Abstract
CHK1 is a crucial DNA damage checkpoint kinase and its activation, which requires ATR and RAD17, leads to inhibition of DNA replication and cell cycle progression. Recently, we reported that SMG7 stabilizes and activates p53 to induce G1 arrest upon DNA damage; here we show that SMG7 plays a critical role in the activation of the ATR-CHK1 axis. Following genotoxic stress, SMG7-null cells exhibit deficient ATR signaling, indicated by the attenuated phosphorylation of CHK1 and RPA32, and importantly, unhindered DNA replication and fork progression. Through its 14-3-3 domain, SMG7 interacts directly with the Ser635-phosphorylated RAD17 and promotes chromatin retention of the 9-1-1 complex by the RAD17-RFC, an essential step to CHK1 activation. Furthermore, through maintenance of CHK1 activity, SMG7 controls G2-M transition and facilitates orderly cell cycle progression during recovery from replication stress. Taken together, our data reveals SMG7 as an indispensable signaling component in the ATR-CHK1 pathway during genotoxic stress response.
Similar content being viewed by others
Introduction
An essential function of the mammalian cell division cycle is to ensure that a cell can duplicate its genome accurately and efficiently during proliferation. This is a daunting task, as cells constantly encounter internal or external conditions that can hinder DNA replication or cause physical damage to DNA1,2. Unsurprisingly, cells employ conserved cell cycle checkpoint mechanisms, which help cope with DNA replication obstacles or damage, in order to maintain genome integrity3. The critical nature of these mechanisms are illustrated by the fact that loss or deficiency of their components can cause various human diseases associated with genome instability including cancer2,4.
The ataxia-telangiectasia mutated (ATM) and ATM- and Rad3-related (ATR) checkpoint kinases, which belong to the phosphatidylinositol 3-kinase-like kinase (PIKK) family, are two key regulators of the DNA damage response3,5,6,7,8. While ATM primarily functions in DNA double-strand break (DSB) signaling, ATR responds to various forms of DNA damage and replication stress caused by a wide range of genotoxic conditions5,6. Activated ATM and ATR phosphorylate and activate downstream effectors to halt cell cycle progression and DNA replication, so cells can repair damaged DNA or resolve any replication stress before entering the next phase of the cell cycle. For example, upon DNA damage (e.g., DSB), ATM activates the tumor suppressor p53, which can cause prolonged arrest of the cell cycle at the G1 phase6. CHK1 is a major downstream effector of ATR, and activation of the ATR-CHK1 axis leads to inhibition of DNA replication in S phase and G2 arrest5.
Activation of ATR is a complex and multi-step process that is initiated in response to stretches of replication protein A (RPA)-coated single-strand DNA (ssDNA)5,9. This aberrant DNA structure arises frequently at stalled replication forks when elongation and unwinding are uncoupled during replication, or at the DSB damage site following resection. The RPA-ssDNA complex serves a recruiting platform to bring together ATR and its activators—topoisomerase II binding protein 1 (TOPBP1)10 and Ewing tumor-associated antigen 1 (ETAA1)11,12,13. ATR does not bind RPA, and its recruitment is mediated by the direct interaction between its binding partner ATR-interacting protein (ATRIP) and RPA14. On the other hand, the recruitment of TOPBP1 occurs independently of ATR and relies on the 5′-ended ssDNA-double-stranded DNA (dsDNA) junction present in the stalled replication fork or resected DSB. Two additional components critical for TOPBP1 recruitment and subsequent ATR activation are the RAD17-RFC complex, which contains the checkpoint protein RAD17 and replication factor C subunits 2–5, and the RAD9-RAD1-HUS1 (9-1-1) checkpoint clamp15,16,17. Facilitated by RPA-ssDNA, RAD17-RFC loads to the 5′-ended ssDNA-dsDNA junction the 9-1-1 clamp complex, which further enables the recruitment of TOPBP118,19,20,21,22,23. Unlike TOPBP1, ETAA1 binds RPA directly, and its recruitment appears to be independent on the 5′-ended ssDNA-dsDNA junction and the 9-1-1 complex11,12,13. Once brought together with ATR-ATRIP, TOPBP1 and ETAA1, both of which contain the ATR-activating domain (AAD), stimulate the kinase activity of ATR in the phosphorylation of its target substrates including downstream effectors (e.g., CHK1)24,25 as well as upstream signaling components.
One such signaling molecule is RAD17, a member of the AAA+ family of ATPases that is critical for 9-1-1 loading in activation of the ATR-CHK1 axis, and is phosphorylated on two serine residues S635 and S645 in the ATM/ATR SQ motif26,27,28. As reported in early studies, while ATM can mediate RAD17 phosphorylation, ATR is the major kinase for S635/645, and loss of these modifications impairs the RAD17 checkpoint function27,28,29. Importantly, phosphorylation of RAD17 on S635/645, which occurs in a cell cycle-dependent manner in undamaged cells, is required to maintain 9-1-1 at the DNA damage site and activate CHK1 in response to replication stress28,29,30. Interestingly, a recent study shows that RAD17 also regulates ATM activation in response to DSBs, involving phosphorylation of RAD17 on T622 by ATM31. Thus, these studies clearly demonstrate that RAD17 plays a key role in checkpoint regulation, and its phosphorylation by ATR and ATM is a pivotal signaling step in shaping the response of a cell to genotoxic stress.
Previously, we identified a critical role for the suppressor with morphological defects in genitalia 7 (SMG7) in the G1/S checkpoint in response to DNA damage response, through promoting ATM-mediated p53 stabilization and transcriptional activation32. Here, based on our initial observation that loss of SMG7 impairs CHK1 activation by ATR, we explored the idea that SMG7 may have a more general role in regulation of the genotoxic stress response. Interestingly, we found that the N-terminal 14-3-3 domain of SMG7 directly binds RAD17 and this interaction is completely dependent on ATR phosphorylation of RAD17 on S635. Sequence analysis revealed at the RAD17 C-terminal a conserved S635-containing SQ motif shared by other SMG7-binding partners including p53 and UPF1, indicating that the SMG7 14-3-3 domain may serve as a common protein-binding module in phospho-SQ-mediated signaling. Furthermore, we show that SMG7 constitutively associates with chromatin and promotes the recruitment of RAD9 to the DNA damage site. Importantly, unlike wild-type cells, the SMG7-deficient cells fail to inhibit S-phase DNA replication after genotoxic treatment, and their recovery from replication stress is significantly compromised because of the inability of cells to maintain CHK1 activity. Taken together, SMG7 has a critical function in activation of the ATR-CHK1 pathway, at least partly through engaging the RAD17-mediated DNA damage signaling.
Results
SMG7 is required for activation of the ATR-CHK1 pathway upon DNA damage
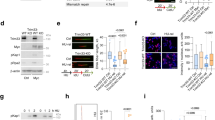
SMG7 is known for its role in nonsense-mediated mRNA decay (NMD), a surveillance pathway that degrades premature stop codon-containing mRNA transcripts including those derived from alternative splicing such as the p53β isoform32,33,34. In our recent studies, we uncovered a novel function of SMG7 in stabilizing the tumor suppressor p53 by promoting ATM phosphorylation of MDM232. Given that SMG7 contains the conserved 14-3-3 protein domain involved in numerous signaling pathways35,36, we reasoned that SMG7 likely has additional roles in the DNA damage response besides regulating the ATM-MDM2-p53 pathway. To explore this idea, we employed HCT116 SMG7 knockout cells (SMG7−/−) generated in our previous studies and examined activation of CHK1 and CHK2, the two checkpoint kinases downstream of ATR and ATM, respectively7. As shown in Fig. 1a, following induction of DSBs (indicated by γ-H2AX) by ionizing radiation (IR), we observed similar levels of CHK2 phosphorylation on the ATM site T68 in both wild type and SMG7−/− cells. Interestingly, loss of SMG7 caused a significant reduction of CHK1 phosphorylation on the ATR site S345—a well-established marker for CHK1 activation24,25 (Fig. 1a, lane 2 vs. lane 4 and Supplementary Fig. 1a), indicating that SMG7 has a specific role in activation of the ATR-CHK1 axis upon DNA damage. To corroborate this finding, we generated SMG7 knockouts in human colorectal adenocarcinoma DLD1 cells using CRISPR-Cas9-mediated gene editing (Supplementary Fig. 1b and 1c). Notably, we made similar observations that deletion of SMG7 impaired ATR-dependent CHK1 activation but had no effect on CHK2 (Fig. 1b, lane 2 vs. lane 4 and Supplementary Fig. 1d). Furthermore, we found that although wild type and SMG7−/− cells exhibited minimal differences in the auto-phosphorylation of ATR on T1989, an ATR activation marker37,38, loss of SMG7 abrogated ATR-mediated phosphorylation of RPA32 on S33 (a direct ATR site) and S4/8 (the DNA-dependent protein kinase sites dependent on S33 phosphorylation)39,40 (Fig. 1c, lane 2 vs. lane 4 and Supplementary Fig. 1e and 1f). Thus, these results show that SMG7 is required for activation of the ATR-CHK1 axis and phosphorylation of some other ATR signaling components such as RPA32 but is dispensable for the intrinsic activity of ATR.
