Abstract
This study analyzed the association of functionally significant SNPs of matrix metalloproteinase (MMP) genes in the development of peptic ulcer disease (PUD) in Caucasians from Central Russia. Ten SNPs of the MMP-1, MMP-2, MMP-3, MMP-8, and MMP-9 genes were analyzed for association with PUD in a cohort of 798 patients with PUD (including 404 H. pylori-positive and 394 H. pylori-negative) and 347 H. pylori-negative controls using logistic regression and assuming the additive, recessive, and dominant genetic models. The variants of MMP-1, MMP-2, MMP-3, and MMP-8 did not manifest any significant associations with the diseases. Five SNPs of the MMP-9 gene demonstrated such association. Allele G of the rs17576 MMP-9 locus conferred a higher risk for PUD (ORadj = 1.31, pperm = 0.016), haplotype AACG of loci rs17576-rs3787268-rs2250889-rs17577 of the MMP-9 gene decreased risk for PUD (ORadj = 0.17, pperm = 0.003). Also, allele C of rs3918249, allele G of rs17576 and haplotype CG of rs3918249-rs17576 of the MMP-9 gene increased risk for H. pylori-positive PUD (ORadj = 1.82, pperm = 0.002; ORadj = 1.53–1.95 pperm = 0.001–0.013 and ORadj = 1.49 pperm = 0.009 respectively). The above loci and 50 linked to them possess significant regulatory effects and may affect the alternative splicing of four genes and the expression of 17 genes in various organs and tissues related to the PUD pathogenesis.
Similar content being viewed by others
Introduction
Peptic ulcer is the cyclical appearance of a limited mucosal defect in the digestive tract (usually the stomach or duodenum) extending deeply beyond the muscular plate of the mucous membrane, with inflammatory infiltration and thrombotic necrosis in adjacent tissues1. The prevalence of peptic ulcer disease (PUD) in the general population is estimated at 5–10%2.
Mucosal defects in patients with the acid peptic disease have been traditionally considered as a result of increased gastric acid secretion in the stomach and degradation of the mucus barrier2,3,4. Risk factors for PUD, including gastric and duodenal ulcers, are infection by H. pylori, alcohol and tobacco consumption, use of non-steroidal anti-inflammatory drugs (NSAIDs) and aspirin, stress, etc.2,3,4,5,6. However, only a relatively small proportion of people infected by H. pylori or using NSAIDs develop PUD that suggests variation in individual susceptibility to the beginning of mucosal damage7. On the other hand, about one-fifth of cases include H. pylori-negative, NSAID-negative, and aspirin-negative PUD collectively classified as an idiopathic ulcer8. This type of ulcer is thought to occur due to the imbalance between factors important for mucosal integrity and aggressive insults, but the exact pathogenic mechanisms of idiopathic peptic ulcer remain unknown3.
Matrix metalloproteinases (MMPs) are endopeptidases playing an important role in the extracellular matrix (ECM) remodeling, cell proliferation, and inflammation. MMPs are synthesized and secreted by gastric and duodenal epithelial cells, macrophages, and neutrophils9. Since ECM degradation is an important factor of gastric and duodenal mucosal damage and subsequent PUD, MMPs play a key role in this process9,10,11. There is evidence that cleaving and remodeling of the ECM by MMPs is one of the factors contributing to gastric ulceration (GU)12,13. The role of several MMPs (MMP-1, MMP-2, MMP-3, MMP-9, and MMP-13) in GU was studied using animal models13,14,15,16. MMP-9 was shown to be important in the early phase of chronic GU16.
Several genes have been reported for their association with peptic ulcers7,17,18,19. Polymorphisms of the MMPs genes (MMP-9, MMP-7, MMP-3) may contribute to a genetic risk profile for gastric and duodenal ulcers in chronic H. pylori infection17,18,19. H. pylori infection can induce the expression of MMP-3, MMP-7, and MMP-9 in the gastric mucosa and sera18,20,21. MMP-9 was significantly up-regulated in H. pylori-positive as compared to H. pylori-negative GU22.
