Abstract
Meta-QTL (MQTL) analysis is a robust approach for genetic dissection of complex quantitative traits. Rice varieties adapted to non-flooded cultivation are highly desirable in breeding programs due to the water deficit global problem. In order to identify stable QTLs for major agronomic traits under water deficit conditions, we performed a comprehensive MQTL analysis on 563 QTLs from 67 rice populations published from 2001 to 2019. Yield and yield-related traits including grain weight, heading date, plant height, tiller number as well as root architecture-related traits including root dry weight, root length, root number, root thickness, the ratio of deep rooting and plant water content under water deficit condition were investigated. A total of 61 stable MQTLs over different genetic backgrounds and environments were identified. The average confidence interval of MQTLs was considerably refined compared to the initial QTLs, resulted in the identification of some well-known functionally characterized genes and several putative novel CGs for investigated traits. Ortho-MQTL mining based on genomic collinearity between rice and maize allowed identification of five ortho-MQTLs between these two cereals. The results can help breeders to improve yield under water deficit conditions.
Similar content being viewed by others
Introduction
Rice is the world’s most important staple food and it is an excellent model crop for plant genetic studies1. Considering climate change scenarios and increasing water deficits, rice breeding programs have invested significant efforts into producing new rice varieties suitable for growing under reduced water inputs2,3,4. Tolerance to water deficit is a highly complex trait controlled by quantitative trait loci (QTLs). QTL mapping based on bi-parental populations is strongly influenced by the choice of marker sets, parents, population size, population types and environments5,6,7,8,9 hampering the transfer of QTLs and associated markers across different breeding programs. A powerful approach to circumvent this issue is Meta-analysis of QTLs (MQTL), which compiles QTL data from independent studies, locations, years and genetic backgrounds in order to detect stable and reliable QTLs10,11,12. An additional benefit of this approach is the reduction of confidence intervals (CIs) in the MQTLs leading to improved genetic resolution for marker-assisted selection (MAS) and identification of candidate genes (CGs). Together, MQTL analysis may increase selection accuracy and efficiency, thus enhancing genetic gains in plants breeding programs5,9,13,14,15. Several MQTL studies for drought stress have been conducted in cereals such as wheat16, maize8,17, and barley5,18. While a recent rice MQTL study considered various traits under unstressed conditions19, relatively few reports address water deficit conditions in rice: MQTL studies by Swamy et al. and Trijatmiko et al. focused on yield integrating data from 15 and 13 experiments20,21, respectively, and Khowaja et al. and Yang et al. reported some MQTLs for plant height and heading date based on QTLs published until 2009 and 2011, respectively22,23.
In the current study, we conducted a comprehensive genome-wide meta-analysis on QTLs reported in the last two decades controlling yield and yield-related traits in rice under water deficit conditions including Yield (YLD), grain weight (GW), heading date (HD), plant height (PH), tiller number (TN) as well as some drought tolerance criteria. Moreover, considering the key role of root architecture in plant responses to water deficit, different root related traits including root dry weight (RDW), root length (RL), root thickness (RT), roots number (RN) and rate of deep rooting (RDR) were subjected to MQTL analyses. We further scanned refined intervals of resulting stable QTLs for CGs related to the aforementioned traits. Additionally, to evaluate transferability of information to other cereals, ortho-MQTLs were investigated based on genomic collinearity between rice and maize24. Results will be applicable to improve selection for yield potential, stability and performance under water deficit conditions in cereal breeding programs.
Results and discussion
Distribution of yield and yield-related QTLs under water deficit conditions on the rice genome
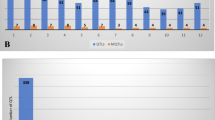
In order to discover consensus genomic regions associated with YLD, PH, TN, HD, GW, RDW, RL, RT, RN and RDR and some drought tolerance-related traits including drought response index (DRI), relative water content (RWC), canopy temperature (CT), leaf rolling (LR), leaf drying (LD) under water deficit conditions in rice, we compiled a total of 563 QTLs derived from 67 QTL populations (57 studies) reported from 2001 to 2019 (Table 1; Fig. 1A). The number of QTLs for each trait and their distribution on 12 rice chromosomes are shown in Fig. 1A,B. The chromosome 1 harbored the highest number of QTLs for all studied traits with 84 initial QTLs followed by chromosome 3 (62 QTLs) and chromosome 4 (58 QTLs). Whereas chromosome 10 harbored the lowest number of QTLs with 23 QTLs (Fig. 1B). The distribution of QTLs on different rice chromosomes with the highest number of QTLs on chromosomes 1 and 3 was similar to previous reports14,19,20. The number of QTLs on each chromosome exhibited a positive correlation (r = 0.73) with the length of chromosome.
The number and distribution of QTLs and MQTLs. (A) The number of initial QTLs used in the MQTL analysis for YLD, HD, PH, GW, TN, DT, RT, RL, RDR, RDW and RN. (B) The distribution of QTLs and MQTLs on the twelve chromosomes of rice shown in dark blue and purple, respectively. (C) The number of MQTLs for different traits on each chromosome of rice.
Among the studied traits, PH, RT and YLD had the highest number of QTLs with 89, 71 and 63 QTLs, respectively (Fig. 1A). The highest number of QTLs for PH was located on chromosome 1 with 24 QTLs, whereas chromosome 3 with 16 and 12 QTLs had the highest number of QTLs for HD and YLD, respectively. The QTLs for TN were mainly situated on chromosome 6 and QTLs for GW were evenly distributed on all chromosomes. For RWC, DRI, CT, LD and LR traits related to drought tolerance, 56 QTLs were distributed all over 12 chromosomes in rice with the highest number of QTLs on chromosome 2.
Detected MQTLs and their distribution on the rice genome
A total of 527 QTLs out of the 563 initial QTLs (93%) were successfully projected on the reference map (Table 2). Consequently, chromosome 1 had the highest (83) and chromosome 5 and 10 had the lowest (21) number of projected QTLs. The meta-analysis greatly summarized the total number of projected QTLs from 527 to 61 MQTLs (11.5%; Fig. 1B,C) supported by at least two QTLs deduced from different populations and considerably reduced the respective confidence intervals (CI) in comparison to the initial QTLs (Table 3). Therefore, MQTL analysis can efficiently confine the number of QTLs and narrow down the genomic regions controlling different traits19.
The number of MQTLs per chromosome ranged from two (chromosome 10) to nine (chromosomes 1 and 9) with an average of 5.08 MQTLs (Fig. 2; Table 3; Additional file 1). Chromosomes 1 and 9 with nine MQTLs and chromosomes 10 and 7 with two MQTLs had the highest and lowest number of MQTLs, respectively (Table 3; Additional file 1). There was a low correlation between the number of initial QTLs and the final number of MQTLs on each chromosome (r = 0.58).
Out of the total number of 61 MQTLs, we detected 10 MQTLs for YLD, five MQTLs for GW, seven MQTLs for HD, 10 MQTLs for PH, two MQTLs for TN, four MQTLs for DT, seven MQTLs for RDW, six MQTLs for RT, four MQTLs for RL, three MQTLs for RN and three MQTLs for RDR. These MQTLs were stable across different environments and genetic backgrounds. MQTL-PH7 and MQTL-RL2 involving the highest number of initial QTLs (6) were considered as the most stable QTLs (Table 3; Additional file 1). Among the identified MQTLs, four MQTLs for HD (MQTL-HD3, MQTL-HD4, MQTL-HD5 and MQTL-HD6) and three MQTLs for YLD (MQTL-YLD2, MQTL-YLD7 and MQTL-YLD10) overlapped with previously reported MQTLs under drought conditions in rice20,23. To the best of our knowledge this is the first MQTL analysis for GW, TN and DT in rice.
