Abstract
In sudden unexpected death in infancy cases, postmortem genetic analysis with next-generation sequencing potentially can extract candidate genes associated with sudden death. However, it is difficult to accurately interpret the clinically significant genetic variants. The study aim was to conduct trio analysis of cases of sudden unexpected death in infancy and their parents to more accurately interpret the clinically significant disease-associated gene variants associated with cause of death. From the TruSight One panel targeting 4813 genes we extracted candidate genetic variants of 66 arrhythmia-, 63 inherited metabolic disease-, 81 mitochondrial disease-, and 6 salt-losing tubulopathy-related genes in 7 cases and determined if they were de novo or parental-derived variants. Thirty-four parental-derived variants and no de novo variants were found, but none appeared to be related to the cause of death. Using trio analysis and an in silico algorithm to analyze all 4813 genes, we identified OBSCN of compound heterozygous and HCCS of hemizygous variants as new candidate genetic variants related to cause of death. Genetic analysis of these deceased infants and their living parents can provide more accurate interpretation of the clinically significant genetic variants than previously possible and help confirm the cause of death.
Similar content being viewed by others
Introduction
Sudden unexpected death in infancy (SUDI) is a term that has been variably used to refer to all cases of sudden and unexpected deaths in infancy, including sudden infant death syndrome (SIDS)1. In 2015, SIDS was the third leading cause of death at age 0 in Japan, with a mortality rate of 9.5 per 100,000 population2. However, SUDI is thought to be more common because of the difficult distinction between SIDS and accidental asphyxia or natural diseases, such as arrhythmias and inherited metabolic disease2. Arrhythmia, inherited metabolic disease, mitochondrial disease, and salt-losing tubulopathy are linked to sudden unexpected death (SUD)3,4,5,6,7,8,9,10,11,12,13. Thus, SUDI is considered to be the severest form of various related diseases. The standard methods of determining the cause of death in SUDI cases include conventional autopsy (macroscopic autopsy and detailed microscopic examination14), toxicology, biochemical tests, and recently, metabolic autopsy15. However, in most SUDI cases, these methods do not reveal the exact cause of death, hence the urgent need to develop novel diagnostic methods.
Recently, some studies have shown that genetic analysis using next-generation sequencing (NGS) of SUDI cases was effective in diagnosing arrhythmias and inherited metabolic disease 9,10. On the other hand, it has been reported that some of the variants in arrhythmia-related genes found in SUD cases were not associated with cause of death16,17. In other words, an accurate interpretation of the association between variants in disease-related genes found in SUD cases and sudden death is important in determining cause of death. However, even if variants in arrhythmia-related genes are found, it is difficult to obtain findings that can be judged as arrhythmia from macroscopic, pathological, and biochemical examinations. Most conventional NGS studies were limited to genetic analysis of sudden death cases only. Recently, familial gene analysis of cardiac sudden death cases and their parents was shown to be effective for diagnosis of cause of death18,19,20,21,22,23,24,25. However, most family analyses have targeted genes for arrhythmia, so it would be useful to target not only those genes but also any other disease-related genes. Furthermore, few studies have reported family genetic analyses with NGS that focused on SUDI. Since SUDI is considered to be the severest form of various diseases, it would be useful to analyze the genes associated not only with arrhythmia but also with other diseases that can cause sudden death. Therefore, we thought that genetic analysis of SUDI cases and their parents would lead to a more accurate interpretation of the clinically significant genetic variants associated with cause of death.
The study aim was to perform trio analysis of SUD infants and their parents to interpret the clinically significant disease-associated gene variants associated with the cause of death more accurately. In addition, we used trio analysis to extract novel candidate gene variants and examined whether these extracted variants were associated with cause of death.
Results
Case collection
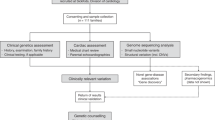
Among the 24 autopsied cases, 17 could not be diagnosed using conventional methods. From the 17 undiagnosed cases, 9 patients’ parents gave consent for whole-genome sequencing. One of the nine cases was diagnosed with carnitine palmitoyltransferase II deficiency by metabolic autopsy26. In another case, we could not extract a sufficient amount of DNA for targeted gene-sequencing panel analysis. A total of seven SUDI cases (two males, five females; age range: 3 months–2 years) were selected (Fig. 1). The case 2 parents had a consanguineous marriage (Table 1).