Despite the apparently significant role of MMPs in PUD pathogenesis, associations of MMP genetic variants with PUD have been poorly analyzed: only a few studies of this problem have been published so far17,18,19,23. Shaimardanova et al.17 reported associations of polymorphic variants of the MMP-1 (rs1799750, rs494379), MMP-3 (rs3025058), MMP-9 (rs17576) genes with PUD in Tatars of Russia but no such association was found for rs3918242 of the MMP-9 gene. Hellmig et al.19 documented rs11568818 of MMP-7 and rs17576 of MMP-9 as risk factors of H. pylori-positive GU in Germans. On the contrary, Yeh et al.18 did not find the association between rs17576 and H. pylori-positive gastric/duodenal ulcer in Taiwanese females. Likewise, no statistically significant associations of rs3918242 MMP-9 with the duodenal ulcer in children in the Chinese population were found23. Overall. this prompts for filling in this gap.
The present study analyzed polymorphisms of MMP-1, MMP-2, MMP-3, MMP-8, and MMP-9 genes for their association with PUD and possible role in the susceptibility to the disease in the Caucasian sample from the Central Region of Russia.
Results
The phenotypic data of the study participants are shown in Table 1. The PUD patients had a more common family history of peptic ulcer (p = 0.0005), alcohol (p = 0.0005) and tobacco (p = 0.0005) consumption, stress (p = 0.0005), the presence of cardiovascular pathology (p = 0.0005) versus the control group. These parameters were used as confounding factors (covariates) in the regression association analyses.
Supplementary Table S1 shows distributions of genotypes and alleles of the ten studied SNPs in the PUD patients and control groups. All analyzed SNPs were in the HWE (p > 0.005, pbonf > 0.05). The analysis yielded no significant associations for all the studied SNPs but one of the MMP-9 gene with PUD (Table 2). Specifically, the increased risk of PUD was associated with allele G of SNP rs17576 MMP-9 (additive model, the odds ratio adjusted for confounding factors ORadj = 1.31, pperm = 0.016, power—82.98%) (Table 2). Besides, two loci of the MMP-9 gene (rs3918249 and rs17576) were individually associated with H. pylori-positive PUD (Table 3). Allele C of SNP rs3918249 showed a significant association with the increased risk of H. pylori-positive PUD (dominant model, ORadj = 1.82, pperm = 0.002, power—96.43%). The increased risk of H. pylori-positive PUD was also associated with a carriage of allele G of loci rs17576 according to the all three genetic models: additive (ORadj = 1.53, pperm = 0.001, power—98.14%), dominant (ORadj = 1.67, pperm = 0.013, power—90.21%), recessive (ORadj = 1.95, pperm = 0.007, power—94.75%).
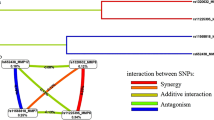
Haplotype AACG defined by rs17576-rs3787268-rs2250889-rs17577 was associated with PUD (ORadj = 0.17, p = 0.001, pperm = 0.003), haplotype CG defined by rs3918249-rs17576 of the MMP-9 gene was associated with H. pylori-positive PUD (ORadj = 1.49, p = 0.004, pperm = 0.009) (Fig. 1). Thus, in total five polymorphisms of the MMP-9 gene were associated with PUD (two individually and three within haplotypes).
Linkage disequilibrium (LD) between SNPs rs3918242, rs3918249, rs17576, rs3787268, rs2250889, and rs17577 of the MMP-9 gene. (A) All PUD patients, (B) H. pylori-positive PUD patients, (C) H. pylori-negative PUD patients, (D) control group. LD values are presented as Lewontin's standardized coefficient D′ (Figure Sects. 1) and the square of the correlation Pearson's coefficient (r2) (Figure Sects. 2) between the SNPs.
Functional SNP
Non-synonymous SNPs
Among the PUD-associated SNPs, three polymorphisms were missense: rs17576 (Gln279Arg), rs2250889 (Arg574Pro), and rs17577 (Arg668Gln) (Supplementary Table S2). According to the SIFT online tool, these loci have prediction values «tolerated» (rs17576 and rs2250889) and «deleterious» (rs17577) (Supplementary Table S2).
