Abstract
Systemic lupus erythematosus (SLE) is a typical autoimmune disease with a strong genetic disposition. Genetic studies have revealed that single-nucleotide polymorphisms (SNPs) in zinc finger protein (ZNF)-coding genes are associated with susceptibility to autoimmune diseases, including SLE. The objective of the current study was to evaluate the correlation between ZNF76 gene polymorphisms and SLE risk in Chinese populations. A total of 2801 individuals (1493 cases and 1308 controls) of Chinese Han origin were included in this two-stage genetic association study. The expression of ZNF76 was evaluated, and integrated bioinformatic analysis was also conducted. The results showed that 28 SNPs were associated with SLE susceptibility in the GWAS cohort, and the association of rs10947540 was successfully replicated in the independent replication cohort (Preplication = 1.60 × 10−2, OR 1.19, 95% CI 1.03–1.37). After meta-analysis, the association between rs10947540 and SLE was pronounced (Pmeta = 9.62 × 10−6, OR 1.29, 95% CI 1.15–1.44). Stratified analysis suggested that ZNF76 rs10947540 C carriers were more likely to develop relatively high levels of serum creatinine (Scr) than noncarriers (CC + CT vs. TT, p = 9.94 × 10−4). The bioinformatic analysis revealed that ZNF76 rs10947540 was annotated as an eQTL and that rs10947540 was correlated with decreased expression of ZNF76. Remarkably, significantly reduced expression of ZNF76 was confirmed by expression data from both our laboratory and an array-based expression database. Taken together, these results suggest that ZNF76 rs10947540 is a possible susceptibility factor associated with SLE susceptibility. The mechanism underlying the relationship between ZNF76 and SLE pathogenesis still requires further investigation.
Similar content being viewed by others
Introduction
Systemic lupus erythematosus (SLE) is a typical autoimmune disease that is characterized by increased generation of apoptotic debris and the presence of autoantibodies specific for nuclear components. Currently, the aetiology of SLE is not fully understood. It has been well documented that genetic factors are important for SLE predisposition.
Zinc finger proteins are DNA-binding proteins that control the transcription of a number of genes. Genetic studies have reported that single-nucleotide polymorphisms in zinc finger protein-coding genes are associated with susceptibility to autoimmune diseases, including SNPs in Eos (also known as Ikaros family zinc finger 4; IKZF4) being associated with alopecia areata1, those in zinc finger 432 (ZNF432) with asthma2, those in zinc finger 193 (ZNF 193) with rheumatoid arthritis3, those in zinc finger 365 (ZNF365) isoform D with Crohn's disease4, and those in zinc finger 3 (IKZF3) with Graves’ disease5. In a large-scale multiracial replication study, rs1453560 located between IKAROS family of zinc finger 3 (AIOLOS; IKZF3) and zona pellucida binding protein 2 (ZPBP2) was identified as a susceptibility locus for systemic lupus erythematosus6. Subsequently, the genetic association between rs907091 in the IKZF3 gene and SLE was validated in a Chinese Han population7, and the association between the IKZF1 5′ UTR variant rs1456896 and lupus nephritis in a northern Han Chinese population was also revealed8.
ZNF76 is a novel transcriptional repressor targeting TATA-binding protein that has been shown to have an inhibitory effect on p53 activity by reporter assays and on endogenous target gene expression9. Integrated analysis of three original datasets, GSE72509, GSE20864, and GSE39088, from the Gene Expression Omnibus (GEO) database identified that the p53 signalling pathway may be implicated in SLE pathogenesis10. Considering both genetic clues concerning variants in zinc finger protein-coding genes contributing to susceptibility to autoimmune diseases, including SLE, and the biological functions of ZNF76, we aimed to explore the role of ZNF76 in the pathogenesis of SLE.
Based on a previous SLE GWAS, we first replicated the tag SNP in our cohort to confirm the genetic association between ZNF76 variants and SLE susceptibility. By using a public database, we explored the functional role of ZNF76 rs10947540 and the differential expression of ZNF76, which was also validated by expression data from our centre.