Target sequence of sudden death cases
On average, ~ 17.5 million total reads were produced and ~ 11.9 million reads mapped to the targeted region in each sample. The mean coverage of the coding sequence was 101.5 ± 33.7 reads, with an overall average gene level coverage ≥ 20 reads of 93.0% ± 10.7%. After the filtering steps, 1897 variants in the 4813 target genes were identified, which corresponded to an average of 271 variants per case.
Detected variants aligned to the 216 target genes
Thirty-four (18 non-synonymous and 16 synonymous) detected variants were aligned with the 216 target genes. Eleven arrhythmia-, two cardiomyopathy-, three metabolic disease-, fifteen mitochondrial disease-, and three salt-losing tubulopathy-related genes were included. All of these genetic variants were heterozygous and did not contain any de novo gene variants (Tables 2 and 3, Supplementary Table 1, and Fig. 2).
Flowchart of the selection process showing candidate genes for cause of death in this study. The frequency of variants in Japanese people was based on ToMMo and HGVD data. (a) The target 216 genes are listed in Table 2. (b) All genes included in the TruSight One panel are shown in Supplementary Text 1. Abbreviations: SUD: sudden unexpected death.
Arrhythmia- and cardiomyopathy-related gene variants
Most cases of genetic arrhythmia have autosomal dominant (AD) inheritance, and a familial history is not always present. Sporadic cases are mostly caused by de novo variant9. Case 1 had Asp85Asn-KCNE1, AD inheritance of a long QT syndrome (LQTS)-related gene variant. This variant was reported to be associated with LQTS and suggested as a possible cause of death in SUDI9,27. The same variant was found in the female infant’s healthy living mother. Case 4 had Arg160Trp-MYBPC3, AD inheritance of a cardiomyopathy-related gene variant suspected to be harmful by in silico analysis. This variant reportedly could not be ruled out as a pathogenic variant28. The female infant’s father had the same variant but did not develop cardiomyopathy. Case 5 had Gln2730Arg-AKAP9, AD inheritance of an LQTS-related gene variant. AKAP9 variants have been reported to be associated with LQTS type 1129, but Gln2730Arg-AKAP9 has not previously been reported to be associated with LQTS type 11. The female infant’s living father had the same variant. Five of seven cases had synonymous variants that were inherited from either of the parents. In silico analysis showed that all of them were benign.
Metabolic disease-, mitochondrial disease-, and salt-losing tubulopathy-related gene variants
Most inherited metabolic disease, mitochondrial disease, and salt-losing tubulopathy have autosomal recessive (AR) or X-chromosomal recessive inheritance. Homozygous amino acid changes or at least two heterozygous amino acid changes are necessary to cause these diseases9,30,31,32,33.
Case 1 had Leu21 = -SDHAF2, AD inheritance of a paraganglioma-related gene variant. TraP Score of Leu21 = -SDHAF2 was 0.089, which meant that the variant was benign. In addition, her mother had the same variant. The diseases associated with the other 20 gene variants are all forms of AR inheritance. Since the variants in the cases were all heterozygous, these diseases were unlikely to have developed and led to death.
Detected de novo, homozygous, compound heterozygous, and hemizygous variants aligned with all 4813 genes in TruSight One
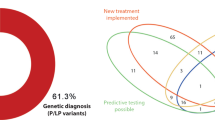
A total of 12 de novo variants in 10 genes, 4 homozygous variants in 4 genes, compound heterozygous variants in 4 genes, and 4 hemizygous variants in 4 genes were found in < 0.1% of the Japanese population. All de novo variants were heterozygous (Table 4, Supplementary Table 2). As of August 2021, a search conducted on PubMed using these 20 gene names and the terms “sudden,” “infant,” and “death” as keywords returned no papers related to SUDI (Fig. 2).
Discussion
First, we examined the significance of variants in disease-associated genes that could cause sudden death by performing trio analysis of SUDI cases and the parents. Arrhythmias, inherited metabolic disease, mitochondrial disease, and salt-losing tubulopathy were considered as candidate diseases that could cause SUDI since these diseases were difficult to diagnose because there are few specific findings at autopsy.