Loss of SMG7 impairs activation of CHK1 upon DNA damage. (a–c) Wild type and SMG7−/− cells (a HCT116, b,c DLD1) were treated with ionizing radiation (a,b 10 Gy; c 20 Gy) and total cell extracts were examined for proteins in the ATM and ATR pathways by western blot analysis using antibodies as indicated including α-SMG7, α-CHK1-pS345, α-CHK1, α-CHK2-pT68, α-CHK2, α-γH2AX, α-H2AX, α-ATM-pS1981, α-ATM, α-ATR-pT1989, α-ATR, α-RPA32-pS4/8, α-RPA32-pS33, α-RPA32, α-Actin and α-Tubulin. Quantitation of relative levels of CHK1-pS345, CHK2-pT68, RPA-pS33, and RPA-pS4/8 in IR-treated cells are shown in Fig. S1a,d,e). (d) Wild type and SMG7−/− HCT116 cells were treated with ionizing radiation (10 Gy, 1 h) and pulsed with 25 μM BrdU for 20 min, and analyzed by immunostaining using α-BrdU antibody (BU1/75). Representative images are shown. (e) Quantification of the percentage of BrdU-positive cells of the total population from (d). Data are presented as Mean ± SEM (n = 3 independent experiments) and were analyzed by ANOVA with Tukey post-test. n.s indicates not significant, P > 0.05. (f) Quantification of the relative intensities of BrdU staining in BrdU + cells from (d) using MetaMorph software. Red bars represent the mean intensities of each population. Data are shown as nuclear BrdU fluorescence intensities of cells pooled from three independent experiments and analyzed by ANOVA with Tukey post-test. **** indicates P < 0.001 and n.s, not significant, P > 0.05. Graphs were generated using GraphPad Prism.
It is well known that the ATR-CHK1 pathway plays a key role in regulation of replication stress, and activation of CHK1 upon DNA damage inhibits DNA replication in S phase41,42. To assess the downstream effects of SMG7 on CHK1, we examined DNA synthesis in SMG7−/− cells after IR. To this end, we exposed HCT116 cells to 10 Gy radiation and labeled them with BrdU to identify replicating cells, which comprise approximately 40% of the total population regardless of SMG7 status (Fig. 1d,e). As expected, IR had no effect on the percentage of S-phase cells (Fig. 1e), but significantly reduced the intensity of BrdU staining in wild type cells (Fig. 1d,f). The lack of reduction of BrdU staining in SMG7−/− cells indicates that DNA synthesis proceeded without hindrance despite DNA damage (Fig. 1f), consistent with impaired CHK1 activation in the absence of SMG7. We made similar observations in DLD1 cells (Supplementary Fig. 1g), which express mutant p5343,44, and importantly, re-introduction of ectopic SMG7 restored CHK1-pS345 phosphorylation and strongly inhibited DNA replication (Supplementary Fig. 1h,i). Taken together, these data indicate that SMG7 is critically important for regulation of the genotoxic stress response through activation of the ATR/CHK1 pathway—a function that is separate from p53-mediated cell cycle arrest following DNA damage.
RAD17 is a binding partner of SMG7
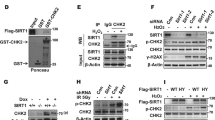
SMG7 contains an N-terminal 14-3-3-like domain and a C-terminal low complex region (LCR), both of which are implicated in mediating protein–protein interactions35,45. To clarify the molecular basis of CHK1 activation by SMG7, we took a proteomic approach to search for SMG7-interacting proteins that potentially regulate the ATR-CHK1 pathway. Thus, we utilized the p53-null human lung carcinoma H1299 cells, which have been widely used to study DNA damage and generate stable lines, to express SMG7 containing N-terminal Flag and HA (hemagglutinin) tags (FH-SMG7) (Supplementary Fig. 2a). Through tandem affinity purification using agarose beads conjugated with α-Flag and α-HA antibodies, followed by mass spectrometric analysis, we identified from the immunoprecipitates RAD17 in addition to several known SMG7-binding proteins including UPF1 and SMG5 (Supplementary Fig. 2b,c)46. By western blot analysis, we verified the presence of RAD17 in the SMG7-specific protein complex (Fig. 2a), indicating that RAD17 is a previously unknown SMG7-interacting protein. RAD17 contains several well-defined domains in its N-terminal region that are critical for chromatin binding and 9-1-1 loading, and two SQ motifs (S635 and S645) at its C-terminal domain (Fig. 2b)47. It is worth noting that SMG7 also interacts with RFC proteins, possibly via RAD17, and exhibits strong binding activity towards phosphorylated RAD17 (Fig. 2c), suggesting that SMG7 may regulate the RAD17-RFC clamp loader.
Identification of RAD17 as an SMG7-binding protein. (a) Verification of SMG7-binding proteins identified from complex purification. The total cell lysates and α-Flag/HA immunoprecipitated materials from H1299 and H1299-FH-SMG7 cells were assayed by western blot analysis using α-RAD17, α-UPF1, α-SMG5, and α-HA (SMG7) antibodies. (b) Schematic presentation of the RAD17 domain structure containing P-loop, Walker B, Sensor 1, Sensor 2 and C-terminal highlighted SQ motifs. (c) As in (a), the total cell lysates and α-Flag immunoprecipitates from H1299 and H1299-FH-SMG7 cells were assayed by western blot analysis using α-RAD17, α-RAD17-pS645, α-SMG7, α-RFC2 and α-RFC3 antibodies. (d) H1299 cells were transfected with SMG7- and RAD17-expressing plasmid DNA, and the cell extracts and α-Flag immunoprecipitated materials were analyzed by western blot with α-RAD17 and α-HA (SMG7) antibodies. (e) The total cell extracts and α-Flag immunoprecipitated materials from H1299 cells transfected with empty vector or FH-RAD17-expression plasmid DNA were examined by western blot analysis using α-SMG7 and α-RAD17 antibodies. (f–g) SMG7 direct binding to RAD17 in vitro. GST, full-length GST-RAD17 (f), or truncated GST-RAD17 fragment (g) fusion proteins were used in pulldown assays with purified FH-SMG7 proteins. FH-SMG7 was detected by western blot analysis using α-SMG7 antibody, and GST proteins visualized by Ponceau S staining in (f) and Supplementary Fig. 2d.
To assess the interaction of SMG7 with RAD17 further, we transiently expressed SMG7 and RAD17 in H1299 cells. As shown in Fig. 2d, RAD17 was only detected in the α-Flag immunoprecipitates from cells expressing FH-SMG7 and HA-Rad17 but not from cells expressing HA-RAD17 alone (lane 3 vs. lane 2), indicating that RAD17 was co-immunoprecipitated with SMG7. Notably, when transiently expressed, RAD17 was able to pull down endogenous SMG7 in the co-immunoprecipitation assays (Fig. 2e), suggesting a strong and stable interaction between RAD17 and SMG7 in cells. To examine whether SMG7 physically interacts with RAD17, we performed in vitro binding assays using purified recombinant proteins. As shown in Fig. 2f, SMG7 was pulled down by immobilized GST-RAD17 but not GST proteins, indicating that SMG7 binds RAD17 directly. Moreover, using various GST-RAD17 fragments, we determined that SMG7 could interact with multiple regions of RAD17 including the N-terminal 1–245 aa fragment containing the P-loop and Walker-B domain (Fig. 2g, lane 4 and Supplementary Fig. 2d) and the middle fragment containing the sensors-1 and 2 (lane 5). Thus, these data demonstrate that RAD17 is a bona fide SMG7-binding protein.
ATR phosphorylation of RAD17 on S635 mediates its interaction with SMG7
To investigate how SMG7 activates the ATR-CHK1 axis through RAD17, several lines of evidence prompted us to look into whether phosphorylation of RAD17 regulates its interaction with SMG7. These include: (1) RAD17 is phosphorylated at two SQ sites S635 and S64527,28; (2) these modifications are enhanced in response to genotoxic stress27; (3) SMG7 contains a 14-3-3 protein domain that binds phosphorylated serine; and (4) SMG7 exhibits strong binding activity towards phosphorylated RAD17 (Fig. 2c). As reported in early studies, ATM and ATR can both phosphorylate RAD1727; thus we tested whether ATM or ATR is required for the interaction of RAD17 with SMG7. To this end, we treated H1299 cells stably expressing FH-SMG7 with the kinase inhibitors KU-55933 (ATM) or VE-822 (ATR)48,49, and found that inhibition of ATR abolished phosphorylation of RAD17 on S635 and its interaction with SMG7 whereas the ATM inhibitor had no detectable effect (Fig. 3a, lane 4 vs. lane 3 and Supplementary Fig. 3a). Moreover, IR enhanced phosphorylation of RAD17 on S635 and its interaction with SMG7, which was completely abrogated by treatment with the ATR inhibitor VE-822 (Fig. 3b, lane 4 vs. lane 3 and Supplementary Fig. 3b). These data indicate that ATR is the major kinase for Rad17 S635 phosphorylation and required for the SMG7-RAD17 interaction.