A total of 10 MQTLs were detected in the same chromosomal regions with similar yield and yield-related traits under well-water condition in rice19. This indicates the same loci might control aforementioned traits under both water deficit and well-water conditions (Additional file 2). They include five MQTLs for PH (MQTL-PH2, PH4, PH7, PH8 and PH9) on chromosomes 1, 2, 4, 7 and 8, two MQTLs for GW (MQTL-GW4 and GW5) on chromosomes 8 and 12, two MQTLs for HD (MQTL-HD1 and HD3) on chromosomes 3 and 5 and one MQTL for YLD (MQTL-YLD3) on chromosome 3 (Fig. 3; Additional file 2).
The distribution pattern of (A) functionally characterized genes on rice chromosomes, (B) gene density on rice chromosomes, (C) MQTLs under water deficit condition for different traits indicated in the color scale on the right side, (D) MQTLs under normal condition for different traits indicated in the color scale on the right side, (E) QTLs density, (F) SNP density shown in white to dark blue scale for the lowest to the highest density, (G) Structural variants (SV) density shown in white to dark red scale for the lowest to the highest density, (H) recombination density shown in white to dark red scale for the lowest to the highest density, (I) rice duplicated regions and rice syntenic regions with maize in light blue, (J) maize chromosomes with orthologous MQTLs with rice, (K) gene density on the maize chromosomes and (L) ortho-MQTLs between rice and maize. The outermost circle represents the rice genome in Mb.
The MQTL analysis considerably narrowed the CI allowing for exploration of a reduced number of candidate genes (CGs) for the investigated traits. The average CI was reduced from 15.57 cM in the initial QTLs to 5.48 cM in the MQTL with 65% of MQTLs having CI < 5 cM (Table 3). In 10 MQTLs, MQTL-GW4, HD4, HD7, PH3, PH5, YLD1, YLD7, DT1, DT3, RL4, RDW1 and RDW7, the CI was reduced to < 1 cM (Table 3). Therefore, MQTL analysis can significantly raise the accuracy of identification of CGs. All the annotated genes located within the CI of each MQTL and the most promising CGs based on their reported function in previous studies are reported in Additional file 3. Some functionally characterized genes such as GRAIN SIZE 2 (GS2), GRAIN WEIGHT 7 (GW7), Early heading date 1 (Ehd1), DWARF 10 (d10) and Grain number, plant height, heading date7 (Ghd7), OsPIN3t, OsSAUR45, and WEG1 were located within MQTL-GW1, GW3, HD6, PH2, PH8, RT1, RL4, RDW5, respectively, and OsAIR1 located at MQTL-RN2 and RDW2, and OsMGT1 located at MQTL-RT2 and RDW1, that are assumed to control the aforementioned traits. Putative novels CGs for each trait are discussed below. In addition, the positions of MQTLs on the rice genome were compared with the gene density, and densities of SNPs, structural variants (SV), recombination and functional variants, and the reported selective sweep regions25 (Fig. 3). Most of the detected MQTLs were located in sub-telomeric regions where generally the gene, SNP, SV and recombination densities are higher (Figs. 2, 3). This is consistent with previous results in barley, maize and rice15,19,26. The regions with high SV frequency could play an effective role in stress response27. A total of 13 MQTLs (MQTL-YLD3, YLD6, GW2, GW5, HD5, TN2, DT2, RT3, RT4, RL3, RL4, RDR3 and RDW6) were co-located with selective sweep regions reported by Huang et al. These MQTLs are likely effective for selection towards drought adaptation during rice breeding and domestication processes25. Five of these MQTLs including MQTL-YLD3, TN2, GW5, RT4 and RDW6 were also co-located with the position of reported functional variants25.
The investigation of collinear regions within the rice genome resulted in identification of five duplicated regions containing MQTLs for the same traits. MQTL-YLD2 and MQTL-YLD4 on chromosomes 2 and 4, and MQTL-YLD8 and MQTL-YLD9 on chromosomes 11 and 12 for yield, MQTL-RWD1 and MQTL-RWD2 on chromosomes 1 and 5, MQTL-RWD4 and MQTL-RWD5 on chromosomes 8 and 9, and MQTL-RN1 and MQTL-RN2 on chromosomes 1 and 5 for root-related traits are co-located at rice genome duplicated regions (Fig. 3). Duplicated genomic regions derived from common ancestors might contain paralogous genes with similar functions that can be considered as promising CGs controlling the trait28. Consequently, we carefully surveyed these regions for detecting possible paralogous CGs in the duplicated regions. In MQTL-RN2, we note the OsABIL3 or PP2C50 gene which has a key role in root architecture and response to drought stress by affecting ABA signaling: overexpression of this gene was reported to lead to the ABA insensitivity along with stomatal density and root architecture29. The paralogous gene Os01g0618200 encoding PP2C07 is also present at the duplicated region on chromosome 1 with MQTL-RN1 for the same root number trait. Moreover, at MQTL-YLD4 interval on chromosome 4, we detected GRAS23 that contributes to drought response in rice30, with paralogues HAM1 and HAM2 colocalizing with the duplicated genome on chromosome 2 with MQTL-YLD2 for the yield under drought stress.
MQTLs and CGs for grain weight
GW is one of the most important components of YLD in rice1,31 and it critically limits YLD during late season drought stress3,32,33. A total of five MQTLs were identified for GW (Table 3). MQTL-GW2 on chromosome 5 is the most stable MQTL for GW with the highest number of initial QTLs from four independent studies. Among the identified MQTLs for GW, MQTL-GW4 and GW5 on chromosomes 8 and 12 were located at the same region of GW MQTLs under well-water conditions19. Therefore, the same genes might control GW under both conditions. MQTL-GW5 and GW2 were co-located with selective sweep regions and functional variants were reported on the former. These MQTLs could be effective for selection towards drought adaptation25. The same source of favorable allele from ‘Tarom Molaei’ parent derived from two independent populations was detected in QTLs located at MQTL-GW3 (Additional file 4).
Some well-known genes controlling GW such as GS234 and GW735 were located within MQTL-GW1 and MQTL-GW3, respectively, suggesting that these genes may play the same role under water deficit conditions. The list of functionally characterized and novel CGs within each MQTL interval are reported (Additional file 3). For instance, the genomic region spanning MQTL-GW3 contains the CYP78A1336 and DEP231 genes that are reported to control grain size and YLD in rice. MQTL-GW4 on chromosome 8 and MQTL-GW5 on chromosome 12 contain Os08g0390000 encoding brassinosteroid receptor kinase (BRI1)31 and the OsVIL237 genes, respectively, which regulate grain size in rice.
MQTLs and CGs for heading date
It is well known that HD is highly correlated with YLD38 and drought adaptation39,40. We detected seven MQTLs for HD under water deficit conditions including two MQTLs on chromosomes 3 and 9 and one MQTL each on chromosomes 5, 10 and 12 (Table 3). MQTL-HD1 on chromosome 3 had the highest number of supporting QTLs with five QTLs from two independent studies (Table 3).