Arrhythmias and inherited metabolic disease have been reported to be associated with SUDI3,4,5,6,7,8,9,10. In this study, we adopted genes related to arrhythmia and inherited metabolic disease that we used in a previous study as candidate genes9. In particular, D85N_KCNE1 has been previously reported as a genetic variant associated with LQTS. In the present study, the same variant was also found in the infant’s mother, which could suggest that this variant alone was not sufficient to cause LQTS.
Mitochondrial disease is a type of inherited metabolic disease that occurs in about 1 in 5000 births34. There have been reports of postmortem diagnosis of mitochondrial disease11. Ohtake et al. reported that about 9% of those diagnosed with mitochondrial disease were cases of SUDI12. In addition, we adopted this gene as a candidate because most mitochondrial diseases are caused by variants in human nuclear DNA35.
Salt-losing tubulopathy is a group of diseases, including Bartter syndrome and Gitelman syndrome, characterized by hypokalemic metabolic alkalosis as a common feature36. Electrolyte abnormalities cannot be established at general autopsy. In one case, Bartter syndrome was diagnosed by postmortem genetic testing13, and salt-losing tubulopathy-related genes were used as candidate genes.
After the filtering step, 34 variants were found aligned with the 31 target genes. All of these variants were heterozygous. Software Implemented Fault Tolerance (SIFT), PolyPhen2, and TraP Score revealed a total of nine harmful missense variants. Genetic analysis of this case alone would have determined that these variants were likely to cause abnormal protein function and were associated with cause of death. In particular, the KCNE1, MYBPC3, and AKAP9 genes that were associated with LQTS or cardiomyopathy would have been suspicious as genes associated with cause of death because of an AD mode of inheritance27,37,38. These were sporadic cases without any family history of sudden death, and none of the AD-inherited arrhythmia-related gene variants in these cases were de novo. In general, if the proband had an AD genetic disease, at least some of the relatives would have the same symptoms. Therefore, these disease-associated genetic variants alone would not have caused the SUDI, as the relatives, including the parents, had not experienced SUD, obviously. These family histories and trio analysis results suggest that some of the variants in SUDI cases could not be involved in the development of the disease, although the same variants were categorized as pathological by in silico analysis or by the results of experimentation.
Second, candidate variants causing sudden infant death were extracted for all 4813 genes in the TruSight One panel. The incidence of SIDS in families is extremely low, and most cases are sporadic39. It is generally believed that rare variants are associated with rare diseases40, and diseases that can cause SUDI are considered to be one of the most severe of rare diseases. If SUDI cases with no family history of SUD manifested genetic variants that caused the sudden death, the variants were considered to be de novo. In addition, the variants of homozygotes or compound heterozygotes in SUDI cases and heterozygotes in their parents were extracted. Similarly, the variants of X-chromosome hemizygotes in SUD in male infancy and heterozygotes in their mothers were extracted.
We found eight de novo heterozygous missense variants in seven genes, three homozygous missense variants in three genes, eleven compound heterozygous missense variants in three genes, and two X-chromosome hemizygous missense variants in two genes (Table 4). These gene names and “sudden,” “death,” and “infant” were entered into PubMed as search terms. No papers described the association between these genes and SUDI. Therefore, some of these variants might be novel variants associated with cause of SUDI. Of these genes, we focused on two as biologically plausible candidate genes potentially associated with cause of death, OBSCN and HCCS.
Obscurin, which is encoded by the OBSCN gene, has an important role in the organization of sarcomeres during myofibril formation and the regular alignment of sarcoplasmic reticulum41. OBSCN variants may be monogenic causes of cardiomyopathy or contribute to the disease phenotype in concert with other variants42. In addition, inhibition of sarcomeric activity may cause arrhythmogenesis. It has also been reported that variants in the sarcomeric gene may have caused sudden cardiac death in a case of infant death without a cardiomyopathy phenotype43. In addition, the association of the OBSCN variant with a case of sudden cardiac death in an 8-year-old girl has also been reported44. Case 4 had maternal Arg1060Gln-OBSCN and paternal Ser5880Asn-OBSCN by compound heterozygotes. We speculated that these two heterozygous variants could be involved in sudden death by compound heterozygotes.