Methods
Study populations
The replication cohort contained 1003 SLE patients and 815 healthy controls from the Henan population in the middle of China. All the patients were diagnosed by at least two experienced physicians according to the revised criteria for the classification of SLE from the American College of Rheumatology (ACR)11. Clinical data were retrospectively collected at the time of diagnosis. Written informed consent was obtained from each participant. This investigation was conducted according to the Declaration of Helsinki. The study was approved by the Medical Ethics Committee of Zhengzhou University First Hospital (2019-KY-134).
SNP selection and genotyping
We focused on the 10-kb upstream and downstream regions of the ZNF76 gene ranging from 35,217,512—35,273,762 on chromosome 6. Thirty-seven SNPs were covered by the ImmunoChip used in the GWAS and are listed in supplementary Table 1. Genetic association results were obtained from a previous publication12.
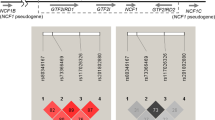
A total of 28 SNPs associated with SLE susceptibility were identified in the GWAS cohort (supplementary Table 1)12. Five intronic polymorphisms within ZNF76, rs10947540, rs9394289, rs2267663, rs1894650, and rs9366883, were the top signals (p = 1.31 × 10−5) and were highly linked (D’ = 1.0, r-square = 1.0, Fig. 1) after analysing genotype data from 103 Chinese Han Beijing (CHB) individuals from the 1000 Genomes Project (Fig. 1). Further, rs10947540 was chosen as the tag SNP and was validated in the replication cohort. The Sequenom MassARRAY platform (Sequenom, Inc., San Diego, California, USA) was used for genotyping the replication cohort, and the genotyping yield was 99.5%.
The LD maps of the 28 identified SLE-associated SNPs in 103 Chinese Han Beijing individuals according to 1000 genome project. The degrees of LD were estimated by CI method using Haploview4.2 (Cambridge, MA, USA) and a standard color scheme (D’/LOD) is used to display the LD pattern. Top signals (p = 1.31 × 10−5), rs10947540, rs9394289, rs2267663, rs1894650, rs9366883 were highlighted by black boxes and were in high linkage (D’ = 1.0, r-square = 1.0). The variant, rs10947540, further replicated in independent cohort was marked with asterisk.
Rigorous quality control of the GWAS cohort was performed previously and provided in a previous publication12. For the replication cohort, the missing genotyping rate of rs10947540 was 0.50% in the cases vs. 0.37% in the controls, and the Hardy–Weinberg equilibrium was p = 0.86 in the cases vs. p = 0.46 in the controls.
Bioinformatic and differential gene expression analyses
Regulatory functions were annotated with rSNPBase13. The summarized expression quantitative trait loci (eQTLs) of rs10947540 were obtained from HaploReg v4.114.
Allele-dependent gene expression was determined by the combined use of the ArrayExpress Archive database (http://www.ebi.ac.uk/arrayexpress) and Ensembl (http://www.ensembl.org). Differential gene expression data for ZNF76, DEF6, and TAF11 was derived from both our in-house test and the E-GEOD-50772 project from the ArrayExpress Archive database15. The E-GEOD-50772 project was conducted with the A-AFFY-44-Affymetrix GeneChip Human Genome U133 Plus 2.0 with RNA from peripheral blood mononuclear cells (PBMCs)15.
Our expression analysis was performed with RNA from whole blood. The cDNA library was prepared according to the previously published protocol16 and sequenced with PE150 (Illumina, San Diego, California, USA). Clean reads were obtained after filtering out reads containing poly-N, sequencing adapters, and low-quality reads. The remaining clean reads were mapped to the reference genome using Hisat2 software. The expression levels were assessed based on the FPKM (Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced) value.