ATR-dependent RAD17 phosphorylation at S635 enhances SMG7 interaction. (a) The FH-SMG7-H1299 cells were treated for 4 h with 10 µM ATM (KU-55933) or ATR (VE-822) inhibitor. The total cell lysates and α-Flag immunoprecipitates were analyzed by western blot using α-SMG7, α-RAD17 and α-RAD17-pS635 antibodies. Relative levels of immunoprecipitated RAD17 are quantitated in Fig. S3a. (b) Cells treated with or without ATR inhibitor (VE-822, 10 µM for 4 h) were exposed to 10 Gy IR and harvested 1 h later. The total cell lysates and α-Flag immunoprecipitates were analyzed by western blot using α-SMG7, α-RAD17 and α-RAD17-pS635 antibodies as in (a). Relative levels of immunoprecipitated RAD17 and RAD17-pS635 are quantitated in Fig. S3b. (c) GST and GST-SMG7 fragment fusion proteins were incubated with lysates from 293FT cells transiently expressing HA-RAD17, and RAD17 pulled down by the GST proteins was analyzed by western blot with α-RAD17 antibody. (d) Alignment of the amino acid sequences adjacent to the serine residues within the SQ motif of RAD17, UPF1 and p53 (top). The FH-RAD17 and FH-RAD17-S635A proteins purified from transfected 293FT cells were analyzed by western blot using α-RAD17-pS635 and α-RAD17 antibodies (bottom). (e) GST or GST-SMG7 fragments were incubated with purified FH-RAD17 or FH-RAD17-S635A protein from (d) in GST pull-down assays. RAD17 and RAD17-pS635 were detected by western blotting with the antibodies indicated. Relative levels of RAD17 binding and RAD17-pS635 binding are quantitated in Fig. S3f. (f) H1299 cells were co-transfected with plasmids expressing FH-SMG7 and HA-RAD17 WT, S635A, S645A or S635/645A, and the total cell extracts and α-Flag immunoprecipitated materials were examined by western blot analysis using α-SMG7, α-RAD17 and α-RAD17-pS635 antibodies. Relative levels of RAD17 and RAD17-pS635 binding to FH-SMG7 are quantitated in Fig. S3g.
Based on the strong correlation between RAD17 phosphorylation and its interaction with SMG7, we reasoned that the phosphorylated serine in the SQ motif possibly serves as a binding site for the 14-3-3 domain of SMG7. To test this hypothesis, we first mapped the RAD17-binding region of SMG7 and found that RAD17 indeed binds the N-terminal fragment containing the 14-3-3 domain as well as the C-terminal LCR (Fig. 3c, lane 3 and lane 6 and Supplementary Fig. 3). Sequence analysis of the two SQ motifs of RAD17 shows that the amino acids adjacent to S635 but not S645 are highly conserved with those of two other known SMG7-binding proteins UPF1 and p53 (Fig. 3d), which is consistent with the finding that loss of S645 phosphorylation had no effect on SMG7 binding (Supplementary Fig. 3d). To examine whether RAD17 S635 contributes to SMG7 binding, we carried out in vitro pulldown assays using purified wild type RAD17 proteins, which are readily phosphorylated on S635 when expressed in cells, and mutant RAD17 with S635 replaced with alanine (Fig. 3d). Interestingly, we found that loss of S635 phosphorylation markedly reduced RAD17 binding to the SMG7 14-3-3 domain (Fig. 3e, lane 3 vs. lane 7 and Supplementary Fig. 3e,f), but only had mild effects on its binding to the LCR region (Fig. 3e. lane 4 vs. lane 8 and Supplementary Fig. 3f). Moreover, when exogenously expressed in cells, wild type RAD17 bound SMG7 much more strongly than the RAD17 S635A mutant (Fig. 3f, lane 2 vs. lane 4 and Supplementary Fig. 3g), and the reduced binding was not further exacerbated by the S645A mutation (lane 4 vs. lane 8). It is worth noting that the S635-phosphorylated RAD17 detected in the immunoprecipitated materials from cells expressing S635A or S635A/S645A mutants is likely endogenous RAD17 (Fig. 3f, lanes 4 and 8), as transiently expressed SMG7 was able to pull down endogenous S635-phosphorylated RAD17 (Supplementary Fig. 3h). Taken together, these data indicate that ATR phosphorylation of RAD17 on S635 is critical for its interaction with SMG7 under both normal and genotoxic stress conditions.
SMG7 promotes chromatin recruitment of RAD9 upon DNA damage
So far, our data suggest that SMG7 activates the ATR-CHK1 axis upon DNA damage through interactions with RAD17, and this protein binding activity appears to be very specific for RAD17, as we did not observe any interactions between SMG7 and ATR, CHK1, or TOPBP1 in HCT116 and DLD1 cells (Fig. 4a, and Supplementary Fig. 4a). A key function of RAD17-RFC in the activation of ATR-CHK1 is to load the 9-1-1 clamp16,21, and given that SMG7 interacts with RFC proteins as well, we examined whether SMG7 regulates the recruitment or retention of RAD9 to the DNA damage site. To this end, we performed immunofluorescence staining of cells following DNA damage, and found that IR induced similar levels of DSBs (~ 90% γ-H2AX positive cells) in wild type and SMG7−/− cells (Fig. 4b and Supplementary Fig. 4b). In addition, compared with wild-type cells, only a very mild reduction in the number of SMG7−/− cells with γ-H2AX/RPA32 foci was observed, which suggests that loss of SMG7 may have, if any, a slight effect on the formation of ssDNA following DSB resection. These data are largely consistent with our observation that loss of SMG7 does not affect activation of ATM (Fig. 1a,b), which controls DSB resection50,51. As formation of RPA-ssDNA is the starting point of ATR activation, the presence of RPA32 foci indicates that the initial molecular signal for ATR-CHK1 activation is present in wild type and SMG7−/− cells. Interestingly, although IR strongly induced RAD9 foci co-localized with the RPA32 foci in wild type cells, the RAD9/RAP32-colocolized foci were much less pronounced in the SMG7−/− cells (Fig. 4c and Supplementary Fig. 4c) and the SMG7−/− cells also had significantly less RAD9/RAP32-colocolized foci fractions than wild type cells (Fig. 4d and Supplementary Fig. 4c), suggesting that SMG7 is important for the recruitment or retention of the RAD9 complex to the DNA damage site.
SMG7 promotes chromatin recruitment of RAD9 upon DNA damage. (a) Cell extracts from DLD1 SMG7−/− cells expressing FH-SMG7 and the α-Flag immunoprecipitates were examined by western blot analysis using α-SMG7, α-RAD17, α-ATR, α-TOPBP1, α-CHK1 and α-Tubulin antibodies. (b,c) Wild type and SMG7−/− cells were treated with ionizing radiation (10 Gy, 1 h), pre-extracted, and immunostained in (b) with α-RPA32 (green) and α-γH2AX (red) antibodies and in (c) with α-RPA32 (green) and α-RAD9 (sc-74464, red). Nuclei were stained with DAPI. Representative images from immunofluorescence microscopy are shown. Wider fields of view at are shown in Fig. S4b-c. (d) Quantification of yH2AX-, yH2AX/RPA- and RPA/RAD9- foci-positive cells from experiments represented in (b,c). Data are presented as Mean ± SEM (n = 3 independent experiments containing pooled data) and were analyzed by one-way ANOVA with Tukey post-test. **** indicates P < 0.001. (e) Wild type and SMG7−/− DLD1 cells were subjected to fractionation, and the total cells extracts, soluble nuclear and chromatin fractions were analyzed by western blot using α-SMG7 and α-RAD17 antibodies. α-alpha-Tubulin and α-Histone H3 antibodies were used as cytoplasmic and chromatin markers, respectively. Relative levels of chromatin-bound SMG7 and RAD17 are quantitated in Fig. S4d. (f) As in (e), the chromatin-bound fractions from the control and irradiated cells were isolated and examined by western blot analysis using α-SMG7, α-RAD17, α-RAD17-pS645, α-RAD9 (A300-890A) and α-H3 antibodies. Relative levels of chromatin-bound RAD9 are quantitated in Fig. S4f.
To corroborate these findings, we carried out chromatin fractionation assays to examine RAD9 recruitment upon DNA damage52. Notably, we found that in unstressed cells, a small fraction of SMG7 was associated with chromatin, and loss of SMG7 had no effect on the levels of the chromatin-bound RAD17 (Fig. 4e, lane 5 vs. lane 6 and Supplementary Fig. 4d,e). Interestingly, the chromatin-bound fraction of RAD9 was substantially reduced in the SMG7−/− cells compared with wild type cells following IR (Fig. 4f, lane 2 vs. lane 4 and Supplementary Fig. 4f), which is consistent with the results from immunofluorescence staining. Thus, these data show that SMG7 plays an important role in recruitment or retention of the RAD9 complex to the DNA damage site and reveals a novel mechanism for SMG7 activation of the ATR-CHK1 axis.