Holocytochrome c-type synthase, encoded by HCCS on the X chromosome, is located on the outer surface of the inner mitochondrial membrane and catalyzes covalent attachment of heme to both Cytc and Cytc145. Cytc transfers electrons from electron-transfer complex III to complex IV in the mitochondria to promote ATP production. The HCCS variant at domain IV reduced expression of HCCS and also impaired release of Cytc46. These suggest that HCCS plays an important role in mitochondrial function. Mitochondrial dysfunction can lead to arrhythmia and sudden death47. In addition, it has been found that HCCS also has an important role in apoptosis and that the HCCS variant is associated with microphthalmia with linear skin defects syndrome, which is an X-linked male-lethal disorder45,48. These results suggest that HCCS is an essential protein for vital function. Case 7, a 6-month-old boy, had hemizygous Ala248Ser-HCCS in domain IV. The in silico algorithms predicted that it was deleterious and probably damaging, so we speculated that this variant was associated with the cause of death. However, there is no report about the clinical significance of Ala248Ser-HCCS, so a functional analysis study is needed.
Trio analysis was thought to be useful for both evaluation of disease-related gene variants that could cause sudden death and extraction of novel candidate genes for sudden death. Trio analysis revealed that some variants in existing candidate genes for cause of death might be insufficient to cause death because these variants were inherited from either of the parents. This means that the postmortem genetic analysis of only SUDI cases could have led to misdiagnosis and was considered insufficient to determine cause of death.
We extracted genes with de novo, homozygous, compound heterozygous, and hemizygous variants and reviewed previous reports that had reported an association between these genes and SUDI. There were no reports of associations between the genes found in this study and SUDI, which suggested that OBSCN and HCCS from these extracted genes are novel candidate genes associated with death caused by gene dysfunction or diseases related to genetic variants. In a large cohort study of whole-exome sequencing for SIDS, no variants were found to be significantly more prevalent in SIDS cases than in the general population49. Therefore, it may be difficult to identify gene variants that are candidates for cause of death by statistical analysis alone even when comparing with a large sample of unrelated healthy subjects. If a sudden death case has an ultra-rare variant, it could be possible to determine the variant with pathological significance by examining whether the parents also have the variant.
Trio-targeted gene-sequencing panel analysis has an advantage over conventional methods for determining the cause of death. Genetic analysis of not only SUD deceased infants but also living parents can be a more exact inquiry for cause of death by SUDI. An autopsy is an investigation of the cause of death that is performed directly from a corpse. However, a genetic analysis of the SUD infants and their parents can provide more accurate interpretation of genetic variants than previously possible and help confirm a diagnosis of the cause of death. Furthermore, trio analysis can be performed in daily medical practice. It is important that the family members of those who have died suddenly should undergo standardized testing, including genetic analysis50,51,52. We were careful to obtain informed consent from the parents before trio analysis. Additionally, if a trio analysis reveals a hereditary disease, it is possible to refer the patient for genetic counseling. Asymptomatic siblings can be provided with important genetic information of the deceased, which may enable detection of diseases before they develop. Furthermore, diagnosed individuals can receive appropriate treatment. Trio analysis data may eventually be utilized for child death reviews.
There were several study limitations that should be considered. Targeted gene-sequencing panel analysis was performed using a panel of 4813 genes; thus, targeted gene-sequencing panel analysis of genes not included in this panel and of the whole-genome sequence was not performed. TruSight One is a 4813-gene-sequencing panel that covers a wide range of known disease-associated genes, so it is helpful in clinical diagnosis53. However, we believe that whole-exome analysis and whole-genome analysis are necessary to search for unknown genetic diseases. Additionally, mitochondrial genome sequencing was not performed. In the 4813 genes analyzed in this study, only limited variants, such as synonymous, missense, insertions, and deletions, were examined, and copy number polymorphisms were not analyzed. In this study, we investigated the association between the cause of death in SUD cases and arrhythmia and cardiomyopathy-related gene variants extracted by trio analysis. However, since incomplete penetrance diseases were identified in which the parents were asymptomatic and the child had a severe phenotype54, further genetic studies including not only the parents, but also other relatives, are necessary.