Among annotated genes within MQTL-HD1, MQTL-HD3 and MQTL-HD6 intervals, OsCCT1141, HBF142 and Ehd138,43,44, respectively, were identified as potential candidates for HD under water deficit conditions. OsCCT11 is considered as a positive regulator of heading date since RNAi-mediated downregulation of this gene delays HD41, while HBF1 is considered as a negative regulator of HD since mutation promotes flowering42. Among genes within the MQTL-HD3 interval, basic region/leucine zipper motif (bZIP), FT-like and circadian clock genes are promising candidates43,45,46. Another CG at this MQTL is OsHAPL1, known to prevent flowering under long-day conditions47. OsTrx148 and OsCCT3141 are potential candidates for MQTL-HD4. In MQTL-HD6, the BRD2 gene49 and the Ehd2 and Ehd1 genes38,43,45 are reported to modify flowering time in rice.
MQTLs and CGs for plant height
Since the Green Revolution, PH has been considered as a major target for YLD improvement50 and it also contributes to drought tolerance40. Among the studied traits, PH and YLD had the highest number of MQTLs; we identified 10 MQTLs for PH including three MQTLs on chromosome 1, two MQTLs on chromosome 2 and one MQTL each on chromosomes 3, 4, 7, 8 and 12. The MQTL-PH7 on chromosome 4 had the largest number of initial QTLs with six QTLs from four independent studies followed by MQTL-PH10 on chromosome 12 with three QTLs reported from three independent studies. These MQTLs are the most stable QTLs for PH under water deficit conditions.
The d1051 and Ghd752 genes are reported to regulate plant height in rice, and they are positioned within MQTL-PH2 and MQTL-PH8 genomic regions, respectively. MQTL-PH4 contains EP3/LP gene, whose mutant shows increased panicle size and PH in rice53. Conversely, mutations in OsKS2 and NAL154,55 at MQTL-PH7 and OsSEC3A56 at MQTL-PH6 decrease PH in rice. In MQTL-PH10, we detected Os12g0271600 that encodes BRI1 and the mutant alleles could act as a dwarfism gene50.
MQTLs and CGs for yield
The maintenance of YLD under drought condition is the ultimate goal in cereal breeding57,58. We identified 10 MQTLs for YLD consisting of two MQTLs on chromosome 12 and one MQTL each on chromosomes 1, 2, 3, 4, 6, 8, 10 and 11 (Table 3). Among them MQTL-YLD3 on chromosome 3 overlapped with a YLD MQTL identified under well-water conditions19. Therefore, the same genes might control YLD under both mentioned conditions at this position.
We detected some genes which affect the photosynthetic rate including Roc5 at MQTL-YLD259, UCL8 at MQTL-YLD360 and OsPTR6 at MQTL-YLD461 that might indirectly contribute to the final YLD. OsALMT7 is located at the MQTL-YLD2 interval with pleiotropic effects on YLD and panicle size62. TAC3 might indirectly regulate YLD through changing HD and tiller angle in rice at MQTL-YLD363. The most likely CGs at MQTL-YLD5 are Os06g0274500 which encodes SERK-like gene and BRI1-associated receptor kinase 1 (BAK1) that affects grain size and number in rice64. The OsNAC5 gene on MQTL-YLD8 is known to have a positive effect on YLD under drought condition65.
MQTLs and CGs for number of tillers
The number of fertile tillers is a major contributor to YLD and its alteration during drought stress can result in drought adaptation4,66,67,68. Tillering is a complex process and highly affected by environmental conditions66. We detected only two MQTLs on chromosomes 5 and 6 which were associated with TN (Table 3). In MQTL-TN2, we identified OsAID1 as a gene associated to TN regulation in rice69.
MQTLs and CGs for drought tolerance
Plant water content is highly affected by water deficit conditions and in turn can contribute to drought tolerance. Plant water content can be measured by different criteria including RWC, CT, LR, LD and DRI3,32,33,70,71. We identified four MQTLs for DT on chromosomes 1, 4, 8 and 11. MQTL-DT1 on chromosome 1 is the most stable MQTL related to rice water content under water deficit conditions with the highest number of initial QTLs (4) from three independent studies. Within the MQTL-DT1 and MQTL-DT4 intervals, we detected several CGs including OsBZ872 and Os6PGDH273 that contribute to abiotic stresses tolerance, respectively.
MQTLs and CGs for root architecture
Root architecture develops through dynamic processes that effectively contribute to water deficit adaptation allowing water and nutrient uptake from deep soil74,75. We studied five major traits related to root architecture including RDW, RL, RN, RT and RDR under water deficit conditions and identified 23 MQTLs including seven MQTLs for RDW, four MQTLs for RL, three MQTLs for RN, six MQTLs for RT and three MQTLs for RDR (Table 3). MQTL-RL2 had the highest number of initial QTLs (six QTLs from three independent studies) and it was considered as the most stable QTL for root architecture (Table 3). Interestingly, MQTL-YLD4 for YLD under water deficit conditions on chromosome 4 overlapped with MQTL-RDR2 and RL1 (Fig. 2).
Overlapping MQTLs for different root architecture traits included MQTL-RDW1 and MQTL-RT2 on chromosome 1, MQTL-RDW2 and MQTL-RN2 on chromosome 5 and MQTL-RN1 and MQTL-RT1 on chromosome 1, suggesting the possible existence of genes with pleiotropic effects on these traits. For example, the genomic region spanning MQTL-RN2 and MQTL-RDW2 harbors OsAIR1, a gene affecting root architecture and contributing to drought tolerance76. In the overlap region between MQTL-RT2 and MQTL-RDW1, noteworthy is OsMGT1 which was shown to affect root architecture during salinity stress77. MQTL-RT1 contains OsPIN3t78 and OsFBK179 genes that are reported to control root architecture under water deficit conditions. The SMOS1 gene within MQTL-RT4 determines root meristem size80, and the cZOGT381 gene within MQTL-RDR2 regulates root architecture. Additionally, AMTR1 (MQTL-RN2) affects root architecture under drought stress82.
The same source of favorable allele from ‘IRAT109’ parent derived from two independent populations was identified in QTLs located at MQTL-RL3 (Additional file 4). The FC1 gene83 on MQTL-RL1 controls root growth and might contribute to drought tolerance under water deficit conditions. Within MQTL-RL4, we detected a cluster of small auxin-up RNA (SAUR) genes. Over-expression of OsSAUR45 regulates root length and other related root traits84.
For RDW, we detected two co-located MQTLs including MQTL-RDW1 and RDW2 co-locating with OsMGT1 and OsAIR1genes, respectively. Additionally, WEG1 at MQTL-RDW5 is a novel gene that regulates root related traits85 and may keep the same role under water deficit conditions.
Ortho-MQTL mining
To investigate ortho-MQTLs for yield and yield-related traits under water deficit conditions between rice and maize as the two most important cereals with generally high water demand, the syntenic regions of all detected rice MQTLs in this study were compared with published maize MQTLs17,86,87. Comparative genomic analyses provide a valuable approach to transfer information across species and identify conserved genes19. Through synteny analysis between rice and maize, we uncovered 5 ortho-MQTLs including 4 ortho-MQTLs for YLD on chromosomes 2, 3, 4 and 8 and 1 ortho-MQTL for PH on chromosome 4 (Table 4; Fig. 3). The genes located at these syntenic regions were further investigated (Additional file 5; Fig. 4).