SMG7 regulates CHK1 activation and fork progression following replication stress
The ATR-CHK1 axis is a key regulator of replication stress, upon which activated CHK1 halts DNA replication in the S phase of the cell cycle by inhibiting replication initiation and progression of the replication fork41,42,53. Thus, our observation that loss of SMG7 impaired activation of CHK1 following IR (Fig. 1) suggests that SMG7 has an important function in the regulation of the cellular response to replication stress induced by DNA lesions. To assess the role of SMG7 in S phase, we utilized camptothecin (CPT), a topoisomerase I inhibitor that specifically induces DSBs on replicating DNA54. As expected, treatment with CPT induced γ-H2AX and RPA32 foci similarly in both wild type and SMG7−/− cells (Supplementary Fig. 5a,b), suggesting that SMG7 does not affect formation of RPA-bound ssDNA. Interestingly, loss of SMG7 significantly reduced CPT-induced phosphorylation of CHK1 on S345 (Fig. 5a, lane 2 vs. lane 4, Supplementary Fig. 5c). Moreover, phosphorylation of RPA32 on S33 and S4/8 following treatment with CPT was nearly abolished in the absence of SMG7 (Fig. 5a, lane 2 vs. lane 4, Fig. 5b,c, and Supplementary Fig. 5c), indicating that SMG7 is critical for activation of the ATR/CHK1 signaling upon replication stress.
SMG7 regulates ATR signaling and CHK1 activation and fork progression following replication stress. (a–c) Wild type and SMG7−/− cells were treated with 1 µM CPT for 1 h. (a) Total cell extracts were examined by western blot analysis using α-SMG7, α-CHK1, α-CHK1-pS345, α-RPA32-pS33, α-RPA32-pS4/8, α-RPA32, and α-Actin antibodies. Relative levels of CHK1-pS345, RPA-pS33, and RPA-pS4/8 in CPT-treated cells are quantitated in Fig. S5c. (b) Cells were immunostained with α-RPA32-pS4/8 (red) and α-RPA32 (green) antibodies. Representative images from immunofluorescence microscopy are shown. (c) Quantification of S4/8-positive cells from (b) using MetaMorph software. Data are presented as Mean ± SEM (n = 3 independent experiments containing pooled data) and analyzed by one-way Anova with Tukey post-test; **** indicates P < 0.001. Graphs were generated using GraphPad Prism. (d–f) DNA fiber analysis of untreated or CPT-treated wild type and SMG7−/− cells as in (a). Cells were first labeled with 25 μM CldU for 30 min and then by 250 μM IdU for 30 min in the absence or presence of CPT. DNA fibers were then spread, stained with α-BrdU antibodies (B44 and BU1/75) and visualized using fluorescence microscopy. More than 500 CIdU-/IdU-labeled tracks from two independent experiments were measured using ImageJ. (d) Quantification of replication fork speed using the lengths of CldU- and IdU-labeled DNA fiber. Fork speed was estimated using 1 μm = 2.6 kbp as described in Methods. Student’s t-test was performed to determine the statistical difference; n.s. not significant. (e) Scheme for the CIdU/IdU labeling and CPT treatment (Top). Representative tracks were shown (Middle). Distribution of the IdU-labeled tracks from the CPT-treated cells (Bottom). (f) Quantification of length ratios of the IdU-labeled tracks to the CldU-labeled tracks per replication fork. Red bars represent the means of each population. Data were analyzed by one-way Anova with Tukey post-test; **** indicates P < 0.001; n.s. not significant, P > 0.05.
To examine how SMG7 regulates replication stress, we performed DNA fiber analysis, a well-established double-labeling approach that uses the BrdU derivatives CldU and IdU to directly assess the replication fork55. This dual labeling strategy makes it possible to (1) identify active replication forks during the labeling times, which proceed bi-directionally from their origins, and (2) determine the effect of replication stress on fork progression by measuring the IdU-labeled DNA tracks. To this end, cells were pulse-labeled with CldU and IdU (with or without CPT), followed by DNA spreading, immunostaining and microscopic analysis for the presence and length of the distinct CIdU and IdU tracks. Under normal conditions, wild type and SMG7−/− cells exhibited similar symmetric CIdU/IdU-labeled tracks (Supplementary Fig. 5d) and showed no significant difference in fork speed (Fig. 5d). Interestingly, in the presence of CPT, wild type cells had much shorter IdU-labeled tracks compared with the SMG7−/− cells (Fig. 5e, distribution within 2–8 µm for wild type and 4–12 µm for the SMG7−/− cells, respectively), which is consistent with previous studies showing that activation of CHK1 inhibits fork progression56. Moreover, by analyzing hundreds of active replication forks individually, we found that treatment with CPT resulted in a significant reduction in the ratio between IdU and CIdU-labeled tracks, which was reversed by inhibition of CHK1 or loss of SMG7 (Supplementary Fig. 5e,f). Thus, these data indicate that by promoting ATR signaling and CHK1 activation, SMG7 has a important role in the control of fork progression in response to replication stress.
SMG7 regulates CHK1-S345 phosphorylation and modulates cell cycle progression during recovery from replication stress
The crucial function of CHK1 in the replication stress response is manifested not only in fork regulation but also during recovery of cells after removal of stress conditions56,57,58,59,60,61. During the process of recovery, the ATR-mediated checkpoint signaling is attenuated in a regulated manner, which ensures coordinated DNA replication and cell cycle progression after replication stress is resolved62. Not surprisingly, deficiency in CHK1 itself or components required for activation of the ATR-CHK1 axis impairs normal cell cycle progression or cell survival after cells are relieved from a short-term replication stress of reversible nature, such as a few hours of treatment with hydroxyurea (HU), a ribonucleotide reductase inhibitor that induces replication stress by depleting cellular deoxyribonucleotides. For example, cells depleted of RAD17 or expressing phosphorylation-defective S635/645A mutant RAD17 are unable to maintain controlled CHK1 activation and lose cell viability during recovery from HU treatment29. Thus, our finding that SMG7 promotes CHK1 activation through robust binding to S635-phosphorylated RAD17 prompted us to examine whether SMG7 is involved in facilitation of replication stress recovery. As expected, HU treatment strongly induced CHK1 activation in a dose- and time-dependent manner, which was markedly reduced in the absence of SMG7 (Supplementary Fig. 6a,b). When we assessed ATR phosphorylation of CHK1 shortly after HU removal, we found that levels of S345-phosphorylated CHK1 decreased over time in wild type cells, and loss of SMG7 further accelerated this downregulation (Supplementary Fig. 6c). Consistent with previous studies29, these data suggest that SMG7 plays an important role in maintaining CHK1 S345 phosphorylation in the early stages following removal of replication stress.
Next, we examined cell cycle profile, and found that when the whole cell population was analyzed in a course of 12-h recovery, the SMG7−/− cells showed deficient cell cycle progression (Supplementary Fig. 6d). To assess the S-phase population specifically, we pulse-labeled cells with BrdU, and then treated them with HU, which arrested BrdU-positive cells throughout the S phase and BrdU-negative cells at G1 (Fig. 6a, control vs. 0 h). Until 4 h after HU removal, we observed little difference in the S-phase progression of the BrdU-positive cells. The gradual increase (from 4 to 8 h) in the wild type BrdU-positive population with 2 N DNA content, which results from completion of mitosis, indicates a regulated and orderly entry into mitosis from G2 after release from HU. Interestingly, the SMG7−/− BrdU-positive cells completed the G2-M transition in an accelerated fashion, and subsequently accumulated at the 2 N position (Fig. 6a; SMG7−/− 34.1% vs. wild type 6.8% by 8 h). The accelerated transition of the SMG7−/− BrdU-positive cells was clearly illustrated when the BrdU-positive population was selected and analyzed by flow cytometry (Supplementary Fig. 6e). To assess the role of SMG7 in the G2-M progression directly during the 12-h course of recovery, we treated cells with the mitosis-arresting agent nocodazole upon HU removal and assayed the presence of the mitotic marker S10-phosphorylated histone 3 (H3-pS10). Notably, the BrdU-positive population of the SMG7−/− cells had a significantly higher H3-pS10-positive fraction compared with that of wild type cells (Fig. 6b,c), suggesting that SMG7 is indeed critical for the G2-M transition during recovery from replication stress. It is worth noting that the BrdU-negative population of wild type cells showed a higher H3-pS10-positive fraction than that of the SMG7−/− cells (Fig. 6d), and this is likely because wild type cells appeared to progress from G1 through S phase more quickly (Fig. 6a).
SMG7 maintains normal attenuation of the ATR-CHK1 axis during recovery from replication stress. (a) Wild type and SMG7−/− HCT116 cells were pulsed with 25 μm BrdU followed by treatment with 5 mM HU for 6 h. After HU treatment, cells were released into fresh normal media, and harvested at the indicated time points. Cells were then fixed, stained with α-BrdU antibody (y-axis) and 7-AAD (x-axis) and analyzed by flow cytometry. The BrdU-positive fractions containing G1-DNA content from total populations are circled (4, 6 and 8 h after HU removal). (b–d) Cells treated as in (a) were released into fresh media containing 1 μg/mL nocodazole for different hours. Cells were then fixed and stained with α-BrdU (BU1/75) (green) and α-Histone H3-pS10 (red) antibodies, and imaged using a fluorescence microscope. Representative images of cells 8 h after HU removal are shown in (b). Cells were counted using ImageJ, and the percentage of BrdU/H3-pS10 double positive (c) cells were quantified. Data are presented as Mean ± SD (n = 3) and analyzed by one-way ANOVA (****P < 0.0001). (d) BrdU-negative/H3-pS10-positive cells were quantified. Data are presented as Mean ± SEM (n = 3) and analyzed by Student’s t-test; P < 0.01. (e) Total cell extracts from wild type and SMG7−/− cells treated as in (b–d) were examined by western blot analysis using α-SMG7, α-CHK1-pS345, α-CHK1, α-RPA32-pS33, α-RPA32, α-RAD17 and α-RAD17-pS635 antibodies.