Statistical analysis
Hardy–Weinberg equilibrium (HWE) among controls was assessed by the goodness-of-fit χ2 test. Allelic association analyses of SLE patients and healthy controls were performed using the chi-square test. The combined result for meta-analysis was from Cochran–Mantel–Haenszel statistics. Genotype-clinical phenotype association analysis was conducted using the chi-square test for categorical variables and Student’s t-test for continuous variables. Spearman’s coefficient was calculated to determine correlations in allele-dependent gene expression analysis. The differences in ZNF76, DEF6, and TAF11 expression between SLE patients and healthy controls were tested using Student's t-test. Statistical analyses were implemented with SPSS 13.0 software. All results with a two-tailed p < 0.05 were considered statistically significant.
Ethics approval
The study was approved by the Medical Ethics Committee of Zhengzhou University First Hospital (2019-KY-134).
Consent to participate
The informed consent was obtained from all participants and/or their legal guardians.
Result
Association of ZNF76 gene polymorphisms with susceptibility to SLE
ZNF76 rs10947540 was selected as the tag SNP, and the association with SLE was successfully replicated (p = 1.60 × 10−2, OR 1.19, 95% CI 1.03–1.37) in a larger cohort with 1003 SLE patients and 815 healthy controls (Table 1). After meta-analysis of both the GWAS cohort from the GWAS and our replication cohort, the association between rs10947540 and SLE was pronounced (Pmeta = 9.62 × 10−6, OR 1.29, 95% CI 1.15–1.44).
Demographics of SLE patients with three rs10947540 genotypes
Nine hundred ninety-eight SLE patients were successfully genotyped for ZNF76 rs10947540 and enrolled for clinical association analysis (Table 2). There were trends toward higher incidences of malar rash, discoid rash, photosensitivity, arthritis, serositis, haematological disorder, anti-dsDNA antibodies, and anti-Sm antibodies (without reaching statistical significance) in patients with the risk C allele than in the other patients. Notably, the SLE patients carrying the risk C allele showed significantly higher levels of serum creatinine (Scr) (CC + CT vs. TT, p = 9.94 × 10−4).
Bioinformatic analysis
ZNF76 rs10947540 was predicted to be a potential regulatory SNP by rSNPBase13. Experimentally supported regulatory elements from ENCODE and other data resources showed that ZNF76 rs10947540 was in LD with other regulatory SNPs (r2 > 0.8) and had potential distal regulation, RNA-binding protein-mediated regulation, and an eQTL effect. Table 3 shows that rs10947540 was correlated with the expression of TCP11, SCUBE3, DEF6, and ZNF76 in certain tissues.
The GTEx Portal provides comprehensive tissue-specific gene expression and regulation data. We inferred from GTEx that rs10947540 is an eQTL (Fig. 2) associated with the expression of 7 genes (DEF6, ZNF76, PPARD, SCUBE3, RPL10A, TCP11, and TAF11) in 27 tissues (supplementary Table 2). Individuals carrying the risk C allele had lower expression of DEF6 (p = 1.1 × 10−49) and ZNF76 (p = 1.1 × 10−19) in whole blood samples (Fig. 3A).
The integrated expression and genotypic analysis of ZNF76 rs10947540. (A) The violin plot for expression of DEF6, ZNF76, and TAF11 in whole blood were obtained from GTEx database. (B) The correlation between ZNF76, DEF6, TCP11, and SCUBE3 and rs10947540 genotypes were adopted in E-MTAB-264 and the significances were tested by spearman's correlation coefficient. The boxplot was generated via boxplot in SPSS.
Furthermore, we validated the eQTL effects in 488 individuals from HapMap projections. Individuals carrying the homozygous CC risk allele showed lower levels of ZNF76 and DEF6 expression and higher levels of TCP11 expression than other patients (Fig. 3B). There was no correlation between the SCUBE3 expression level and rs10947540 genotypes, which might be due to tissue-specific expression.