Regulatory effects
The data on the regulatory effects of the PUD-associated loci of the MMP-9 gene are presented in Supplementary Table S3. According to the HaploReg database, three SNPs (rs17576, rs2250889, and rs17577) are located in evolutionarily conserved regions, all five polymorphisms—in the region of DNA binding with modified histone (H3K4me3, H3K9ac) marking promoters and hypersensitivity region to DNAse-1 in various tissues, four SNPs (rs17576, rs3918249, rs3787268, and rs2250889)—in the region of DNA binding with modified histone (H3K4me1, H3K27ac) marking enhancers and two polymorphisms (rs17577 and rs2250889)—in the protein-bound region. Importantly, the PUD-associated SNPs manifest their regulatory effects in the tissues and organs related to the pathogenesis of the disease (fetal stomach and small intestine, adult gastric and small intestine, adult stomach, and duodenum mucosa, etc.).
In addition to the five PUD-associated SNPs, regulatory significance was estimated for 50 polymorphisms linked to them (Supplementary Table S3). Three synonymous SNPs were located in exons of the MMP-9 gene, 28 SNPs were in 5'-UTR of the MMP9, ZNF335, and SLC12A5 genes, 19 were in introns. Ten loci were located in evolutionarily conserved regions. The in silico analysis of the linked SNPs suggested several polymorphisms with pronounced regulatory effects (Supplementary Table S3). For example, rs3848722, rs3848721, and rs13969 (were in linkage disequilibrium with SNPs rs3918249 and rs17576) are located in the hypersensitive region to DNAase-I (19, 20, and 24 tissues, respectively), in the region of DNA binding with modified histone marking promoters and enhancers (5 and 14 tissues respectively for rs3848722; 4 and 12 tissues respectively for rs3848721; 12 and 14 tissues respectively for rs13969), and a putative transcription factor binding sites (Pax-6, HNF4, ZID, NRSF for rs3848722; SP1, Zfp281, STAT for rs3848721; ATF3, E2F, XBP-1, p300 for rs13969). Also, the SNP rs13969 is situated in the protein-bound region (with this DNA region interact seven regulatory proteins—SMC3, CCNT2, HAE2F1, RAD21, ZEB1, CTCF, ZNF263) (Supplementary Table S3).
Expression QTLs
In silico analysis for the eQTL impact of the PUD-associated SNPs shows their might affect the expression of 17 genes (MMP9, CD40, NTTIP1, NEURL2, PCIF1, PLTP, RP11-465L10.10, RP3-337O18.9, RPL13P2, SLC12A5, SNX21, SPATA25, SYS1, WFDC10B, WFDC3, ZNF335, ZSWIM1) in more than 20 tissues and organs (Supplementary Table S4). For example, rs3918249 and rs17576 correlate with the transcription levels of various genes in the digestive organs (esophagus, colon) and other tissues and organs related to the pathophysiology of PUD: thyroid (NEURL2), adrenal gland (PCIF1, SLC12A5, RP11-465L10.10), whole blood (ZNF335), adipose tissue (visceral and subcutaneous) (SPATA25, NEURL2, PLTP, CD40, RP3-337O18.9, ZSWIM1), etc. (Supplementary Table S4) The PUD risk alleles G rs17576 and C rs3918249 determined in the present study downregulate the affected genes in most eQTL (Supplementary Table S4). The PUD-associated loci were also in strong LD with the 48 SNPs affecting the expression of the above 17 genes in various organs and tissues (Supplementary Table S5).
Splicing QTLs
The PUD-associated SNPs possessed sQTL with the potential impact on alternative splicing and might affect four genes (PLTP, ACOT8, SNX21, SLC12A5) (Supplementary Table S6). These loci were tightly linked to 48 polymorphisms affecting sQTL of the above four genes in more than 20 tissues and organs (Supplementary Table S7). Noteworthy is the data that independently associated with PUD and/or H. pylori-positive PUD SNPs, rs3918249 and rs17576, correlate with genes alternative splicing in various parts of the brain (cortex and substantia nigra of brain, pituitary, etc.) implicated in the pathophysiology of the disease. According to the results of the present study, allelic variants rs17576 and rs3918249) (alleles G and C respectively) may have a multidirectional effect in different parts of the brain (Supplementary Table S6). For example, allele C rs3918249 is associated with a low level of alternative splicing of the SLC12A5 gene in the brain cortex (effect size β = − 0.44, p = 2.9e−7) and a high level of sQTL of the same gene in substantia nigra of brain (β = 0.61, p = 5.6e−7) and pituitary (β = 0.63, p = 3.0e−12). Similarly, allele G rs17576 correlates with a low sQTL value of the SLC12A5 gene in brain cortex (β = −0.45, p = 3.3e−7) and a high sQTL value of this gene in the pituitary (β = 0.63, p = 9.3e−12) (Supplementary Table S6).
