceptibility of Klebsiella pneumoniae from community settings in Taiwan, a trend analysis. Sci. Rep. 6, 36280 (2016).
Aktaş, Z. et al. Carbapenem-hydrolyzing oxacillinase, OXA-48, persists in Klebsiella pneumoniae in Istanbul Turkey. Chemotherapy 54, 101–106 (2008).
Poirel, L., Walsh, T. R., Cuvillier, V. & Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123 (2011).
Doyle, D. et al. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J. Clin. Microbiol. 50, 3877–3880 (2012).
Woodford, N. et al. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A β-lactamase, KPC-3, in a New York medical center. Antimicrob. Agents Chemother. 48, 4793–4799 (2004).
Clinical & Institute, L. S. (Clinical and Laboratory Standards Institute Wayne, PA, 2017).
Control, C. f. D. & Prevention. Standard operating procedure for PulseNet PFGE of Escherichia coli O157: H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. Centers for Disease Control and Prevention, Atlanta (2013).
Wirth, T. et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151 (2006).
Diancourt, L., Passet, V., Verhoef, J., Grimont, P. A. & Brisse, S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43, 4178–4182 (2005).
Zhong, X., Droesch, J., Fox, R., Top, E. M. & Krone, S. M. On the meaning and estimation of plasmid transfer rates for surface-associated and well-mixed bacterial populations. J. Theor. Biol. 294, 144–152 (2012).
Bankevich, A. et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Carattoli, A. et al. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228 (2005).
Johnson, T. J. et al. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68, 43–50 (2012).
Acknowledgements
Authors thank to staffs helping the collection of specimen at Veterinary Medical Teaching Hospital, Seoul National University.
Funding
This study was supported by National Research Foundation (NRF- 2020R1A2C2008794), BK21 FOUR Future Veterinary Medicine Leading Education and Research Center and Research Institute for Veterinary Science, Seoul National University, Seoul, Republic of Korea.
Author information
Authors and Affiliations
Contributions
S.M.K., S.S. and H.S.Y. conceived and designed the experiments. N.L. and C.H. maintained the collections. S.M.K., S.W.C. and Y.B.L. performed the experiments. S.M.K., D.K. and J.L. analyzed the data. S.M.K., S.K. and H.S.Y. corrected and discussed. S.M.K. wrote the paper. H.S.Y. reviewed and edited the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kyung, S.M., Choi, SW., Lim, J. et al. Comparative genomic analysis of plasmids encoding metallo-β-lactamase NDM-5 in Enterobacterales Korean isolates from companion dogs. Sci Rep 12, 1569 (2022). https://doi.org/10.1038/s41598-022-05585-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-05585-1
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.