To determine the underlying mechanism by which SMG7 regulates the G2-M transition, we examined the status of CHK1 phosphorylation during recovery, as previous studies have shown that the mitotic entry from G2 is controlled by CHK157,59,60. While treatment with a higher dose of HU (5 mM) for a prolonged period of time (6 h) induced similar CHK1 S345 phosphorylation regardless of SMG7 status, importantly, wild type cells retained markedly higher levels of S345-phosphorylated CHK1 than SMG7−/− cells after removal of HU (Fig. 6e, lanes 3–4 vs. 7–8). The further reduced CHK1 S345 phosphorylation in the absence of SMG7 during recovery is consistent with the observation that SMG7−/− cells showed an accelerated G2-M transition. It is also interesting to note that loss of SMG7 abolished HU-induced phosphorylation of RPA32 on S33 but had no effect on RAD17 phosphorylation on S635 before or after HU removal. Thus, these data indicate that SMG7 plays an important role in regulating CHK1 S635 phosphorylation and modulating cell cycle progression during recovery from replication stress.
Discussion
Here, we report a previously unknown function of SMG7 in regulation of the ATR-CHK1 pathway in response to DNA damage and replication stress. For the first time, we have identified SMG7 as a binding partner to the RAD9-RAD1-HUS1 clamp loader protein RAD17 and shown that SMG7 promotes the recruitment of RAD9 to RPA-coated ssDNA in the activation of CHK1. The interaction of SMG7 with RAD17 is highly dynamic and tightly controlled by the ATR-dependent phosphorylation of RAD17 on S635, a conserved SQ site that is required for RAD17 checkpoint function. Importantly, loss of SMG7 impairs ATR signaling and CHK1 activation upon genotoxic stress and disables intra-S checkpoint function in control of replication fork progression. Furthermore, SMG7 has an important role in maintaining CHK1 S345 phosphorylation and cell cycle progression. In summary, our present studies identify SMG7 as an indispensable signaling component in activation of the ATR-CHK1 pathway through RAD17 binding and RAD9 recruitment, and suggests that by engaging phospho-SQ-mediated signaling through its 14-3-3 domain, SMG7 has a more general and direct role in regulation of the genotoxic stress response.
Activation of the ATR-CHK1 axis upon DNA damage is a complex process, which necessitates recruitment of the 9-1-1 complex to the damage site by RAD17-RFC2,5,8. Given that loss of SMG7 markedly impairs the formation of RAD9 foci in regions of RPA-bound ssDNA, our data provide an important insight into how SMG7 engages in the ATR-CHK1 signaling pathway (Fig. 4c). Although the precise mechanism by which SMG7 promotes RAD9 recruitment/retention remains to be fully elucidated, SMG7 binding to RAD17 and other RFC subunits likely has a direct impact on 9-1-1 association and/or recruitment (Fig. 2). For example, studies have shown that the N-terminal P-loop and Walker-B domains of RAD17 are critical for 9-1-1 binding and recruitment47,63; thus the direct binding of SMG7 to the RAD17 N-terminal region may facilitate 9-1-1 recruitment or stabilize the RAD17 interaction with 9-1-1. Additionally, SMG7 may promote the recruitment/retention of the RAD9 complex through interaction with the S635-phosphorylated RAD17. While early reports suggest that ATR phosphorylation of RAD17 is not required for initial RAD9 loading, recent studies show that phosphorylation of the RAD17 SQ motif indeed plays an important role in maintaining DNA damage-induced RAD9 foci22,30. Therefore, it is likely that through interacting with S635-phosphorylated RAD17, SMG7 may stabilize RAD9 foci following its recruitment, which allows full activation of the ATR-CHK1 axis. Besides acting as a RAD9-RAD1-HUS1 clamp loader protein, RAD17 also functions in CHK1 recruitment by CLASPIN, which is critical for ATR phosphorylation and activation of CHK129,64. Intriguingly, SMG7 and CLASPIN both engage with RAD17 in a phosphorylation-dependent manner via interactions with phosphorylated RAD17, but it is not known whether S635, S645 or both are required for the RAD17-CLASPIN interaction. While we show that loss of SMG7 reduces RAD9 recruitment/retention (Fig. 4), our data does not rule out the possibility that SMG7 may be involved in CLASPIN regulation of CHK1. It is possible that SMG7 may compete with CLASPIN in RAD17 binding; however, given that both SMG7 and CLASPIN positively regulate CHK1, it is intriguing how these two RAD17 binding proteins may affect each other with respect to different aspects of CHK1 activation. For example, CLASPIN is critically important for the attenuation of the ATR-CHK1 signaling during recovery57,59 and it remains to be elucidated whether and how SMG7/phosphorylated RAD17 are involved in this regulation.
RAD17 is an important checkpoint regulator, and its phosphorylation by ATR and ATM appears to be a pivotal signaling step in shaping the response of a cell to genotoxic stress27,29,31. As reported here and previously, ATR is the major kinase that phosphorylates RAD17 on the SQ sites S635/645, and loss of these modifications impairs the RAD17 checkpoint function27,29. Moreover, a recent study has shown that RAD17 activates ATM in response to DSB through binding to Nijmegen breakage syndrome (NBS1) of the MRE11-RAD50-NBS1 (MRN) complex, which involves the phosphorylation of RAD17 on T622 by ATM31. Thus, our finding that SMG7 interacts with RAD17 in an ATR-dependent but ATM-independent manner reveals phosphorylation-mediated SMG7 binding to RAD17 as a potential underlying mechanism by which SMG7 exerts its function in response to genotoxic stress (Fig. 3a,b). Indeed, our data show that loss of SMG7 abrogates CHK1 activation but has no effect on the ATM-CHK2 pathway, and support a model in which through binding to S635-phosphorylated RAD17 in an ATR-dependent fashion, SMG7 acts on ATR-CHK1 signaling following DNA damage (Figs. 1a–c and 3e,f). Importantly, consistent with its role in activation of the ATR-CHK1 axis, loss of SMG7 disables CHK1-dependent intra-S checkpoint function in control of replication fork progression in response to CPT-induced replication stress (Fig. 5). However, it appears that SMG7 is not significantly involved in regulation of CHK1 under basal conditions, as indicated by normal DNA replication and fork progression in SMG7−/− cells during S phase (Figs. 1d,e and 5). Moreover, unlike RAD17-null cells, which rapidly lose viability following deletion of RAD1765, SMG7−/− cells are able to grow and proliferate normally, consistent with the phenotype observed in cells expressing the phosphorylation-defective S635/645A RAD17 mutant29. Thus, these data support the notion that SMG7 functions in the ATR-CHK1 pathway mainly, if not exclusively, through interaction with the S635-phosphorylated RAD17.
In addition to being indispensable for CHK1 activation upon DNA damage, our data further show that SMG7 may have an important role in regulating CHK1 and cell cycle progression during recovery from replication stress. Unlike wild type cells, SMG7−/− cells fails to retain CHK1 S345 phosphorylation shortly after release from replication stress, consistent with a previous study showing that loss of RAD17 phosphorylation on S635/645A leads to a defect in maintaining Chk1 activation after HU withdrawal29. Furthermore, SMG7−/− cells exhibit an accelerated transition from G2 to M after release from prolonged treatment (6 h) with a higher dose of HU (5 mM), which is supported by data that SMG7−/− cells have even lower S345-phosphorylated CHK1 compared to wild-type cells during recovery (Fig. 6 and Supplementary Fig. 6). As S635/645-phosphorylated RAD17 engages both SMG7 and CLASPIN, whose degradation is critical for CHK1 inactivation during recovery from replication stress29, it is possible that SMG7 may regulate CHK1 by modulating CLASPIN function during recovery from replication stress. It is important to note that in these experiments HU treatment (5 mM for 6 h) induced similar CHK1 S345 phosphorylation regardless of SMG7 status, suggesting that wild-type and SMG7−/− cells have the same levels of initial CHK1 activation following HU treatment. As our data show that loss of SMG7 does not abolish CHK1 S345 phosphorylation after HU treatment and only attenuates it in a dose- and time-dependent manner, indicating that the mechanism for CHK1 activation still exists in SMG7−/− cells (Supplementary Fig. 6a,b). It is worth noting that the level of CHK1 phosphorylation in SMG7−/− cells treated with 2 mM HU for 1 h is even higher than that in wild-type cells treated with 0.5 mM HU. A recent study shows that the amount of HU and the time of its treatment has very dynamic effects on the recruitment pattern of several critical proteins involved in CHK1 activation such as TOPBP1 and the 9-1-1 complex66. Thus, it is conceivable that under specific conditions such as prolonged treatment with a high concentration (5 mM) of HU, CHK1 S345 phosphorylation may be maximized in both wild-type and SMG7−/− cells, which leads to similar levels of S345-phosphorylated CHK1.