Comparative maps of ortho-MQTLs between rice and maize. (A) ortho-MQTL-PH7, (B) ortho-MQTL-YLD2, (C) ortho-MQTL-YLD3, (D) ortho-MQTL-YLD4 and (E) ortho-MQTL-YLD6. The chromosome number, genomic position in Mb and the original name of MQTLs are indicated. The orthologous genes in rice and maize are indicated in green color with the corresponding rice gene name.
MQTL-PH7 and MQTL-YLD4 on chromosome 4 of rice were co-linear with mQTL_PEH_10 and mQTL_GY_10b on chromosome 10 in maize, respectively (Table 4). Three rice MQTLs including MQTL-YLD2, YLD3 and YLD6 on chromosomes 2, 3 and 8, respectively, were situated in syntenic regions of maize yield MQTLs on chromosome 5, 1 and 6, respectively (Table 4; Fig. 3).
The orthologous genes located at these ortho-MQTLs in both rice and maize are shown in the Additional file 5 and Fig. 4. The rice genomic region subtending MQTL-PH7 harbors the NAL1 gene as a regulator of PH55: we identified the ortholog of this gene (Zm00001d026296) in the maize ortho-MQTL. In the syntenic region of rice MQTL-YLD2 on chromosome 5 of maize, there were two MQTLs (mQTL_GY_586, MQTL5.717) containing the orthologs of OsALMT7 and SID1 genes (Zm00001d017571 and Zm00001d017560), known to affect YLD in rice62,88. The orthologous gene of TAC3 in maize (Zm00001d033857) in the syntenic region of MQTL-YLD3 in maize (mQTL_GY_1b) regulates tiller angle that might affect YLD under water deficit conditions63. In the syntenic region of rice MQTL-YLD6, there was a MQTL (MQTL6.117) on chromosome 6 of maize. This rice MQTL contains Ehd3 gene regulating flowering and consequently YLD38 and its orthologous (Zm00001d035008) was detected in its ortho-MQTL in maize, likely to have similar functions. This approach provided better understanding of genes controlling investigated traits under water deficit conditions with similar evolutionary history and conserved function between these cereals. These results can benefits breeders by tracing CGs and using marker-assisted selection in breeding programs of cereals under water deficit conditions.
Conclusions
Through MQTL analysis this study provides an overview of genomic regions controlling YLD, yield-related traits, root architecture and plant water content including GW, HD, PH, TN, RDW, RL, RT, RN, RDR and DT under water deficit conditions in rice. This approach is useful in overcoming some limitations of single QTL mapping studies on different genetic backgrounds and environments and greatly facilitates the identification of CGs and robust flanking markers for MAS in breeding programs. The results offer a framework for future genetic studies of yield under drought conditions, e.g. through fine mapping, positional cloning, producing chromosome substitution lines, as well as validation of CGs by genome editing using Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and similar approaches. This study also demonstrates the value of ortho-MQTL mining among evolutionarily close crop species for identification of genomic regions and CGs controlling complex quantitative traits.
Materials and methods
QTL studies used for MQTL analysis
An exhaustive bibliographic review was carried out on rice QTLs related to yield and yield-related traits under water deficit conditions published from 2001 to 2019. All QTL studies except those lacking proper genetic map information or QTL-related information were used in the MQTL analysis. Consequently, a total of 563 QTLs for YLD, PH, TN, HD, GW, RDW, RL, RT, RN, RDR and traits related to water content of plant under water deficit conditions including DRI, RWC, CT, LR and LD from 67 biparental rice populationsextracted from 57 studies, including all the five major subpopulations of rice—Indica, tropical japonica, temperate japonica, aus, aromatic and also one wild species O. rufipogon IRGC 105491 and landraces, were implemented for the MQTL analysis (Table 1). The size of mapping populations varied from 72 to 4560 progenies of various types including 7 DH, 17 F2, 15 BC and 28 RIL populations phenotyped in different locations and years (Table 1). Moreover, 56 QTLs related to water content of plant under water deficit conditions including DRI, RWC, CT, LR, LD were subjected to MQTL analysis and resulting MQTLs were reported as drought tolerance (DT). Detailed information on the used QTLs including parents, population type and size, number of markers, map density and evaluated traits are reported in Table 1.
Projection of QTLs on the reference map
A rice reference map of Wu et al. which is the most comprehensive available genetic map integrated from six identified and saturated maps in rice was chosen based on its high marker density and inclusion of different marker types including SSR, RFLP and AFLP markers. It consists of 6969 markers with an average distance of 0.25 cM between markers, and the average chromosome length is 147.65 cM for a total length of 1771.8 cM89. In order to incorporate SNP markers of those initial QTLs with SNP markers (Table 1) into the reference map, we applied our previous approach19 in which the genomic position of SNP markers on the rice genome were determined and in consequence the closest markers based on the physical position were used to project them on the reference map.
QTL position, CI, proportion of phenotypic variance (R2), log of odds ratio (LOD score), additive effects and favorable alleles were compiled for each QTL in the 67 populations (Additional file 4). In order to calculate 95% CI for QTLs, we used the following formulas: CI = 530/(N × R2) for BC and F2 lines, CI = 287/(N × R2) for DH lines and CI = 163/(N × R2) for RLLs lines90,91, where N is the population size and R2 is the proportion of phenotypic variance of the QTL. MQTL analysis was carried out using BioMercator V4.211,92.
Meta-QTL analysis and ortho-MQTL mining
The MQTL analysis was conducted on integrated and re-positioned QTLs on the reference map using BioMercator V4.211,12,92. The best model of MQTLs was chosen according to the prevalent value among Akaike Information Criterion (AIC), corrected Akaike Information Criterion (AICc and AIC3), Bayesian Information Criterion (BIC) and Average Weight of Evidence (AWE) criteria. Therefore, the consensus QTL from the best model was reported as a “real” QTL/MQTL12,92. Considering the known correlations among RWC, CT, LR, LD and DRI3,32,33,70, the respective QTLs were analyzed together as one trait named as DT in BioMercator V4.2.12,92. Mapchart V.2.32 software93 was used to show the MQTLs and related QTLs on the reference map.
The distribution of MQTLs on the rice genome (IRGSP-1.0) compared to the position of centromeric and telomeric regions and the gene density along each chromosome were surveyed and shown by heatmap using pheatmap and R94,95. Centromere position, gene density, SNP and structural variations (SV) and recombination rate density of each chromosome, as well as rice genome duplications were retrieved from EnsemblPlants (https://plants.ensembl.org/index.html) database. Additionally, the position of identified MQTLs were compared with selective sweep regions and functional variants in coding regions with strong alteration in allele frequency between cultivated and wild rice reported by Huang et al.25. The distribution of aforementioned factors, number of MQTL under water deficit conditions and number of MQTLs under well-water conditions19 over the rice genome were shown by using Circos96.
To detect ortho-MQTLs between rice and maize, syntenic regions between the two species were identified by using EnsemblPlants database97. MQTLs identified for yield and yield-related traits under drought conditions in maize17,86,87 were compared with MQTLs detected for similar traits in our study.