In this study, we showed that trio analysis enabled more accurate interpretation of the clinically significant gene variants in SUD infants, which could help prevent misdiagnosis and extract novel genetic variants associated with SUDI. Trio analysis is a novel method that can be useful for determining cause of death because it also analyzes genetic information from living parents or relatives as well as from deceased victims. In addition, trio analysis may enable investigations of the cause of death even in hospitals. Trio analysis can identify unknown genetic variants that may be related to cause of death and help prevent SUDI in the future.
Methods
Study design and participants
The SUD cases of infants autopsied at ≤ 2 years of age were selected between April 2013 and March 2017. Informed consent for whole-genome sequencing and trio analysis were obtained from parents. A comprehensive forensic investigation, including a thorough examination of the death scene, a review of the clinical history, and performance of an autopsy that included macroscopic and microscopic examinations and a toxicology examination, were performed in all cases.
Extraction of genomic DNA and genetic analysis
As reported previously9, genomic DNA of SUDI deceased infants and their living parents was isolated from blood leukocytes and buccal mucosa, respectively, by using the QIAamp DNA Blood Mini Kit (Qiagen, Tokyo, Japan) in accordance with the manufacturer’s standard methods. A TruSight One sequencing panel (Illumina, San Diego, CA, USA) targeting 4813 disease-associated genes was used, and the sequencing was performed on an Illumina MiSeq (Illumina) (Supplementary Text 1).
Filtering steps for extraction of variants
As reported previously9, the sequencing reads were mapped to the hg19 human reference genome sequence by using Variant Studio (version 3.0) software (Illumina). The variants with a low Q30 score or a read depth of < 30 were excluded. Copy number variation was not analyzed in this study. To identify putatively pathogenic variants, those with allele frequencies < 1% in East Asian ethnic subgroups were retained and listed by using data from the dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP), the 1000 Genomes Project (http://www.1000genomes.org), the NHLBI Exome Sequencing Project (http://evs.gs.washington.edu/EVS), the Exome Aggregation Consortium (http://exac.broadinstitute.org). In silico algorithms, SIFT (http://sift.jcvi.org), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2), and TraP Score (http://trap-score.org/index.jsp)55, were used to predict whether the detected variants would affect the function of each protein. For the synonymous variants, the pathogenicity of the variant was confirmed by ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/).
Detected variants aligned with the target 216 genes
As reported previously9, from the TruSight One panel targeting 4813 genes, we extracted sequence information from the 216 genes for inherited arrhythmia, cardiomyopathy, metabolic diseases, mitochondrial disease, and salt-losing tubulopathy: 19 Brugada Syndrome genes, 15 LQTS genes, 6 short QT syndrome genes, 6 progressive cardiac conduction disturbance genes, 5 catecholaminergic polymorphic ventricular tachycardia genes, 5 arrhythmogenic right ventricular cardiomyopathy genes, 24 other cardiac genes, 63 inherited metabolic disease genes (such as fatty acid oxidation, amino acid, and organic acid disorders), 81 mitochondrial disease genes30, and 6 salt-losing tubulopathy genes (such as Bartter syndrome and Gitelman syndrome)31,32,33. The gene list and criteria of gene selection are provided in Table 2. Single-nucleotide variations causing synonymous substitutions, non-synonymous substitutions, nonsense substitutions, and insertions/deletions occurring in the coding regions were retrieved.
Extraction of de novo, homozygous, compound heterozygous, hemizygous variants of SUDI
After filtering steps, to extract < 0.1% of Japanese allele variant frequencies, the data from the Genome Cohort Study of Tohoku Medical Megabank Organization (ToMMo) (http://ijgvd.megabank.tohoku.ac.jp) and the Human Genetic Variation Database (HGVD) in the Human Genetic Variation Browser (http://www.genome.med.kyoto-u.ac.jp/SnpDB/index.html) were referred to. Trio analysis was a comparative examination of genetic variants in SUDI deceased infants and their living parents. De novo variants, found in the SUD infants but not in their parents, were extracted by trio analysis. In the SUDI cases, homozygous variants or compound heterozygous variants that were heterozygous in their parents were extracted. In addition, in boy cases, hemizygous variants in X chromosome were extracted. Of these extracted variants, allele frequencies ≥ 0.1% in the Japanese subgroup in ToMMo and HGVD were excluded. Especially for the compound heterozygous variants, both allele frequencies of < 0.1% each were adopted. Sudden infant death candidate gene names and the terms “sudden,” “infant,” and “death” were combined and searched in PubMed.