In response to genotoxic stress, ATR phosphorylates a plethora of proteins, and there is enormous interest in understanding the mechanism for ATR activation towards the different substrates3,5,8. By examining CHK1 and RPA32, a recent study has identified two distinct ATR activation modes, which are orchestrated by RAD17 and NBS1 in phosphorylation of CHK1 and RPA32, respectively67. Interestingly, our present data show that loss of SMG7 abolishes ATR phosphorylation of CHK1 and RPA32 but has no effect on RAD17 (Figs. 1a–c, 4f, 5a and 6a,f), suggesting that SMG7 may regulate phosphorylation of ATR substrates differentially, and likely engages either directly or indirectly other components (e.g., NBS1) of the ATR pathway besides RAD17. Future studies aimed to elucidate the precise mechanisms of SMG7-mediated signaling will further our understanding of the role of SMG7 in the regulation of genotoxic stress response.
Methods
Cell culture
The human colorectal adenocarcinoma HCT116, DLD1 and their SMG7-null derivative cell lines were cultured in McCoy’s 5A medium (Corning, Manassas, VA, USA) supplemented with 10% fetal bovine serum (Sigma, F2442). The H1299 and 293FT cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Corning/Cellgro) supplemented with 10% fetal bovine serum and combined antibiotics (100 I.U/ml penicillin and 100 ug/ml streptomycin).
Generation of DLD1 SMG7 −/− cell line
We carried out CRISPR-Cas9-mediated gene targeting in DLD1 cells based on the protocol described in the previous study68. Briefly, two sgRNAs (target sequence: 5′- ATGTTCGAATAATCAGATTG-3′ and 5′-GATTATAGGTGATCAATCCC-3′) designed using the online CRISPR Design Tool (http://tools.genome-engineering.org) were cloned into the Cas9-expressing plasmid PX458 (Addgene, #48138). After transfection with the sgRNA-expressing plasmid DNA and pCin4-puro (Clontech, #6031-1), cells were treated with 3 µg/ml puromycin for 24 h and plated in 96-well in the absence of puromycin. Two weeks later, clones were expanded and screened for SMG7 targeting, which was further verified by DNA sequencing.
Plasmids construction
pCin4-FH-SMG7, pCin4-FH-RAD17 and pCin4-HA-RAD17 plasmids were constructed by cloning the cDNA encoding SMG7 or RAD17 generated by RT-PCR from mRNA extracted from HCT116 cells into pCin4-Flag-HA or pCin4-HA vectors. pCin4-FH-RAD17-S635A, pCin4-HA-RAD17-S635A, S645A and S635A/645A plasmids were generated by site-directed mutagenesis. GST-RAD17 full-length, GST-RAD17 fragments and GST-SMG7 fragments expressing plasmids were generated by cloning of each DNA fragments into pGEX-2TL(+) vector. All the plasmids were confirmed by DNA sequence analysis.
Antibodies and inhibitors
The following antibodies were from Bethyl: α-H2AX (A300-081A, 1:1000), α-H2AX (A303-837A, 1:2000), α-RAD9 (A300-890A, WB 1:500, IF 1:50), α-RAD17-pSer645 (A300-153A, 1:1000), α-RFC2 (A300-142A, 1:1000), α-RFC3 (A300-188A, 1:1000), α-RPA32-pS4/8 (A300-245A, 1:1000), α-RPA32-pS33 (A300-246A, 1:1000), α-RPA32 (A300-244A, 1:3000), α-SMG7 (A302-170A, 1:1000). The following antibodies were from Cell Signaling Technology: α-ATM-pS1981 (5883, 1:1000), α-ATM (2873, 1:1000), α-CHK1-pS345 (2348, 1:1000), α-CHK2-pT68 (2661, 1:1000), α-CHK2 (2662, 1:1000), α-RAD17-pS635 (13404S, 1:1000), α-RAD17 (8561, 1:1000), α-RPA32 (2208S, IF 1:1000), α-Histone H3 (3638, 1:2000), α-rabbit IgG-HRP (7074, 1:5000). The following antibodies were from Santa Cruz Technology: α-Actin-beta (sc-47778, 1:1000), α-ATR (sc-1887, 1:1000), α-CHK1 (sc-8408, 1:100), α-Histone H3 p-Ser10 (sc-8656, IF 1:200), α-RAD9 (sc-74464, IF 1:100), α-RAD17 (sc-17761, 1:500), α-Tubulin-alpha (sc-53029, 1:1000). Goat α-rat IgG-HRP (3050-05, 1:5000) and donkey α-goat IgG-HRP (6420-05, 1:5000) were from Southern Biotech. α-ATR-pT1989 (GTX128145, 1:1000), α-HA (MMS-101P, 1:1000), α-SMG5 (12694-1-P, 1:1000), α-SMG7 (LS-C353204-100, 1:1000) and Sheep α-mouse IGG-HRP (NA931, 1:5000) were from GeneTex, Bio-Rad, Covance, Proteintech, LifeSpan Biosciences and GE healthcare, respectively. Antibodies used for BrdU staining include α-BrdU clone B44 (347580, 1:200; BD Biosciences) and α-BrdU clone BU1/75 (MCA2060, 1:200 BioRad). The following inhibitors were used: ATR inhibitor (VE-822; MedKoo Biosciences) and ATM inhibitor (KU-55933; Sigma-Aldrich).
Purification of the SMG7-associated protein complex
The H1299-FH-SMG7 stable cell lines were generated by transfection with pCin4-FH-SMG7, followed by selection with 1 mg/ml G418 for 2–3 weeks. To purify the SMG7 protein complex, freshly grown H1299-FH-SMG7 cells (~ 5 × 108) were harvested in cold PBS. The cell pellets were suspended in 20 times volume of ice cold BC100 buffer (20 mM Tris–HCl pH 7.9, 100 mM NaCl, 10% glycerol, 0.2 mM EDTA and 0.5% Triton X-100) with fresh proteinase inhibitor cocktail and incubated on ice for 1 h with several times of brief vertex. After centrifugation at 15,000 rpm for 30 min at 4 °C, the supernatants were cleared by passing through 0.45 μm syringe filters and the final cell extracts were subjected to immunoprecipitation with anti-Flag antibody-conjugated M2 agarose beads (Sigma). The bound polypeptides eluted with the Flag peptide were further affinity purified by anti-HA antibody-conjugated agarose, and the final elutes from the HA-beads with HA peptides were resolved on an 8% SDS-PAGE gel for silver staining or subjected to mass spectrometric analysis (by MS Bioworks LLC).
Immunoprecipitation and western blotting
Cells were washed twice with PBS, and then lysed in BC100 lysis buffer supplemented with protease inhibitor cocktail and phosphatase inhibitors (Sigma) for 1 h on ice with occasional vortex. Cell lysates were cleared by centrifugation at 14000 rpm for 20 min at 4 °C. For IP of Flag-tagged proteins, cell lysates with or without Flag-tagged proteins were incubated with anti-Flag M2 beads (Sigma) overnight at 4 °C. Beads were washed five times in lysis buffer and bound proteins were eluted with 1% SDS or Flag peptides (Sigma) in lysis buffer. For direct western blotting, the cells were washed twice with PBS and lysis in BC100 buffer containing 0.5% SDS and protease inhibitor cocktail and phosphatase inhibitors and sonicated for 20 s on power setting 1 using a sonic dismembrator (Model 100, Fisher Scientific). After measuring protein concentration using the Bio-Rad protein assay, equal amounts of proteins were loaded and separated on a SDS-PAGE gel ranging from 8–12%, and transferred to 0.45 μM nitrocellulose membranes (GE healthcare). Transfer quality was verified by Ponceau staining, followed by blocking with 5% milk/TBST/0.5 h, followed by overnight incubation with primary antibody diluted in 5% milk or BSA. The membranes were washed and incubated for 1 h in horseradish peroxidase (HRP)-conjugated secondary antibody diluted 1:1000–1:10,000. Clarity western ECL substrate (Bio-Rad) and blue film (Crystalgen) were used to detect the proteins. Densitometry of Western blots was performed using ImageJ software (https://imagej.net/Fiji). Western blot images were prepared using Photoshop CS6 (https://www.adobe.com/products/photoshop.html).
Chromatin fractionation
Cell fractionation was performed largely according to the protocol described in the previous study69. Cells from 10-cm dishes were washed with PBS twice, and suspended in 200 μl of Buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 10 mM NaF, 1 mM DTT, protease inhibitor cocktail and phosphatase inhibitors), and Triton X-100 was added to 0.1%, then the cells were left on ice for 5 min. The cytoplasmic fractions were separated from the nuclei by centrifugation at 1300 g for 4 min at 4 °C, and the cytoplasmic fractions were further clarified by centrifugation at 20,000 g for 5 min. The nuclei were washed one time with Buffer A, and lysed in 100 μl of Buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, protease inhibitor cocktail and phosphatase inhibitors) on ice for 30 min. The soluble nuclear fractions were separated from the chromatin fractions by centrifugation at 1700 g for 4 min. The chromatin fractions were washed one time with Buffer B and centrifuged at 10,000 g for 1 min, and lysed in 100 μl of BC100 buffer with 0.5% Triton and 0.5% SDS with sonication.