Identification of candidate genes
CGs related to YLD, PH, TN, HD, GW, RDW, RL, RT, RN, RDR and DT traits located in the CI of each detected MQTL were investigated on the rice genome (IRGSP-1.0) using EnsemblPlants and Gramene (http://archive.gramene.org/qtl/). In case of flanking markers without genomic positions, the closest markers were applied for detecting the genomic coordinates of MQTL. Gene annotations within MQTL genomic regions were carefully explored by EnsemblPlants (https://plants.ensembl.org/index.html) and FunRiceGenes (https://funricegenes.github.io/)98 databases.
Data availability
The relevant data and additional information are available in the supplementary files and also from the corresponding author on reasonable request.
Abbreviations
- AIC:
-
Akaike information content
- AICs:
-
AIC correction
- AIC3:
-
AIC 3 candidate models
- AWE:
-
Average weight of evidence
- BIC:
-
Bayesian information criterion
- CGs:
-
Candidate genes
- CT:
-
Canopy temperature
- CI:
-
Confidence interval
- DRI:
-
Drought response index
- DT:
-
Drought tolerance
- GW:
-
Grain weight
- HD:
-
Heading date
- LOD:
-
The log of odds ratio
- LR:
-
Leaf rolling
- LD:
-
Leaf drying
- MQTL:
-
Meta-QTL
- MAS:
-
Marker-assisted selection
- PH:
-
Plant height
- QTL:
-
Quantitative trait loci
- R2 :
-
The proportion of phenotype variance
- RWC:
-
Relative water content
- RDW:
-
Root dry weight
- RL:
-
Root length
- RT:
-
Root thickness
- RN:
-
Roots number
- RDR:
-
Rate of deep rooting
- TN:
-
Tiller number
- YLD:
-
Yield
References
Xing, Y. & Zhang, Q. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442 (2010).
Zou, G. et al. Grain yield responses to moisture regimes in a rice population: Association among traits and genetic markers. Theor. Appl. Genet. 112(1), 106–113 (2005).
Sellamuthu, R., Liu, G. F., Ranganathan, C. B. & Serraj, R. Genetic analysis and validation of quantitative trait loci associated with reproductive-growth traits and grain yield under drought stress in a doubled haploid line population of rice (Oryza sativa L.). Field Crops Res. 124(1), 46–58 (2011).
Sandhu, N., Jain, S., Kumar, A., Mehla, B. S. & Jain, R. Genetic variation, linkage mapping of QTL and correlation studies for yield, root, and agronomic traits for aerobic adaptation. BMC Genet. 14(1), 104 (2013).
Li, W. T. et al. Meta-analysis of QTL associated with tolerance to abiotic stresses in barley. Euphytica 189(1), 31–49 (2013).
Van, K. & McHale, L. K. Meta-analyses of QTLs associated with protein and oil contents and compositions in soybean [Glycine max (L.) Merr] .seed. Int. J. Mol. Sci. 18(6), 1180 (2017).
Lei, L. et al. Genetic dissection of rice (Oryza sativa L.) tiller, plant height, and grain yield based on QTL mapping and metaanalysis. Euphytica 214(7), 109 (2018).
Zhao, X., Peng, Y., Zhang, J., Fang, P. & Wu, B. Identification of QTLs and meta-QTLs for seven agronomic traits in multiple maize populations under well-watered and water-stressed conditions. Crop Sci. 58(2), 507–520 (2018).
Izquierdo, P. et al. Meta-QTL analysis of seed iron and zinc concentration and content in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 131(8), 1645–1658 (2018).
Goffinet, B. & Gerber, S. Quantitative trait loci: A meta-analysis. Genetics 155(1), 463–473 (2000).
Arcade, A. et al. BioMercator: Integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20(14), 2324–2326 (2004).
Sosnowski, O., Charcosset, A. & Joets, J. BioMercator V3: An upgrade of genetic map compilation and quantitative trait loci meta-analysis algorithms. Bioinformatics 28(15), 2082–2083 (2012).
Zhang, L. Y. et al. Genomic distribution of quantitative trait loci for yield and yield-related traits in common wheat. J. Integr. Plant Biol. 52(11), 996–1007 (2010).
Swamy, B. M. & Sarla, N. Meta-analysis of yield QTLs derived from inter-specific crosses of rice reveals consensus regions and candidate genes. Plant Mol. Biol. Rep. 29(3), 663–680 (2011).
Khahani, B., Tavakol, E. & Shariati, V. Genome-wide meta-analysis on yield and yield-related QTLs in barley (Hordeum vulgare L.). Mol. Breed. 39(4), 56 (2019).
Acuña-Galindo, M. A., Mason, R. E., Subramanian, N. K. & Hays, D. B. Meta-analysis of wheat QTL regions associated with adaptation to drought and heat stress. Crop Sci. 55(2), 477–492 (2015).
Semagn, K. et al. Meta-analyses of QTL for grain yield and anthesis silking interval in 18 maize populations evaluated under water-stressed and well-watered environments. BMC Genom. 14(1), 313 (2013).
Zhang, X., Shabala, S., Koutoulis, A., Shabala, L. & Zhou, M. Meta-analysis of major QTL for abiotic stress tolerance in barley and implications for barley breeding. Planta 245(2), 283–295 (2017).
Khahani, B., Tavakol, E., Shariati, V. & Fornara, F. Genome wide screening and comparative genome analysis for Meta-QTLs, ortho-MQTLs and candidate genes controlling yield and yield-related traits in rice. BMC Genom. 21, 1–24 (2020).
Swamy, B. M., Vikram, P., Dixit, S., Ahmed, H. & Kumar, A. Meta-analysis of grain yield QTL identified during agricultural drought in grasses showed consensus. BMC Genom. 12(1), 319 (2011).
Trijatmiko, K. R. et al. Meta-analysis of quantitative trait loci for grain yield and component traits under reproductive-stage drought stress in an upland rice population. Mol. Breed. 34(2), 283–295 (2014).
Khowaja, F. S., Norton, G. J., Courtois, B. & Price, A. H. Improved resolution in the position of drought-related QTLs in a single mapping population of rice by meta-analysis. BMC Genom. 10(1), 276 (2009).
Yang, L. et al. QTL mapping for heading date, leaf area and chlorophyll content under cold and drought stress in two related recombinant inbred line populations (Japonica rice) and meta-analysis. Plant Breed. 137(4), 527–545 (2018).
Hirsch, C. N. et al. Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26(1), 121–135 (2014).
Huang, X. et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 490(7421), 497 (2012).
Martinez, A. K. et al. Yield QTLome distribution correlates with gene density in maize. Plant Sci. 242, 300–309 (2016).
Fuentes, R. R. et al. Structural variants in 3000 rice genomes. Genome Res. 29(5), 870–880 (2019).
Pont, C. et al. Paleogenomics: Reconstruction of plant evolutionary trajectories from modern and ancient DNA. Genome Biol. 20(1), 29 (2019).
Li, C., Shen, H., Wang, T. & Wang, X. ABA regulates subcellular redistribution of OsABI-LIKE2, a negative regulator in ABA signaling, to control root architecture and drought resistance in Oryza sativa. Plant Cell Physiol. 56(12), 2396–2408 (2015).
Xu, K. et al. OsGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes. BMC Plant Biol. 15(1), 1–3 (2015).
Huang, R. et al. Genetic bases of rice grain shape: So many genes, so little known. Trends Plant Sci. 18(4), 218–226 (2013).
Subashri, M. et al. Trait identification and QTL validation for reproductive stage drought resistance in rice using selective genotyping of near flowering RILs. Euphytica 166(2), 291–305 (2009).