Sanger sequencing
Sanger sequencing was performed to confirm the detected candidate variants (OBSCN and HCCS), as previously described. The polymerase chain reaction primers were designed by using Primer3 version 0.4.0 (https://bioinfo.ut.ee/primer3-0.4.0). (Supplementary Table 3.)
Ethical approval
Written informed consent was obtained from the parents for the use of the samples in this study, which was approved by the ethics committees of the Nagasaki University Graduate School of Medicine (20170504-3, 20200801-2). This study was performed in accordance with the Declaration of Helsinki.
Data availability
The main data supporting the findings of this study are available within the article and its Supplementary Information. Additional data are available from the corresponding author upon reasonable request.
References
Blair, P. S., Byard, R. W. & Fleming, P. J. Sudden unexpected death in infancy (SUDI): suggested classification and applications to facilitate research activity. Forensic Sci. Med. Pathol. 8, 312–315 (2012).
Osawa, M. et al. Circumstances and factors of sleep-related sudden infancy deaths in Japan. PLoS ONE 15, 1–12 (2020).
Ackerman, M. J. et al. Postmortem molecular analysis of SCN5A defects in sudden infant death syndrome. JAMA 286, 2264–2269 (2001).
Cheng, J. et al. α1-syntrophin mutations identified in sudden infant death syndrome cause an increase in late cardiac sodium current. Circ. Arrhythm. Electrophysiol. 2, 667–676 (2009).
Millat, G. et al. Contribution of long-QT syndrome genetic variants in sudden infant death syndrome. Pediatr. Cardiol. 30, 502–509 (2009).
Franciosi, S. et al. The role of the autonomic nervous system in arrhythmias and sudden cardiac death. Auton. Neurosci. 205, 1–11 (2017).
Nakano, Y. & Shimizu, W. Genetics of long-QT syndrome. J. Hum. Genet. 61, 51–55 (2016).
Kaku, N. et al. Diagnostic potential of stored dried blood spots for inborn errors of metabolism: A metabolic autopsy of medium-chain acyl-CoA dehydrogenase deficiency. J. Clin. Pathol. 71, 885–889 (2018).
Oshima, Y. et al. Postmortem genetic analysis of sudden unexpected death in infancy: neonatal genetic screening may enable the prevention of sudden infant death. J. Hum. Genet. 62, 989–995 (2017).
Neubauer, J. et al. Post-mortem whole-exome analysis in a large sudden infant death syndrome cohort with a focus on cardiovascular and metabolic genetic diseases. Eur. J. Hum. Genet. 25, 404–409 (2017).
Shimura, M. et al. Sudden death in baby sling carrier of infant due to mitochondrial respiratory chain complex I activity deficiency. J. Tokyo Med. Univ. 76, 343–346 (2018).
Ohtake, A. et al. Diagnosis and molecular basis of mitochondrial respiratory chain disorders: Exome sequencing for disease gene identification. Biochim. Biophys. Acta 1840, 1355–1359 (2014).
Lopez, H. U., Haverfield, E. & Chung, W. K. Whole-exome sequencing reveals CLCNKB mutations in a case of sudden unexpected infant death. Pediatr. Dev. Pathol. 18, 324–326 (2015).
Dettmeyer, R. B. & Kandolf, R. Cardiomyopathies-misdiagnosed as sudden infant death syndrome (SIDS). Forensic Sci. Int. 194, 21–24 (2010).
Yamamoto, T. et al. Metabolic autopsy with postmortem cultured fibroblasts in sudden unexpected death in infancy: Diagnosis of mitochondrial respiratory chain disorders. Mol. Genet. Metab. 106, 474–477 (2012).
Yamamoto, T., Matsusue, A., Umehara, T., Kubo, S. I. & Ikematsu, K. No association between cardiac ion channel variants and sudden infant death. Pediatr. Int. 60, 483–484 (2018).