Reduced level of ZNF76 was observed in SLE
Considering the eQTL effect of rs10947540, we recruited 75 SLE patients and 24 healthy controls to ascertain whether ZNF76 is differentially expressed. In accordance with the association of the risk C allele with lower levels of gene expression, our expression data for whole blood showed that lower levels of ZNF76 expression were observed in the patients with SLE (Fig. 4A). Moreover, mRNA expression data from the E-GEOD-50772 project were consistent with our finding that the ZNF76 expression in peripheral blood mononuclear cells from 61 SLE patients was significantly lower than that in PBMCs from 20 healthy controls (Fig. 4B).
The mRNA expression of ZNF76 was decreased in SLE patients comparing with controls. The expression levels of ZNF76 were compared between SLE patients with healthy controls in our cohort shown as FPKM (Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced) (A) and E-GEOD-50772 project shown as expression values (B). The differences of ZNF76 expression between SLE patients and healthy controls were calculated by Student's t-test. Abbreviation: PBMC, peripheral blood mononuclear cells; FPKM, fragments per kilobase of transcript sequence per millions base pairs sequenced.
Associated with rs10947540 in whole blood, we perform additional gene expression for DEF6 and TAF11. The expression of TAF11 were significantly lower in SLE patients comparing with healthy controls both in our cohort and E-GEOD-50772 project (Supplement Fig. 1). However, there were no difference in DEF6 expression between SLE patients and healthy controls (Supplement Fig. 1).
Discussion
To determine whether SNPs in the ZNF76 gene predispose patients to SLE, we conducted a genetic replication of a previous GWAS genetic association result in an independent Chinese Han population. Our results showed that rs10947540 in ZNF76 predisposed patients to susceptibility to SLE. In the stratified analysis, we found that patients carrying the risk C allele (CC + CT genotypes) had a more evident risk and higher Scr levels.
ZNF76, a novel transcriptional repressor targeting TATA-binding protein, has a strong inhibitory effect on p53 in various cell lines, including the HeLa, U2OS, MCF-7, and H1299 cell lines9. Expression data from both our laboratory and the ArrayExpress Archive database demonstrated that the expression of ZNF76 was lower in patients with SLE than in healthy controls. P53 might be deregulated by the reduced expression of ZNF76. The presence of autoantibodies, increased cell apoptosis, and overactivation of type I IFN signalling pathways are prominent characteristics of SLE. In patients with SLE, elevated levels of p53 are detected in fibroblasts, bone marrow-derived mesenchymal stem cells, peripheral blood mononuclear cells, and renal tissues; they are also found in the skin of discoid lupus erythematosus patients21,22,23,24,25. Researchers have reported that increased apoptosis is associated with p53 upregulation in p21-/- lupus mice26. Moreover, p53 activation in SLE patients may stimulate type I IFN activity, promoting innate immune signalling directly27,28. The production of autoantibodies requires p53 in B6/lpr lupus mice29. We hypothesized that the reduced expression of ZNF76 promoted the pathogenesis of SLE, which might be due to deregulation of p53.
DEF6 is required for maintaining T cell effector functions and lymphocyte homeostasis and preventing systemic autoimmunity30. Mice deficient in DEF6 can spontaneously develop a lupus-like syndrome with increased levels of autoantibodies and glomerulonephritis30. The bioinformatic analysis revealed that ZNF76 rs10947540 was annotated as an eQTL associated with the expression of DEF6. Furthermore, genotyping and expression data from a HapMap population confirmed that the risk allele of rs10947540 was correlated with decreased expression of DEF6. We could not rule out the possibility that rs10947540 might promote the development of SLE by affecting DEF6 expression (supplementary Fig. 1). ZNF76 functions as a transcriptional repressor. Whether the reduced expression of DEF6 was due to the regulation of ZNF76 remains to be elucidated.
The risk allele was associated with decreased ZNF76 expression levels according to data for individuals from the GTEx database and the expression data of HapMap3 projections. However, the data from the GTEx database and the expression data of HapMap3 projections were from healthy individuals. Thus, a limitation of this study was that we lacked data from SLE patients to explore the association between the risk allele and ZNF76 expression.