Prince, S. J., Beena, R., Gomez, S. M., Senthivel, S. & Babu, R. C. Mapping consistent rice (Oryza sativa L.) yield QTLs under drought stress in target rainfed environments. Rice 8(1), 25 (2015).
Hu, J. et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant. 8(10), 1455–1465 (2015).
Wang, S. et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47(8), 949–954 (2015).
Xu, F. et al. Variations in CYP78A13 coding region influence grain size and yield in rice. Plant Cell Environ. 38(4), 800–811 (2015).
Yang, J. et al. Chromatin interacting factor OsVIL2 increases biomass and rice grain yield. Plant Biotechnol. J. 17(1), 178–187 (2019).
Hori, K., Matsubara, K. & Yano, M. Genetic control of flowering time in rice: Integration of Mendelian genetics and genomics. Theor. Appl. Genet. 129(12), 2241–2252 (2016).
Tavakol, E. et al. Genetic dissection of heading date and yield under Mediterranean dry climate in barley (Hordeum vulgare L.). Euphytica 212, 343–353 (2016).
Xu, Y. et al. Further QTL mapping for yield component traits using introgression lines in rice (Oryza sativa L.) under drought field environments. Euphytica 214(2), 33 (2018).
Zhang, L. et al. Three CCT domain-containing genes were identified to regulate heading date by candidate gene-based association mapping and transformation in rice. Sci. Rep. 5, 7663 (2015).
Brambilla, V. et al. Antagonistic transcription factor complexes modulate the floral transition in Rice. Plant Cell 29(11), 2801–2816 (2017).
Tsuji, H., Taoka, K.-I. & Shimamoto, K. Regulation of flowering in rice: Two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 14(1), 45–52 (2011).
Galbiati, F. et al. Hd3a, RFT1 and Ehd1 integrate photoperiodic and drought stress signals to delay the floral transition in rice. Plant Cell Environ. 39(9), 1982–1993 (2016).
Brambilla, V. & Fornara, F. Molecular control of flowering in response to day length in rice. J Integr. Plant Biol. 55(5), 410–418 (2013).
Brambilla, V., Gomez-Ariza, J., Cerise, M. & Fornara, F. The importance of being on time: Regulatory networks controlling photoperiodic flowering in cereals. Front. Plant Sci. 8, 665 (2017).
Zhu, S. et al. The OsHAPL1-DTH8-Hd1 complex functions as the transcription regulator to repress heading date in rice. J. Exp. Bot. 68(3), 553–568 (2017).
Choi, S. C. et al. Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date3. Plant Physiol. 164(3), 1326–1337 (2014).
Liu, X. et al. Brassinosteroid (BR) biosynthetic gene lhdd10 controls late heading and plant height in rice (Oryza sativa L.). Plant Cell Rep. 35(2), 357–368 (2016).
Sakamoto, T. & Matsuoka, M. Identifying and exploiting grain yield genes in rice. Curr. Opin. Plant Biol. 11(2), 209–214 (2008).
Yano, K. et al. Isolation of a novel lodging resistance QTL gene involved in strigolactone signaling and its pyramiding with a QTL gene involved in another mechanism. Mol. Plant. 8(2), 303–314 (2015).
Weng, X. et al. Ghd7 is a central regulator for growth, development, adaptation and responses to biotic and abiotic stresses. Plant Physiol. 20, 20 (2014).
Li, M. et al. Mutations in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice. Plant Biotechnol. J. 9, 1002–1013 (2011).
Ji, S. H., Gururani, M. A., Lee, J. W., Ahn, B. O. & Chun, S. C. Isolation and characterisation of a dwarf rice mutant exhibiting defective gibberellins biosynthesis. Plant Biol. 16(2), 428–439 (2014).
Qi, J. et al. Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 147(4), 1947–1959 (2008).
Ma, J. et al. Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. J. Exp. Bot. 69(5), 1051–1064 (2018).
Solis, J. et al. Genetic mapping of quantitative trait loci for grain yield under drought in rice under controlled greenhouse conditions. Front. Chem. 5, 129 (2018).
Bilgrami, S. S. et al. Evaluation of agro-morphological traits related to grain yield of Iranian wheat genotypes in drought-stress and normal irrigation conditions. Aust. J. Crop Sci. 12(05), 738–748 (2018).
Zou, L. P. et al. Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice. Plant Physiol. 156(3), 1589–1602 (2011).
Zhang, J. P. et al. MiR408 regulates grain yield and photosynthesis via a phytocyanin protein. Plant Physiol. 175(3), 1175–1185 (2017).
Fang, Z. et al. The rice peptide transporter OsNPF7.3 is induced by organic nitrogen, and contributes to nitrogen allocation and grain yield. Front. Plant Sci. 8, 1338 (2017).
Heng, Y. et al. OsALMT7 maintains panicle size and grain yield in rice by mediating malate transport. Plant Cell 30(4), 889–906 (2018).
Dong, H. et al. A novel tiller angle gene, TAC3, together with TAC1 and D2 largely determine the natural variation of tiller angle in rice cultivars. PLoS Genet. 12(11), e1006412 (2016).
Yuan, H. et al. 08SG2/OsBAK1 regulates grain size and number, and functions differently in Indica and Japonica backgrounds in rice. Rice 10(1), 1–2 (2017).
Jeong, J. S. et al. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 11(1), 101–114 (2013).
Hussien, A. et al. Genetics of tillering in rice and barley. Plant Genome 7, 1 (2014).
Tavakol, E. et al. The barley Uniculme4 gene encodes a BLADE-ON-PETIOLE-like protein that controls tillering and leaf patterning. Plant Physiol. 168(1), 164–174 (2015).
Bilgrami, S. S. et al. Detection of genomic regions associated with tiller number in Iranian bread wheat under different water regimes using genome-wide association study. Sci. Rep. 10(1), 1–17 (2020).
Zhu, Q. H., Ramm, K., Shivakkumar, R., Dennis, E. S. & Upadhyaya, N. M. The ANTHER INDEHISCENCE1 gene encoding a single MYB domain protein is involved in anther development in rice. Plant Physiol. 135(3), 1514–1525 (2004).
Manickavelu, A., Nadarajan, N., Ganesh, S., Gnanamalar, R. & Babu, R. C. Drought tolerance in rice: Morphological and molecular genetic consideration. Plant Growth Regul. 50(2–3), 121–138 (2006).
Srinivasan, S. et al. QTLs linked to leaf epicuticular wax, physio-morphological and plant production traits under drought stress in rice (Oryza sativa L.). Plant Growth Regul. 56(3), 245–256 (2008).
Paul, A., Dasgupta, P., Roy, D. & Chaudhuri, S. Comparative analysis of Histone modifications and DNA methylation at OsBZ8 locus under salinity stress in IR64 and Nonabokra rice varieties. Plant Mol. Biol. 95(1–2), 63–88 (2017).
Hou, F. Y., Huang, J., Yu, S. L. & Zhang, H. S. The 6-phosphogluconate dehydrogenase genes are responsive to abiotic stresses in rice. J. Integr. Plant Biol. 49(5), 655–663 (2007).
Courtois, B. et al. Rice root genetic architecture: Meta-analysis from a drought QTL database. Rice 2(2–3), 115–128 (2009).