Paludan-Müller, C. et al. Reappraisal of variants previously linked with sudden infant death syndrome: Results from three population-based cohorts. Eur. J. Hum. Genet. 27, 1427–1435 (2019).
Shanks, G. W., Tester, D. J., Nishtala, S., Evans, J. M. & Ackerman, M. J. Genomic triangulation and coverage analysis in whole-exome sequencing-based molecular autopsies. Circ. Cardiovasc. Genet. 10, 1–10 (2017).
Hofman, N. et al. Contribution of inherited heart disease to sudden cardiac death in childhood. Pediatrics 120, e967–e973 (2007).
Tan, H. L., Hofman, N., Van Langen, I. M., Van Der Wal, A. C. & Wilde, A. A. M. Sudden unexplained death: Heritability and diagnostic yield of cardiological and genetic examination in surviving relatives. Circulation 112, 207–213 (2005).
Tester, D. J. & Ackerman, M. J. Postmortem long QT syndrome genetic testing for sudden unexplained death in the young. J. Am. Coll. Cardiol. 49, 240–246 (2007).
Altmann, H. M. et al. Homozygous/compound heterozygous triadin mutations associated with autosomal-recessive long-QT syndrome and pediatric sudden cardiac arrest: elucidation of the triadin knockout syndrome. Circulation 131, 2051–2060 (2015).
Behr, E. R. et al. Sudden arrhythmic death syndrome: Familial evaluation identifies inheritable heart disease in the majority of families. Eur. Heart J. 29, 1670–1680 (2008).
Van Der Werf, C. et al. Diagnostic yield in sudden unexplained death and aborted cardiac arrest in the young: The experience of a tertiary referral center in the Netherlands. Hear. Rhythm 7, 1383–1389 (2010).
Quenin, P. et al. Clinical yield of familial screening after sudden death in young subjects: The French experience. Circ. Arrhythm. Electrophysiol. 10, 1–10 (2017).
Yamamoto, T. et al. Metabolic autopsy with next generation sequencing in sudden unexpected death in infancy: Postmortem diagnosis of fatty acid oxidation disorders. Mol. Genet. Metab. Rep. 5, 26–32 (2015).
Nishio, Y. et al. D85N, a KCNE1 polymorphism, is a disease-causing gene variant in long QT syndrome. J. Am. Coll. Cardiol. 54, 812–819 (2009).
Kadota, C. et al. Screening of sarcomere gene mutations in young athletes with abnormal findings in electrocardiography: Identification of a MYH7 mutation and MYBPC3 mutations. J. Hum. Genet. 60, 641–645 (2015).
De Villiers, C. P. et al. AKAP9 is a genetic modifier of congenital long-QT syndrome type 1. Circ. Cardiovasc. Genet. 7, 599–606 (2014).
Ng, Y. S. & Turnbull, D. M. Mitochondrial disease: Genetics and management. J. Neurol. 263, 179–191 (2016).
Nozu, K. et al. The pharmacological characteristics of molecular-based inherited salt-losing tubulopathies. J. Clin. Endocrinol. Metab. 95, E511–E518 (2010).
Nozu, K. et al. Molecular analysis of digenic inheritance in Bartter syndrome with sensorineural deafness. J. Med. Genet. 45, 182–186 (2008).
Birkenhäger, R. et al. Mutation of BSND causes Bartter syndrome with sensorineural deafness and kidney failure. Nat. Genet. 29, 310–314 (2001).
Skladal, D., Halliday, J. & Thorburn, D. R. Minimum birth prevalence of mitochondrial respiratory chain disorders in children. Brain 126, 1905–1912 (2003).
Koopman, W. J. H., Willems, P. H. G. M. & Smeitink, J. A. Monogenic mitochondrial disorders. N. Engl. J. Med. 366, 1132–1141 (2012).
Konrad, M. et al. Mutations in the chloride channel gene CLCNKB as a cause of classic Bartter syndrome. J. Am. Soc. Nephrol. 11, 1449–1459 (2000).
Arif, M. et al. Genetic, clinical, molecular, and pathogenic aspects of the South Asian–specific polymorphic MYBPC3 Δ25bp variant. Biophys. Rev. 12, 1065–1084 (2020).