Discussion
The present study reports for the first time the association of MMP-9 gene polymorphisms with PUD in Caucasians from Central Russia: allele G of SNPs rs17576 locus increased risk for PUD (ORadj = 1.31) whereas haplotype AACG of rs17576-rs3787268-rs2250889-rs17577 decreased the risk (ORadj = 0.17). Also, allele C of rs3918249, allele G of the rs17576 and haplotype CG of rs3918249-rs17576 increased risk for the H. pylori-positive PUD (ORadj = 1.82, ORadj = 1.53–1.95 and ORadj = 1.49 respectively). The PUD-associated loci appeared to possess significant regulatory effects and influence the expression of 17 genes and alternative splicing of four genes.
One of the PUD-associated loci, rs17576, was previously shown as a candidate for H. pylori-positive gastric ulcer19, peptic ulcer, and H. pylori-positive peptic ulcer17. However, the data about the risk alleles of this locus were contradictory, Specifically, Shaimardanova et al.17 reported allele G (i.e., the same as determined in the present study) as the risk factor for PUD and H. pylori-positive PUD in Tatars from the Bashkortostan Republic of Russia, whereas Hellmig et al.19 determined allele A as the risk factor for H. pylori-positive gastric ulcer in Germans. On the other hand, Yeh et al.18 did not find any association of rs17576 with either gastric or duodenal ulcer after H. pylori infection in Taiwanese. Okada et al.24 ported the association of rs17576 MMP-9 c gastric cancer both individually and within haplotype CAA rs3918242-rs17576-rs17577 of the MMP-9 gene.
The MMP-9 protein (gelatinase B) cleaves denatured collagen and plays a significant role in ECM modification25. MMPs can be induced by both H. pylori bacterial products and proinflammatory cytokines26. Overexpression of MMPs may result in extracellular matrix breakdown and tissue disintegration. Li et al.22 reported higher MMP-9 expression in the gastric mucosa at the boundary of the gastric ulcer. Significantly elevated expression of pro-MMP9 (about 12-fold) was documented in the indomethacin-induced gastric ulcer as compared to unaffected tissues. Ethanol produced an even stronger effect and increased pro-MMP-9 expression in rat gastric tissues up to 22-fold14. Overexpression of MMP-9 in indomethacin-induced gastric ulcer in mice correlated with up-regulation of activator protein-1 and preceded oxidative stress27.
During PUD, the gastric and duodenal mucosa is infiltrated by monocytes, lymphocytes, neutrophils, and plasma cells. Inflammatory cells produce multiple pro-inflammatory cytokines and growth factors (e.g., epidermal growth factor, transforming growth factor-β, platelet-derived growth factor, vascular endothelial growth factor, etc.)2,22. Pro-inflammatory cytokines can elevate the expression of MMPs26. Chronic inflammation precedes oxidative stress and increases the expression of MMP-922.
We determined associations of the MMP-9 gene polymorphisms with H. pylori-positive PUD but did not find the association of any of the analyzed MMP genes with H. pylori-negative PUD. The polymorphisms of the MMP-9 gene may contribute to a complex genetic risk profile of PUD in chronic H. pylori infection17,19. Our results are in agreement with the previous reports about more significant contribution of MMP-9 to the development of H. pylori-positive gastric ulcer and gastritis as compared to the other MMP genes22,28,29,30. Li et al.22 showed that MMP-9 expression levels in the gastric mucosa were significantly elevated in H. pylori-positive gastric ulcer patients as compared to the H. pylori-negative ones and correlated with the histologically determined activity level and inflammation at the boundary of the ulcer. Epithelium of the H. pylori-induced gastric ulcer manifested higher MMP-9 expression than that of the NSAID-related gastric ulcer28. Significantly higher serum levels of MMP-9 were determined in patients with H. pylori-positive gastritis as compared to H. pylori-negative controls29. Antral mucosa of H. pylori-infected patients with gastritis demonstrated a 19-fold higher MMP-9 protein activity and tenfold increase of the MMP-9 gene expression than that in uninfected individuals30. Successful treatment of the H. pylori infection lowered the MMP-9 expression levels, whereas the elevated levels remain unchanged when the treatment failed31.