Darzi-Ramandi, H. et al. Detection of consensus genomic regions associated with root architecture of bread wheat on groups 2 and 3 chromosomes using QTL meta-analysis. Aust. J. Crop Sci. 11(7), 777 (2017).
Hwang, S. G., Park, H. M., Han, A. R. & Jang, C. S. Molecular characterization of Oryza sativa arsenic-induced RING E3 ligase 1 (OsAIR1): Expression patterns, localization, functional interaction, and heterogeneous overexpression. J. Plant Physiol. 191, 140–148 (2016).
Chen, Z. C. et al. A magnesium transporter OsMGT1 plays a critical role in salt tolerance in rice. Plant Physiol. 174(3), 1837–1849 (2017).
Zhang, Q. et al. The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance. Plant J. 72(5), 805–816 (2012).
Borah, P. et al. Analysis of drought-responsive signalling network in two contrasting rice cultivars using transcriptome-based approach. Sci. Rep. 7, 42131 (2017).
Li, J. et al. SHOEBOX modulates root meristem size in rice through dose-dependent effects of gibberellins on cell elongation and proliferation. PLoS Genet. 11(8), e1005464 (2015).
Kudo, T., Makita, N., Kojima, M., Tokunaga, H. & Sakakibara, H. Cytokinin activity of cis-zeatin and phenotypic alterations induced by overexpression of putative cis-zeatin-O-glucosyltransferase in rice. Plant Physiol. 160(1), 319–331 (2012).
Kothari, K. S., Dansana, P. K., Giri, J. & Tyagi, A. K. Rice stress associated protein 1 (OsSAP1) interacts with aminotransferase (OsAMTR1) and pathogenesis-related 1a protein (OsSCP) and regulates abiotic stress responses. Front. Plant Sci. 7, 1057 (2016).
Minakuchi, K. et al. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 51(7), 1127–1135 (2010).
Xu, Y. X. et al. The small auxin-up RNA OsSAUR45 affects auxin synthesis and transport in rice. Plant Mol. Biol. 94(1–2), 97–107 (2017).
Lucob Agustin, N. et al. WEG1, which encodes a cell wall hydroxyproline rich glycoprotein, is essential for parental root elongation controlling lateral root formation in rice. Physiol. Plant. 20, 20 (2020).
Almeida, G. D. et al. QTL mapping in three tropical maize populations reveals a set of constitutive and adaptive genomic regions for drought tolerance. Theor. Appl. Genet. 126(3), 583–600 (2013).
Almeida, G. D. et al. Molecular mapping across three populations reveals a QTL hotspot region on chromosome 3 for secondary traits associated with drought tolerance in tropical maize. Mol. Breed. 34(2), 701–715 (2014).
Deng, L. et al. Suppressor of rid1 (SID1) shares common targets with RID1 on florigen genes to initiate floral transition in rice. PLoS Genet. 13(2), e1006642 (2017).
Wu, Y. et al. Quantitative trait loci identification and meta-analysis for rice panicle-related traits. Mol. Genet. Genom. 291(5), 1927–1940 (2016).
Darvasi, A. & Soller, M. A simple method to calculate resolving power and confidence interval of QTL map location. Behav. Genet. 27(2), 125–132 (1997).
Guo, B. et al. QTLs associated with resistance to soybean cyst nematode in soybean: Meta-analysis of QTL locations. Crop Sci. 46(2), 595–602 (2006).
Veyrieras, J. B., Goffinet, B. & Charcosset, A. MetaQTL: A package of new computational methods for the meta-analysis of QTL mapping experiments. BMC Bioinform. 8(1), 49 (2007).
Voorrips, R. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 93(1), 77–78 (2002).
Kolde, R. pheatmap: Pretty Heatmaps. R package version 0.7. 7. (2013).
R Core Team. R: A Language and Environment For Statistical Computing. R Foundation for Statistical Computing.
Krzywinski, M. et al. Circos: An information aesthetic for comparative genomics. Genome Res. 19(9), 1639–1645 (2009).
Bolser, D. M., Staines, D. M., Pritchard, E. & Kersey, P. Ensembl plants: Integrating tools for visualizing, mining, and analyzing plant genomics data. Plant Bioinform. 20, 115–140 (2016).
Yao, W., Li, G., Yu, Y. & Ouyang, Y. funRiceGenes dataset for comprehensive understanding and application of rice functional genes. Giga Sci. 7(1), gix119 (2017).
Moncada, P. et al. Quantitative trait loci for yield and yield components in an Oryza sativa× Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor. Appl. Genet. 102(1), 41–52 (2001).
Kamoshita, A. et al. Mapping QTLs for root morphology of a rice population adapted to rainfed lowland conditions. Theor. Appl. Genet. 104(5), 880–893 (2002).
Price, A. H., Steele, K. A., Moore, B. J. & Jones, R. G. Upland rice grown in soil-filled chambers and exposed to contrasting water-deficit regimes: II. Mapping quantitative trait loci for root morphology and distribution. Field Crops Res. 76(1), 25–43 (2002).
Courtois, B. et al. Locating QTLs controlling constitutive root traits in the rice population IAC 165× Co39. Euphytica 134(3), 335–345 (2003).
Babu, R. C. et al. Genetic analysis of drought resistance in rice by molecular markers. Crop Sci. 43(4), 1457–1469 (2003).
Cui, K. et al. Molecular dissection of the genetic relationships of source, sink and transport tissue with yield traits in rice. Theor. Appl. Genet. 106(4), 649–658 (2003).
Kobayashi, S., Fukuta, Y., Sato, T., Osaki, M. & Khush, G. Molecular marker dissection of rice (Oryza sativa L.) plant architecture under temperate and tropical climates. Theor. Appl. Genet. 107(8), 1350–1356 (2003).
Zheng, B. S. et al. Mapping QTLs and candidate genes for rice root traits under different water-supply conditions and comparative analysis across three populations. Theor. Appl. Genet. 107(8), 1505–1515 (2003).
Cui, K., Peng, S., Ying, Y., Yu, S. & Xu, C. Molecular dissection of the relationships among tiller number, plant height and heading date in rice. Plant Prod. Sci. 7(3), 309–318 (2004).
Lanceras, J. C., Pantuwan, G., Jongdee, B. & Toojinda, T. Quantitative trait loci associated with drought tolerance at reproductive stage in rice. Plant Physiol. 135(1), 384–399 (2004).
Li, Z. et al. QTL mapping of root traits in a doubled haploid population from a cross between upland and lowland japonica rice in three environments. Theor. Appl. Genet. 110(7), 1244–1252 (2005).
Gomez, M. S. et al. Mapping QTLs linked to physio-morphological and plant production traits under drought stress in rice (Oryza sativa L.) in the target environment. Am. J. Biochem. Biotechnol. 2(4), 161–169 (2006).
Horii, H., Nemoto, K., Miyamoto, N. & Harada, J. Quantitative trait loci for adventitious and lateral roots in rice. Plant Breed. 125(2), 198–200 (2006).
Steele, K. A., Price, A. H., Shashidhar, H. E. & Witcombe, J. R. Marker-assisted selection to introgress rice QTLs controlling root traits into an Indian upland rice variety. Theor. Appl. Genet. 112(2), 208–221 (2006).
Yue, B. et al. Genetic basis of drought resistance at reproductive stage in rice: Separation of drought tolerance from drought avoidance. Genetics 172(2), 1213–1228 (2006).