GST pull-down assay
Recombinant proteins of GST-RAD17 full-length and different fragments, or of GST-SMG7 different fragments were expressed in Rosetta 2(DE3) cells (EMD, San Diego, CA, USA) and bound to glutathione agarose (Pierce, Rockford, lL, USA). GST-proteins beads were incubated with either purified proteins or cell lysates overnight at 4 °C and washed five times in BC100 washing buffer (20 mM Tris.Cl pH 7.3, 100 mM NaCl, 10% glycerol, 0.2 mM EDTA, 0.4% Triton X-100) supplemented with protease inhibitor cocktail. Bound materials were eluted with 2 × SDS-PAGE sample buffer and resolved by SDS-PAGE, transferred to nitrocellulose membranes, stained with ponceau for GST-proteins, and analyzed by western blotting.
Silver staining
Proteins were separated on a polyacrylamide gel using electrophoresis. The gel fixed with 45% methanol/10% acetic acid for 30 min, followed by fixation with 10% glutaraldehyde for 10 min, thorough washing in water for 2 h, and treatment with 5ug/mL DTT for 30 min. The gel was incubated with 0.1% silver nitrate for 30 min, and quickly rinsed with water followed by carbonate developing solution. The gel was developed with carbonate developing solution, and the reaction was stopped using 5 mL of 2.3 M citric acid per 100 mL developing solution.
Immunofluorescence
Cells were plated and grown on coverslips in 12-well plates for 20–24 h. For visualization of DNA damage foci, cells were treated with IR or CPT followed by pre-extraction with CSK buffer (10 mM Pipes pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA) with 0.7% Triton X-100 plus protein inhibitor cocktail for 5 min on ice. After washing once with CSK buffer without Triton X-100 and twice with PBS, cells were fixed (4% paraformaldehyde, 15 min, room temperature) and permeabilized (0.7% Triton X-100 in PBS, 10 min), blocked (5% BSA in PBS, 1 h), and incubated with primary antibody overnight. After extended wash in PBS-Triton buffer, incubated with fluorophore-conjugated secondary antibodies. After counterstaining with DAPI, coverslips were washed briefly with water and mounted in Prolong Diamond Antifade mounting media (#P36961, Invitrogen). Cells were visualized under an Olympus BX61 upright microscope using a 40 × oil objective. DNA damage foci were analyzed using ImageJ software and were defined by positive staining of distinct yH2AX or chromatin-bound RPA nuclear puncta.
Cell number and fluorescence intensity for RPA32/p-RPA32 S4/8 staining was quantified using the MetaMorph Multi-Wavelength Cell Scoring Application (https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy) using cells pooled from three independent experiments. Cells were imaged using a 20 × air objective. Quantitation of RPA32-pS4/8 positive cells was performed as follows: DAPI staining was used to define the region of the nucleus of each cell, and this mask was used to measure the fluorescence intensity of RPA-pS4/8 in the TxRed channel. Untreated cells were used to establish the level of background fluorescence. Cells above this threshold were defined as RPA-pS4/8-positive, and cells below this threshold were defined as RPA-pS4/8-negative.
For BrdU detection, cells grown on coverslips were pulse labeled with 25 μM BrdU for 20 min and then washed with ice-cold PBS, fixed with 70% ethanol for 5 min and methanol (-20 °C) for 5 min and stored at 4 °C. DNA was denatured with 2.5 N HCl for 30 min and wash in PBS for 30 min. After blocking in blocking buffer (5% BSA in PBS with 0.5% tween-20) for 1 h, incubated with rat anti-BrdU (#MCA2060, Bio-Rad, 1:200) for 1 h, following washing in PBS and blocking buffer, incubated with fluorophore-conjugated anti-rat secondary antibody (#A11006, Invitrogen, 1:1000). DNA were counterstained with DAPI. Cells were visualized under an Olympus BX61 upright microscope using a 20 × air objective. Fluorescence intensity was quantified using the MetaMorph Multi-Wavelength Cell Scoring Application using cells pooled from three independent experiments. Data were analyzed as follows: DAPI staining was used to define the region of the nucleus. Unstained cells were used to establish the level of background fluorescence, and BrdU-stained cells above this threshold were defined as BrdU-positive. Using the mask created by DAPI, the fluorescence intensity of BrdU (of BrdU-positive cells) was measured in the FITC channel.
For BrdU/p-H3 S10 staining, cells were plated in log phase on poly-L-lysine-coated glass coverslips. Cells were pulsed with 25 μM BrdU for 30 min and treated with 5 mM hydroxyurea for 6 h. After treatment, the cells were washed and allowed to recover in fresh media in the presence of 1 μg/mL nocodazole. The cells were fixed with 2% PFA/10 min at 4 °C and permeabilized with 100% methanol/5 min at -20 °C. The cells were treated with 3 M HCl/30 min to denature the DNA and blocked with 2% BSA/0.5% Triton-X-100 before overnight incubation with primary antibodies (1:1000 rabbit anti-pH3-S10 (Santa Cruz #sc-8656); 1:200 rat anti-BrdU (clone BU1/75 (ICR1); AbD Serotec #MCA2060)) and 2-h incubation with secondary antibodies conjugated to AlexaFluor-488 and AlexaFluor-568. The cells were stained with DAPI and mounted in ProLong Diamond Antifade (Molecular Probes) before they were imaged using a 20 × air objective on a Olympus BX61 upright microscope. Cell number and fluorescence intensity were quantified using the MetaMorph Multi-Wavelength Cell Scoring Application using cells pooled from three independent experiments. Data were analyzed and graphs were generated using GraphPad Prism (https://www.graphpad.com/scientific-software/prism/). Fluorescent images were prepared using Photoshop CS6 (https://www.adobe.com/products/photoshop.html).
Flow cytometry cell cycle analysis
Cells were plated and grown in 12-well plates overnight to log phase. Cells were pulsed with 25 μM BrdU for 20 min at 37 °C. The cells were washed with PBS × 2 and treated with media containing 5 mM hydroxyurea (Sigma-Aldrich) for 6 h at 37 °C. The HU was removed, the cells were washed with PBS × 2, and fresh media was added to allow recovery for 0, 4, 6, 8, and 12 h. After recovery, the cells were washed with PBS and harvested using 0.25% trypsin. The cells were fixed with 70% ethanol at -20 °C, pelleted at 1000 g, and rehydrated with 2% BSA/0.1% Tween20/PBS. The DNA was denatured with 3 M HCl for 30 min/RT, and the cells were blocked with 2% BSA/0.1% Tween20/PBS for 30 min/RT. BrdU was stained using a rat anti-BrdU antibody (1:200 rat α-BrdU (clone BU1/75; #MCA2060 BioRad)) for 1 h/RT, followed by incubation with 1:500 goat α-rat AlexaFluor-488 (Life Technologies) in the presence of RNAse for 2 h/RT. Equal numbers of cells were stained with 7-AAD (5ug/mL) and analyzed using a Becton Dickinson FACSCalibur flow cytometer and FlowJo software (https://www.flowjo.com/).
DNA fiber assay
Cells were plated in log phase in 24-well plates. The cells were pulsed with media containing 25 μM 5-chloro-2′-deoxyuridine (CldU) for 30 min at 37 °C. The media was then removed, and the cells were pulsed with 250 μM iododeoxyuridine (IdU) + /- 2.5 μMcamptothecin (CPT) for 30 min at 37 °C. The cells were washed with PBS, harvested with 0.25% trypsin, and suspended in PBS to a concentration of 200,000 cells/mL. 2 μL cells were plated on a glass slide and allowed to dry slightly. The cells were lysed with 10 μL spreading buffer (200 mM Tris pH 7.4, 50 mM EDTA, 0.5% SDS) for 3 min, and the slides were tilted at a 20–30° angle to allow the DNA to spread. Excess lysate was blotted off the bottom edge of the slide, and the slides were allowed to dry for 1 h at room temperature. The DNA was fixed with 3:1 methanol/acetic acid for 3 min at − 20 °C. The DNA was denatured with 2.5 M HCl for 60 min at room temperature. The slides were blocked with 2% BSA/0.1% Tween20/PBS, and incubated with a-BrdU antibodies specific to CldU and IdU (1:100 rat α-BrdU (clone BU1/75; #MCA2060 BioRad), 1:100 ms α-BrdU (clone B44; #347,580 BD Biosciences)) and secondary (1:100 goat α-rat AlexaFluor 488; 1:100 goat α-ms AlexaFluor 568 (Life Technologies)) antibodies in a humidified chamber at 37 °C with washes in 0.1% Tween20/PBS in between. The slides were mounted with Prolong Diamond Antifade (Molecular Probes) and imaged with a 40 × air objective using an Olympus BX61 upright microscope. Data were analyzed using ImageJ software using fibers pooled from three independent experiments. Fork speed was calculated using the estimation that 1 µm corresponds to 2.6 kb as reported in a previous study70.
Statistical analysis
Data were analyzed using Student’s t-test and one-way ANOVA with Tukey post-test using GraphPad Prism software (https://www.graphpad.com/scientific-software/prism/) and Microsoft Excel (https://www.microsoft.com/en-us/microsoft-365/excel). *** indicates P < 0.001, * indicates P < 0.05, n.s. indicates not significant (P > 0.05).
References
Zeman, M. K. & Cimprich, K. A. Causes and consequences of replication stress. Nat. Cell Biol. 16, 2–9 (2014).
Ciccia, A. & Elledge, S. J. The DNA damage response: Making it safe to play with knives. Mol. Cell 40, 179–204 (2010).