It should be noted that the current study is somewhat limited because only one ethnic population was analyzed. The well-known ethnic disparities in the prevalence of complex diseases warrant validation studies of the determined associations of the MMP genes and PUD in other ethnic populations.
Conclusions
Genetic variants of the gene are associated with PUD in a population of Central Russia. However, the data about the possible role of the MMP genes polymorphic variants in the susceptibility to PUD in different ethnic populations remain inconsistent that warrants further studies to identify possible causative variants for the disease.
Methods
Study subjects
In total, 1145 participants, including 798 patients with PUD (434 with gastric ulcer and 364 with the duodenal ulcer), and 347 controls, were recruited for the study. The inclusion criteria were as follows: Russian ethnicity (self-reported) and birthplace in Central Russia32,33, age of 20 and above, voluntary consent to participate in the study, a positive diagnosis of PUD (case group) or absence of the gastrointestinal disease (control group)34. PUD and complications (if any) were determined on the basis of conventional clinical and endoscopic findings. They were not examined by endoscopy because, apart from ethical reasons, the chance of finding an active ulcer in patients without symptoms was very low35. Individuals with chronic diseases of the vital organs (cardiovascular, respiratory, or kidney insufficiency), severe autoimmune disorders, and taking NSAIDs, corticosteroids, and aspirin for a long-term treatment were excluded from the study34.
The H. pylori infection in patients was diagnosed histologically (Giemsa stain36) in biopsies taken from the antrum and corpus of the stomach by the endoscopic procedure35. Among 798 patients with PUD, 404 were H. pylori-positive and 394 were H. pylori-negative. In the controls, the presence of H. pylori was diagnosed by the serological test using a commercial IgG ELISA kit (Plate Helicobacter IgG, Roche). Control group volunteers diagnosed with H. pylori infection were excluded from the study.
The study protocol was approved by the Medical Institution Ethics Committee of Belgorod State University. All participants signed an informed consent prior to enrolment in the study. All methods were performed following the relevant guidelines and regulations. The participants took the medical examination at the Department of Gastroenterology of St. Joasaph Belgorod Regional Clinical Hospital.
Isolation of DNA and genotyping
A blood sample (4–5 ml) was collected by venipuncture from all study participants in EDTA-coated tubes (Vacutainer®). Genomic DNA was isolated from the buffy coat using a standard phenol/chloroform procedure (as described earlier37).
Ten SNPs of the MMP genes (rs1799750 MMP-1, rs243865 MMP-2, rs679620 MMP-3, rs1940475 MMP-8, rs3918242, rs3918249, rs3787268, rs2250889, rs17576, and rs17577 MMP-9) were selected for the analysis according to the following criteria38,39: previously reported associations with digestive diseases (PUD, gastric cancer, etc.), regulatory potential, and MAF > 0.05.
All selected SNPs had significant regulatory potential as evidenced by the HaploReg online tools40 (Supplementary Table S8); eight polymorphisms were associated with digestive diseases (PUD, gastric and esophageal cancer, digestive cancers, gastritis) (including two SNPs associated with PUD) in previously published candidate gene association studies (Supplementary Table S9). Two SNPs (rs3918249 and rs3787268 MMP-9) did not demonstrate a significant association with digestive diseases but had significant regulatory potential (according to HaploReg).
The polymorphisms were genotyped using the MALDI‐TOF mass spectrometry iPLEX platform (Agena Bioscience Inc, San Diego, CA). The quality was controlled by genotyping of blind replicates41. Regenotyping of 5% of the studied samples, selected on a random basis, showed 100% reproducibility of the original results.
Statistical analysis
The observed allele and genotype frequencies were assessed for correspondence to the Hardy–Weinberg equilibrium using the chi-square test42. Associations of the SNPs with PUD were analyzed by logistic regression according to three main genetic models, additive, recessive, and dominant43. The regression analysis was adjusted for covariates: family history of peptic ulcer, alcohol and tobacco consumption, stress, the presence of cardiovascular pathology were used as qualitative variables (Table 1). The haplotype blocks were constructed for MMP-9 gene variants using the «Solid Spine» algorithm (D′ > 0.8) by HaploView program44. The logistic regression analyses and adaptive permutation test to adjust for multiple comparisons45 were calculated by using the PLINK software46. Pperm ≤ 0.017 was set to be statistically significant (after the Bonferroni correction based on the numbers of paired comparisons, n = 3: PUD—control, H. pylori-positive PUD—control, and H. pylori-negative PUD—control).