Our present study demonstrates that the rs10947540 polymorphism of the ZNF76 gene is a possible susceptibility factor associated with SLE susceptibility. The mechanism underlying the association between ZNF76 and the pathogenesis of SLE still requires further investigation.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Petukhova, L. et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature 466, 113–117. https://doi.org/10.1038/nature09114 (2010).
Wu, A. C. et al. Inhaled corticosteroid treatment modulates ZNF432 gene variant’s effect on bronchodilator response in asthmatics. J. Allergy Clin. Immunol. https://doi.org/10.1016/j.jaci.2013.09.037 (2014).
Xie, G. et al. Identification of the NF-κB activating protein-like locus as a risk locus for rheumatoid arthritis. Ann. Rheum. Dis. 72, 1249–1254. https://doi.org/10.1136/annrheumdis-2012-202076 (2013).
Haritunians, T. et al. Variants in ZNF365 isoform D are associated with Crohn’s disease. Gut 60, 1060–1067. https://doi.org/10.1136/gut.2010.227256 (2011).
Li, L. et al. Polymorphisms of IKZF3 gene and autoimmune thyroid diseases: associated with graves’ disease but not with hashimoto’s thyroiditis. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 45, 1787–1796. https://doi.org/10.1159/000487870 (2018).
Lessard, C. J. et al. Identification of IRF8, TMEM39A, and IKZF3-ZPBP2 as susceptibility loci for systemic lupus erythematosus in a large-scale multiracial replication study. Am. J. Hum. Genet. 90, 648–660. https://doi.org/10.1016/j.ajhg.2012.02.023 (2012).
Cai, X. et al. Association between polymorphisms of the IKZF3 gene and systemic lupus erythematosus in a Chinese Han population. PLoS ONE 9, e108661. https://doi.org/10.1371/journal.pone.0108661 (2014).
Zhang, Y. M. et al. Association of the IKZF1 5’ UTR variant rs1456896 with lupus nephritis in a northern Han Chinese population. Scand. J. Rheumatol. 46, 210–214. https://doi.org/10.1080/03009742.2016.1194458 (2017).
Zheng, G. & Yang, Y.-C. ZNF76, a novel transcriptional repressor targeting TATA-binding protein, is modulated by sumoylation. J. Biol. Chem. 279, 42410–42421 (2004).
Yang, F. et al. Bioinformatics identification of key candidate genes and pathways associated with systemic lupus erythematosus. Clin. Rheumatol. 39, 425–434. https://doi.org/10.1007/s10067-019-04751-7 (2020).
Hochberg, M. C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 40, 1725 (1997).
Sun, C. et al. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat. Genet. 48, 323–330. https://doi.org/10.1038/ng.3496 (2016).
Guo, L., Du, Y., Qu, S. & Wang, J. rVarBase: an updated database for regulatory features of human variants. Nucl. Acids Res. 44, D888–D893. https://doi.org/10.1093/nar/gkv1107 (2016).
Ward, L. D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucl. Acids Res. 40, D930-934. https://doi.org/10.1093/nar/gkr917 (2012).
Kennedy, W. P. et al. Association of the interferon signature metric with serological disease manifestations but not global activity scores in multiple cohorts of patients with SLE. Lupus Sci. Med. 2, e000080. https://doi.org/10.1136/lupus-2014-000080 (2015).
Parkhomchuk, D. et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucl. Acids Res. 37, e123. https://doi.org/10.1093/nar/gkp596 (2009).
Ardlie, K. G. Human genomics: the genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660. https://doi.org/10.1126/science.1262110 (2015).
Lappalainen, T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511. https://doi.org/10.1038/nature12531 (2013).
Westra, H. J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243. https://doi.org/10.1038/ng.2756 (2013).