Blackford, A. N. & Jackson, S. P. ATM, ATR, and DNA-PK: The trinity at the heart of the DNA damage response. Mol. Cell 66, 801–817 (2017).
Jackson, S. P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Saldivar JC, Cortez D, Cimprich KA. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. (2017).
Shiloh, Y. & Ziv, Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14, 197–210 (2013).
Smith, J., Tho, L. M., Xu, N. & Gillespie, D. A. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 108, 73–112 (2010).
Marechal, A. & Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harbor Perspect. Biol. 5, 2 (2013).
Toledo, L., Neelsen, K. J. & Lukas, J. Replication catastrophe: When a checkpoint fails because of exhaustion. Mol. Cell 66, 735–749 (2017).
Kumagai, A., Lee, J., Yoo, H. Y. & Dunphy, W. G. TopBP1 activates the ATR-ATRIP complex. Cell 124, 943–955 (2006).
Lee, Y. C., Zhou, Q., Chen, J. & Yuan, J. RPA-binding protein ETAA1 Is an ATR activator involved in DNA replication stress response. Curr. Biol. CB 26, 3257–3268 (2016).
Haahr, P. et al. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat. Cell Biol. 18, 1196–1207 (2016).
Bass, T. E. et al. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat. Cell Biol. 18, 1185–1195 (2016).
Zou, L. & Elledge, S. J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 (2003).
Shiomi, Y. et al. Clamp and clamp loader structures of the human checkpoint protein complexes, Rad9-1-1 and Rad17-RFC. Genes Cells 7, 861–868 (2002).
Parrilla-Castellar, E. R., Arlander, S. J. & Karnitz, L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair 3, 1009–1014 (2004).
Venclovas, C. & Thelen, M. P. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 28, 2481–2493 (2000).
Lee, J., Kumagai, A. & Dunphy, W. G. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J. Biol. Chem. 282, 28036–28044 (2007).
Delacroix, S., Wagner, J. M., Kobayashi, M., Yamamoto, K. & Karnitz, L. M. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 21, 1472–1477 (2007).
Zou, L., Liu, D. & Elledge, S. J. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. U.S.A. 100, 13827–13832 (2003).
Chaudhury, I. & Koepp, D. M. Recovery from the DNA replication checkpoint. Genes 7, 2 (2016).
Zou, L., Cortez, D. & Elledge, S. J. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16, 198–208 (2002).
Ellison, V. & Stillman, B. Biochemical characterization of DNA damage checkpoint complexes: Clamp loader and clamp complexes with specificity for 5’ recessed DNA. PLoS Biol. 1, E33 (2003).
Liu, Q. et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 14, 1448–1459 (2000).
Zhao, H. & Piwnica-Worms, H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 21, 4129–4139 (2001).
Hanson, P. I. & Whiteheart, S. W. AAA+ proteins: Have engine, will work. Nat. Rev. Mol. Cell Biol. 6, 519–529 (2005).
Bao, S. et al. ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature 411, 969–974 (2001).
Post, S. et al. Phosphorylation of serines 635 and 645 of human Rad17 is cell cycle regulated and is required for G(1)/S checkpoint activation in response to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 98, 13102–13107 (2001).
Wang, X. et al. Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol. Cell 23, 331–341 (2006).
Medhurst, A. L. et al. ATR and Rad17 collaborate in modulating Rad9 localisation at sites of DNA damage. J. Cell Sci. 121, 3933–3940 (2008).
Wang, Q. et al. Rad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellular response to DNA double-strand breaks. EMBO J. 33, 862–877 (2014).
Luo, H., Cowen, L., Yu, G., Jiang, W. & Tang, Y. SMG7 is a critical regulator of p53 stability and function in DNA damage stress response. Cell Discov. 2, 15042 (2016).
Unterholzner, L. & Izaurralde, E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell 16, 587–596 (2004).
Nasif, S., Contu, L. & Muhlemann, O. Beyond quality control: The role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin. Cell Dev. Biol. 75, 78–87 (2018).
Fukuhara, N. et al. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol. Cell 17, 537–547 (2005).
Gardino, A. K. & Yaffe, M. B. 14-3-3 proteins as signaling integration points for cell cycle control and apoptosis. Semin. Cell Dev. Biol. 22, 688–695 (2011).
Nam, E. A. et al. Thr-1989 phosphorylation is a marker of active ataxia telangiectasia-mutated and Rad3-related (ATR) kinase. J. Biol. Chem. 286, 28707–28714 (2011).
Liu, S. et al. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol. Cell 43, 192–202 (2011).
Liu, S. et al. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 40, 10780–10794 (2012).
Olson, E., Nievera, C. J., Klimovich, V., Fanning, E. & Wu, X. RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. J. Biol. Chem. 281, 39517–39533 (2006).
Iyer, D. R. & Rhind, N. The intra-S checkpoint responses to DNA damage. Genes 8, 2 (2017).
Smits, V. A. & Gillespie, D. A. DNA damage control: Regulation and functions of checkpoint kinase 1. FEBS J. 282, 3681–3692 (2015).
Liu, Y. & Bodmer, W. F. Analysis of P53 mutations and their expression in 56 colorectal cancer cell lines. Proc. Natl. Acad. Sci. U.S.A. 103, 976–981 (2006).
Rodrigues, N. R. et al. p53 mutations in colorectal cancer. Proc. Natl. Acad. Sci. U.S.A. 87, 7555–7559 (1990).
Coletta, A. et al. Low-complexity regions within protein sequences have position-dependent roles. BMC Syst. Biol. 4, 43 (2010).
Ohnishi, T. et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell 12, 1187–1200 (2003).
Lee, J. & Dunphy, W. G. Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol. Biol. Cell 21, 926–935 (2010).
Hickson, I. et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Can. Res. 64, 9152–9159 (2004).
Charrier, J. D. et al. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J. Med. Chem. 54, 2320–2330 (2011).
Shiotani, B. & Zou, L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol. Cell 33, 547–558 (2009).
Jazayeri, A. et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 8, 37–45 (2006).
Yang, X. H. & Zou, L. Recruitment of ATR-ATRIP, Rad17, and 9-1-1 complexes to DNA damage. Methods Enzymol. 409, 118–131 (2006).
Iyer, D. R. & Rhind, N. Checkpoint regulation of replication forks: global or local?. Biochem. Soc. Trans. 41, 1701–1705 (2013).
Pommier, Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. Cancer 6, 789–802 (2006).
Merrick, C. J., Jackson, D. & Diffley, J. F. Visualization of altered replication dynamics after DNA damage in human cells. J. Biol. Chem. 279, 20067–20075 (2004).
Seiler, J. A., Conti, C., Syed, A., Aladjem, M. I. & Pommier, Y. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol. Cell. Biol. 27, 5806–5818 (2007).
Mailand, N., Bekker-Jensen, S., Bartek, J. & Lukas, J. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell 23, 307–318 (2006).
Zachos, G., Rainey, M. D. & Gillespie, D. A. Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J. 22, 713–723 (2003).
Peschiaroli, A. et al. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell 23, 319–329 (2006).
Mamely, I. et al. Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr. Biol. CB 16, 1950–1955 (2006).
Bartek, J. & Lukas, J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19, 238–245 (2007).
Shaltiel, I. A., Krenning, L., Bruinsma, W. & Medema, R. H. The same, only different—DNA damage checkpoints and their reversal throughout the cell cycle. J. Cell Sci. 128, 607–620 (2015).
Fukumoto, Y., Ikeuchi, M., Nakayama, Y. & Yamaguchi, N. The KYxxL motif in Rad17 protein is essential for the interaction with the 9-1-1 complex. Biochem. Biophys. Res. Commun. 477, 982–987 (2016).
Smits, V. A. J., Cabrera, E., Freire, R. & Gillespie, D. A. Claspin—checkpoint adaptor and DNA replication factor. FEBS J. 286, 441–455 (2019).
Wang, X. et al. Genomic instability and endoreduplication triggered by RAD17 deletion. Genes Dev. 17, 965–970 (2003).
Dungrawala, H. et al. The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol. Cell 59, 998–1010 (2015).
Shiotani, B. et al. Two distinct modes of ATR activation orchestrated by Rad17 and Nbs1. Cell Rep. 3, 1651–1662 (2013).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Mendez, J. & Stillman, B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612 (2000).
Jackson, D. A. & Pombo, A. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140, 1285–1295 (1998).
Acknowledgements
We thank Bert Vogelstein for HCT116 cells and Fred Bunz for DLD1 cells. We are grateful for Lee Zou and Stephanie Yazinski for their help with the DNA fiber assays, Ken Norman and Gang Liu for their help with microscopy studies, and Cheryl Zajd for her help with Flow cytometry. This work was supported by NIH/NIGMS grant R01GM112085 to YT.
Author information
Authors and Affiliations
Contributions
Y.T. conceived the study; Y.T., H.L., and K.H. designed the experiments, analyzed and assembled the data; H.L., K.H., W.Z., and W.J. performed the experiments; Y.T. and K.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ho, K., Luo, H., Zhu, W. et al. Critical role of SMG7 in activation of the ATR-CHK1 axis in response to genotoxic stress. Sci Rep 11, 7502 (2021). https://doi.org/10.1038/s41598-021-86957-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-86957-x
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.