Functional SNPs
The polymorphisms associated with PUD and those strongly linked to them (r2 ≥ 0.8) were analyzed for their functional significance (non-synonymous SNPs, regulatory potential, eQTLs, and sQTLs)47. SNPs in strong linkage disequilibrium (LD) with the PUD-associated variants were identified using HaploReg40. Non-synonymous SNPs and their functional predictions were analyzed using the SIFT online tool48. The regulatory impact of the candidate MMP loci for PUD was evaluated by using HaploReg40. The effects of the investigated SNPs on the mRNA levels and splicing QTLs was estimated using the GTEx project data49 and the FDR ≤ 0.05 as the significance level. Likewise, eQTL and sQTL values of polymorphisms in strong LD (r2 ≥ 0.8) with the PUD-associated loci were estimated50.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Narayanan, M., Reddy, K. M. & Marsicano, E. Peptic ulcer disease and Helicobacter pylori infection. Mo. Med. 115, 219–224 (2018).
Lanas, A. & Chan, F. K. L. Peptic ulcer disease. Lancet 390, 613–624. https://doi.org/10.1016/S0140-6736(16)32404-7 (2017).
Søreide, K. et al. Perforated peptic ulcer. Lancet 386, 1288–1298. https://doi.org/10.1016/S0140-6736(15)00276-7 (2015).
Kuna, L. et al. Peptic ulcer disease: A brief review of conventional therapy and herbal treatment options. J. Clin. Med. 8, 179. https://doi.org/10.3390/jcm8020179 (2019).
Levenstein, S., Rosenstock, S., Jacobsen, R. K. & Jorgensen, T. Psychological stress increases risk for peptic ulcer, regardless of Helicobacter pylori infection or use of nonsteroidal anti-inflammatory drugs. Clin. Gastroenterol. Hepatol. 13, 498-506.e1. https://doi.org/10.1016/j.cgh.2014.07.052 (2015).
Huang, J. Q., Sridhar, S. & Hunt, R. H. Role of Helicobacter pylori infection and non-steroidal anti-inflammatory drugs in peptic-ulcer disease: A meta-analysis. Lancet 359, 14–22. https://doi.org/10.1016/S0140-6736(02)07273-2 (2002).
De Datta, D. & Roychoudhury, S. To be or not to be: The host genetic factor and beyond in Helicobacter pylori mediated gastro-duodenal diseases. World J. Gastroenterol. 21, 2883–2895. https://doi.org/10.3748/wjg.v21.i10.2883 (2015).
Charpignon, C. et al. Peptic ulcer disease: One in five is related to neither Helicobacter pylori nor aspirin/NSAID intake. Aliment. Pharmacol. Ther. 38, 946–954. https://doi.org/10.1111/apt.12465 (2013).
Shahin, M. et al. Remodeling of extracellular matrix in gastric ulceration. Microsc. Res. Tech. 53, 396–408 (2001).
Tarnawski, A. S. & Ahluwalia, A. Molecular mechanisms of epithelial regeneration and neovascularization during healing of gastric and esophageal ulcers. Curr. Med. Chem. 19, 16–27 (2012).
Ganguly, K., Kundu, P., Banerjee, A., Reiter, R. J. & Swarnakar, S. Hydrogen peroxide-mediated downregulation of matrix metalloprotease-2 in indomethacin-induced acute gastric ulceration is blocked by melatonin and other antioxidants. Free. Radic. Biol. Med. 41, 911–925 (2006).
Chakraborty, S. et al. The use of nano-quercetin to arrest mitochondrial damage and MMP-9 upregulation during prevention of gastric inflammation induced by ethanol in rat. Biomaterials 33, 2991–3001 (2012).
Kim, S. J., Park, Y. S., Paik, H. D. & Chang, H. I. Effect of anthocyanins on expression of matrix metalloproteinase-2 in naproxen-induced gastric ulcers. Br. J. Nutr. 106, 1792–1801 (2011).