Chen, L. et al. Mutation of an A-kinase-anchoring protein causes long-QT syndrome. Proc. Natl Acad. Sci. U.S.A. 104, 20990–20995 (2007).
Beal, S. M. & Blundell, H. K. Recurrence incidence of sudden infant death syndrome. Arch. Dis. Child. 63, 924–930 (1988).
Manolio, T. A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
Kontrogianni-Konstantopoulos, A. et al. Obscurin modulates the assembly and organization of sarcomeres and the sarcoplasmic reticulum. FASEB J. 20, 2102–2111 (2006).
Marston, S. Obscurin variants and inherited cardiomyopathies. Biophys. Rev. 9, 239–243 (2017).
Brion, M. et al. Sarcomeric gene mutations in sudden infant death syndrome (SIDS). Forensic Sci. Int. 219, 278–281 (2012).
Jaouadi, H. et al. Multiallelic rare variants support an oligogenic origin of sudden cardiac death in the young. Herz 46, 94–102 (2021).
Indrieri, A. et al. The impairment of HCCS leads to MLS syndrome by activating a non-canonical cell death pathway in the brain and eyes. EMBO Mol. Med. 5, 280–293 (2013).
Babbitt, S. E., San Francisco, B., Bretsnyder, E. C. & Kranz, R. G. Conserved residues of the human mitochondrial holocytochrome c synthase mediate interactions with heme. Biochemistry 53, 5261–5271 (2014).
Yang, K. C., Bonini, M. G. & Dudley, S. C. Mitochondria and arrhythmias. Free Radic. Biol. Med. 71, 351–361 (2014).
Wimplinger, I. et al. Mutations of the mitochondrial holocytochrome c-type synthase in X-linked dominant microphthalmia with linear skin defects syndrome. Am. J. Hum. Genet. 79, 878–889 (2006).
Tester, D. J. et al. Exome-wide rare variant analyses in sudden infant death syndrome. J. Pediatr. 203, 423-428.e11 (2018).
Stattin, E. L. et al. Genetic screening in sudden cardiac death in the young can save future lives. Int. J. Legal Med. 130, 59–66 (2016).
Semsarian, C., Ingles, J. & Wilde, A. A. M. Sudden cardiac death in the young: The molecular autopsy and a practical approach to surviving relatives. Eur. Heart J. 36, 1290–1296 (2015).
Behr, E. et al. Cardiological assessment of first-degree relatives in sudden arrhythmic death syndrome. Lancet 362, 1457–1459 (2003).
Pajusalu, S. et al. Large gene panel sequencing in clinical diagnostics-results from 501 consecutive cases. Clin. Genet. 93, 78–83 (2018).
Møller, R. S. et al. Mutations in KCNT1 cause a spectrum of focal epilepsies. Epilepsia 56, e114–e120 (2015).
Gelfman, S. et al. Annotating pathogenic non-coding variants in genic regions. Nat. Commun. 8, 1–10 (2017).
Acknowledgements
We are grateful to the patients and their families for participating in this study and the pediatricians who provided an initial response to these cases at Nagasaki University Hospital, National Hospital Organization Nagasaki Medical Center, and Sasebo City General Hospital. We also would like to thank Enago (www.enago.jp) for the English language review.
Author information
Authors and Affiliations
Contributions
K.S., T.Y., and K.I. conceived the study and experimental design. K.S. and T.Y. performed the targeted gene-sequencing panel analysis and data analysis and drafted the manuscript. K.S. performed Sanger sequencing. T.Y., Y.A., Y.S., M.M., T.M., T.U., and H.Y. contributed to manuscript writing. All authors reviewed the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shingu, K., Murase, T., Yamamoto, T. et al. Accurate interpretation of genetic variants in sudden unexpected death in infancy by trio-targeted gene-sequencing panel analysis. Sci Rep 11, 21532 (2021). https://doi.org/10.1038/s41598-021-00962-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-00962-8
This article is cited by
-
I536T variant of RBM20 affects splicing of cardiac structural proteins that are causative for developing dilated cardiomyopathy
Journal of Molecular Medicine (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.