Kabakchiev, B. & Silverberg, M. S. Expression quantitative trait loci analysis identifies associations between genotype and gene expression in human intestine. Gastroenterology https://doi.org/10.1053/j.gastro.2013.03.001 (2013).
Günther, C. et al. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J. Clin. Investig. 125, 413–424. https://doi.org/10.1172/JCI78001 (2015).
Gao, L. et al. Bone marrow-derived mesenchymal stem cells from patients with systemic lupus erythematosus have a senescence-associated secretory phenotype mediated by a mitochondrial antiviral signaling protein-interferon-β feedback loop. Arthritis Rheumatol. 69, 1623–1635. https://doi.org/10.1002/art.40142 (2017).
Zamolo, G. et al. Expression of p53 and apoptosis in discoid lupus erythematosus. Croat. Med. J. 46, 678–684 (2005).
El-Sayed, Z. A., Farag, D. H. & Eissa, S. Tumor suppressor protein p53 and anti-p53 autoantibodies in pediatric rheumatological diseases. Pediatr. Allergy Immunol. 14, 229–233 (2003).
Wang, J. S., Tseng, H. H., Shih, D. F., Jou, H. S. & Ger, L. P. Expression of inducible nitric oxide synthase and apoptosis in human lupus nephritis. Nephron 77, 404–411 (1997).
Lawson, B. R. et al. Deficiency of the cyclin kinase inhibitor p21(WAF-1/CIP-1) promotes apoptosis of activated/memory T cells and inhibits spontaneous systemic autoimmunity. J. Exp. Med. 199, 547–557 (2004).
Brzostek-Racine, S., Gordon, C., Van Scoy, S. & Reich, N. C. The DNA damage response induces IFN. J. Immunol. 187, 5336–5345. https://doi.org/10.4049/jimmunol.1100040 (2011).
Muñoz-Fontela, C. et al. Transcriptional role of p53 in interferon-mediated antiviral immunity. J. Exp. Med. 205, 1929–1938. https://doi.org/10.1084/jem.20080383 (2008).
Kuan, A. P. & Cohen, P. L. p53 is required for spontaneous autoantibody production in B6/lpr lupus mice. Eur. J. Immunol. 35, 1653–1660 (2005).
Fanzo, J. C. et al. Loss of IRF-4-binding protein leads to the spontaneous development of systemic autoimmunity. J. Clin. Invest. 116, 703–714. https://doi.org/10.1172/jci24096 (2006).
Acknowledgments
We thank all the members of our laboratory for their technical assistance. We also thank the patients, their families and healthy donors for their cooperation and for giving consent to participate in this study. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from: the GTEx Portal on 08/15/20.
Funding
This work was supported by the National Science Foundation of China [Grant Number 81900643, 81873611]; the China Postdoctoral Science Foundation Grant [Grant Number 2019M652592]; the Postdoctoral Research Grant in Henan Province [Grant Number 1902005, 1901004]; the Science and Technology Innovation Team of Henan [Grant Number 17IRTSTHN020]; the Foundation for Leading Personnel of Central Plains of China [Grant Number 194200510006]; the Foundation for Medical Science and Technology Program of Henan [Grant Number 11195, 11272]; and the Henan Science and Technology Research Program [Grant Number 201901048, 2018020102, 2018020142]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiment: Y-Y.Q., Z-Z.Z.; Performed the experiments: Y-Y.Q., Y.C., X-R.L., Y-F.Z. and X-H.N.; Analyzed the data: Y-Y.Q., H.L., X-Y.W., Y-L.Z., and X-X.Z.; Interpretation of the findings: Y-Y.Q., H.L., Y.C. and Z-Z.Z.; All the authors contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Qi, Yy., Cui, Y., Lang, H. et al. The ZNF76 rs10947540 polymorphism associated with systemic lupus erythematosus risk in Chinese populations. Sci Rep 11, 5186 (2021). https://doi.org/10.1038/s41598-021-84236-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-84236-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.