Singh, L. P., Mishra, A., Saha, D. & Swarnakar, S. Doxycycline blocks gastric ulcer by regulating matrix metalloproteinase-2 activity and oxidative stress. World J. Gastroenterol. 17, 3310–3321 (2011).
Pradeepkumar Singh, L., Vivek Sharma, A. & Swarnakar, S. Upregulation of collagenase-1 and -3 in indomethacin-induced gastric ulcer in diabetic rats: Role of melatonin. J. Pineal. Res. 51, 61–74 (2011).
Kim, S. J. et al. Antiulcer activity of anthocyanins from Rubus coreanus via association with regulation of the activity of matrix metalloproteinase-2. J. Agric. Food Chem. 59, 11786–11793 (2011).
Shaymardanova, EKh. et al. Role of allelic genes of matrix metalloproteinases and their tissue inhibitors in the peptic ulcer disease development. Genetika 52, 364–375 (2016) ((Russian)).
Yeh, Y. C., Cheng, H. C., Chang, W. L., Yang, H. B. & Sheu, B. S. Matrix metalloproteinase-3 promoter polymorphisms but not dupA-H. pylori correlate to duodenal ulcers in H. pylori-infected females. BMC Microbiol. 10, 218. https://doi.org/10.1186/1471-2180-10-218 (2010).
Hellmig, S. et al. Genetic variants in matrix metalloproteinase genes are associated with development of gastric ulcer in H. pylori infection. Am. J. Gastroenterol. 101, 29–35 (2006).
Mori, N. et al. Helicobacter pylori induces matrix metalloproteinase-9 through activation of nuclear factor kappaB. Gastroenterology 124, 983–992. https://doi.org/10.1053/gast.2003.50152 (2003).
Crawford, H. C. et al. Helicobacter pylori strain-selective induction of matrix metalloproteinase-7 in vitro and within gastric mucosa. Gastroenterology 125, 1125–1136. https://doi.org/10.1016/S0016-5085(03)01206-X (2003).
Li, S. L. et al. Increased expression of matrix metalloproteinase-9 associated with gastric ulcer recurrence. World J. Gastroenterol. 19, 4590–4595. https://doi.org/10.3748/wjg.v19.i28.4590 (2013).
Shan, Q. W. et al. Relationship between gene polymorphisms in MMP-9 and Helicobacter pylori-related upper gastrointestinal disease in children. Zhongguo Dang Dai Er Ke Za Zhi 12, 262–266 (2010).
Okada, R. et al. Matrix metalloproteinase 9 gene polymorphisms are associated with a multiple family history of gastric cancer. Gastric Cancer 20, 246–253. https://doi.org/10.1007/s10120-016-0608-2 (2017).
Cui, N., Hu, M. & Khalil, R. A. Biochemical and biological attributes of matrix metalloproteinases. Prog. Mol. Biol. Transl. Sci. 147, 1–73. https://doi.org/10.1016/bs.pmbts.2017.02.005 (2017).
Bergin, P. J. et al. Gastric gelatinase B/matrix metalloproteinase-9 is rapidly increased in Helicobacter felis-induced gastritis. FEMS Immunol. Med. Microbiol. 52, 88–98. https://doi.org/10.1111/j.1574-695X.2007.00349.x (2008).
Ganguly, K. & Swarnakar, S. Chronic gastric ulceration causes matrix metalloproteinases-9 and -3 augmentation: Alleviation by melatonin. Biochimie 94, 2687–2698 (2012).
Cheng, H. C. et al. Expressions of MMPs and TIMP-1 in gastric ulcers may differentiate H. pylori-infected from NSAID-related ulcers. Sci. World J. 2012, 539316 (2012).
Rautelin, H. I. et al. Enhanced systemic matrix metalloproteinase response in Helicobacter pylori gastritis. Ann. Med. 41, 208–215 (2009).
Bergin, P. J. et al. Increased production of matrix metalloproteinases in Helicobacter pylori-associated human gastritis. Helicobacter 9, 201–210. https://doi.org/10.1111/j.1083-4389.2004.00232.x (2004).
Kubben, F. J. et al. Eradication of Helicobacter pylori infection favourably affects altered gastric mucosal MMP-9 levels. Helicobacter 12, 498–504 (2007).
Litovkina, O. et al. Genes involved in the regulation of vascular homeostasis determine renal survival rate in patients with chronic glomerulonephritis. Gene 546, 112–116. https://doi.org/10.1016/j.gene.2014.04.020 (2014).