Ikeda, H., Kamoshita, A. & Manabe, T. Genetic analysis of rooting ability of transplanted rice (Oryza sativa L.) under different water conditions. J. Exp. Bot. 58(2), 309–318 (2007).
Lin, M. H. et al. Tagging rice drought-related QTL with SSR DNA markers. Crop Environ. Bioinform. 4(1), 65–76 (2007).
Kumar, R., Venuprasad, R. & Atlin, G. N. Genetic analysis of rainfed lowland rice drought tolerance under naturally-occurring stress in eastern India: Heritability and QTL effects. Field Crops Res. 103(1), 42–52 (2007).
Bernier, J., Kumar, A., Ramaiah, V., Spaner, D. & Atlin, G. A large-effect QTL for grain yield under reproductive-stage drought stress in upland rice. Crop Sci. 47(2), 507–516 (2007).
Liu, L. et al. Localization of QTL for basal root thickness in japonica rice and effect of marker-assisted selection for a major QTL. Euphytica 164(3), 729–737 (2008).
Qu, Y. et al. Mapping QTLs of root morphological traits at different growth stages in rice. Genetica 133(2), 187–200 (2008).
Uga, Y., Okuno, K. & Yano, M. QTLs underlying natural variation in stele and xylem structures of rice root. Breed. Sci. 58(1), 7–14 (2008).
Yue, B., Xue, W., Luo, L. & Xing, Y. Identification of quantitative trait loci for four morphologic traits under water stress in rice (Oryza sativa L.). J. Genet. Genom. 35(9), 569–575 (2008).
Zheng, B., Yang, L., Mao, C., Huang, Y. & Wu, P. Comparison of QTLs for rice seedling morphology under different water supply conditions. J. Genet. Genom. 35(8), 473–484 (2008).
Gomez, S. M. et al. Molecular mapping and location of QTLs for drought-resistance traits in indica rice (Oryza sativa L.) lines adapted to target environments. Acta Physiol. Plant. 32(2), 355–364 (2010).
Li, J. et al. Development of upland rice introgression lines and identification of QTLs for basal root thickness under different water regimes. J. Genet. Genom. 38(11), 547–556 (2011).
Srividya, A. et al. Molecular mapping of QTLs for drought related traits at seedling stage under PEG induced stress conditions in rice. Am. J. Plant Sci. 2(02), 190 (2011).
Uga, Y., Okuno, K. & Yano, M. Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J. Exp. Bot. 62(8), 2485–2494 (2011).
Suji, K. K. et al. Evaluation of rice (Oryza sativa L.) near iso-genic lines with root QTLs for plant production and root traits in rainfed target populations of environment. Field Crops Res. 137, 89–96 (2012).
Yun, W. et al. Identification of genetic overlaps for salt and drought tolerance using simple sequence repeat markers on an advanced backcross population in rice. Crop Sci. 52(4), 1583–1592 (2012).
Lang, N. T., Nha, C. T., Ha, P. T. & Buu, B. C. Quantitative trait loci (QTLs) associated with drought tolerance in rice (Oryza sativa L.). SABRAO J. Breed. Genet. 45(3), 409–421 (2013).
Uga, Y. et al. A major QTL controlling deep rooting on rice chromosome 4. Sci Rep. 3, 3040 (2013).
Uga, Y. et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45(9), 1097–1102 (2013).
Wang, Y. et al. Background-independent quantitative trait loci for drought tolerance identified using advanced backcross introgression lines in rice. Crop Sci. 53(2), 430–441 (2013).
Dixit, S. et al. Multiple major QTL lead to stable yield performance of rice cultivars across varying drought intensities. BMC Genet. 15(1), 16 (2014).
Xing, W., Zhao, H. & Mei, D. Detection of main-effect and epistatic QTL for yield-related traits in rice under drought stress and normal conditions. Can. J. Plant Sci. 94(4), 633–641 (2014).
Kitomi, Y. et al. QTLs underlying natural variation of root growth angle among rice cultivars with the same functional allele of DEEPER ROOTING 1. Rice 8(1), 16 (2015).
Li, J. et al. qRT9, a quantitative trait locus controlling root thickness and root length in upland rice. J. Exp. Bot. 66(9), 2723–2732 (2015).
Lou, Q. et al. Quantitative trait locus mapping of deep rooting by linkage and association analysis in rice. J. Exp. Bot. 66(15), 4749–4757 (2015).
Niones, J. M., Inukai, Y., Suralta, R. R. & Yamauchi, A. QTL associated with lateral root plasticity in response to soil moisture fluctuation stress in rice. Plant Soil 391(1–2), 63–75 (2015).
Uga, Y. et al. A QTL for root growth angle on rice chromosome 7 is involved in the genetic pathway of DEEPER ROOTING 1. Rice 8(1), 1–8 (2015).
Sandhu, N. et al. Rice root architectural plasticity traits and genetic regions for adaptability to variable cultivation and stress conditions. Plant Physiol. 171(4), 2562–2576 (2016).
Catolos, M. et al. Genetic loci governing grain yield and root development under variable rice cultivation conditions. Front. Plant Sci. 8, 1763 (2017).
Bhattarai, U. & Subudhi, P. K. Identification of drought responsive QTLs during vegetative growth stage of rice using a saturated GBS-based SNP linkage map. Euphytica 214(2), 38 (2018).
Sabar, M. et al. Identification and mapping of QTLs associated with drought tolerance traits in rice by a cross between Super Basmati and IR55419-04. Breed. Sci. 69(1), 169–178 (2019).
Yun, Y. T., Kim, H. J. & Tai, T. H. Identification of QTLs controlling seedling traits in temperate Japonica Rice under different water conditions. Plant Breed. Biotechnol. 7(2), 106–122 (2019).
Acknowledgements
This work has been supported by the Shiraz University, Iran.
Funding
This work has been supported by the Center for International Scientific Studies and collaboration (CISSC), Ministry of Science, Research and Technology, Iran and Shiraz University, Iran.
Author information
Authors and Affiliations
Contributions
B.K. performed the initial analyses and wrote the draft of manuscript, E.T. conceived and designed the project and complemented the analyses and writing of the manuscript, V.S. enriched the analyses and provided critical advices on the project and the manuscript, L.R. provided critical advices on the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Khahani, B., Tavakol, E., Shariati, V. et al. Meta-QTL and ortho-MQTL analyses identified genomic regions controlling rice yield, yield-related traits and root architecture under water deficit conditions. Sci Rep 11, 6942 (2021). https://doi.org/10.1038/s41598-021-86259-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-86259-2
This article is cited by
-
Meta-QTL analysis reveals the important genomics regions for biotic stresses, nutritional quality and yield related traits in pearl millet
CABI Agriculture and Bioscience (2024)
-
Uncovering the Genomic Regions Associated with Yield Maintenance in Rice Under Drought Stress Using an Integrated Meta-Analysis Approach
Rice (2024)
-
Integrated meta-analysis and transcriptomics pinpoint genomic loci and novel candidate genes associated with submergence tolerance in rice
BMC Genomics (2024)
-
Meta-analysis of identified genomic regions and candidate genes underlying salinity tolerance in rice (Oryza sativa L.)
Scientific Reports (2024)
-
Meta-QTLs, ortho-meta QTLs and related candidate genes for yield and its component traits under water stress in wheat (Triticum aestivum L.)
Physiology and Molecular Biology of Plants (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.