Reshetnikov, E. A. et al. The insertion-deletion polymorphism of the ACE gene is associated with increased blood pressure in women at the end of pregnancy. J. Renin Angiotensin Aldosterone Syst. 16, 623–632. https://doi.org/10.1177/1470320313501217 (2015).
Minyaylo, O. N. Allele distribution and haploblock structure of matrix metalloproteinase gene polymorphism in patients with H. pylori-negative gastric ulcer and duodenal ulcer. Res. Results Biomed. 6, 488–502. https://doi.org/10.18413/2658-6533-2020-6-4-0-5 (2020) (Russian).
García-González, M. A. et al. Association of interleukin 1 gene family polymorphisms with duodenal ulcer disease. Clin. Exp. Immunol. 134, 525–531 (2003).
Lee, J. Y. & Kim, N. Diagnosis of Helicobacter pylori by invasive test: Histology. Ann. Transl. Med. 1, 10. https://doi.org/10.3978/j.issn.2305-5839.2014.11.03 (2015).
Ponomarenko, I. et al. Candidate genes for age at menarche are associated with endometriosis. Reprod. Biomed. Online 41, 943–956. https://doi.org/10.1016/j.rbmo.2020.04.016 (2020).
Starikova, D., Ponomarenko, I., Reshetnikov, E., Dvornyk, V. & Churnosov, M. Novel data about association of the functionally significant polymorphisms of the MMP-9 gene with exfoliation glaucoma in the Caucasian population of Central Russia. Ophthalmic Res. https://doi.org/10.1159/000512507 (2020).
Ponomarenko, I. V. et al. Association of genetic polymorphisms with age at menarche in Russian women. Gene 686, 228–236. https://doi.org/10.1016/j.gene.2018.11.042 (2019).
Ward, L. D. & Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. D1, D877–D881 (2016).
Golovchenko, O. et al. Functionally significant polymorphisms of ESR1and PGR and risk of intrauterine growth restriction in population of Central Russia. Eur. J. Obstet. Gynecol. Reprod. Biol. 253, 52–57. https://doi.org/10.1016/j.ejogrb.2020.07.045 (2020).
Reshetnikov, E. et al. Genetic markers for inherited thrombophilia are associated with fetal growth retardation in the population of Central Russia. J. Obstet. Gynaecol. Res. 7, 1139–1144. https://doi.org/10.1111/jog.13329 (2017).
Ponomarenko, I. et al. Candidate genes for age at menarche are associated with endometrial hyperplasia. Gene 757, 144933. https://doi.org/10.1016/j.gene.2020.144933 (2020).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265. https://doi.org/10.1093/bioinformatics/bth457 (2005).
Che, R., Jack, J. R., Motsinger-Reif, A. A. & Brown, C. C. An adaptive permutation approach for genome-wide association study: Evaluation and recommendations for use. BioData Min. 7, 9. https://doi.org/10.1186/1756-0381-7-9 (2014).
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. https://doi.org/10.1086/519795 (2007).
Ponomarenko, I. et al. Candidate genes for age at menarche are associated with uterine leiomyoma. Front. Genet. 11, 512940. https://doi.org/10.3389/fgene.2020.512940 (2021).
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 7, 1073–1081 (2009).
The GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Moskalenko, M., Ponomarenko, I., Reshetnikov, E., Dvornyk, V. & Churnosov, M. Polymorphisms of the matrix metalloproteinase genes are associated with essential hypertension in a Caucasian population of Central Russia. Sci. Rep. 11, 5224. https://doi.org/10.1038/s41598-021-84645-4 (2021).
Author information
Authors and Affiliations
Contributions
O.M., V.D., M.C. substantial contributions to conception and design. O.M., E.R. acquisition of data. I.P., V.D. analysis and interpretation of data. O.M., I.P., drafting the article. E.R., V.D., M.C. revising it critically for important intellectual content. All authors final approval of the version to be published. All authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Minyaylo, O., Ponomarenko, I., Reshetnikov, E. et al. Functionally significant polymorphisms of the MMP-9 gene are associated with peptic ulcer disease in the Caucasian population of Central Russia. Sci Rep 11, 13515 (2021). https://doi.org/10.1038/s41598-021-92527-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-92527-y
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.