Abstract
In response to environmental stress the model cyanobacterium, Synechocystis sp. PCC6803 can switch from a planktonic state to autoaggregation and biofilm formation. The precise mechanism of this transition remains unknown. Here we investigated the role of a candidate two-component regulatory system (TCS) in controlling morphological changes, as a way to understand the intermediate molecular steps that are part of the signaling pathway. A bacterial two-hybrid assay showed that the response regulator Rre6 formed a TCS together with a split histidine kinase consisting of Hik36 and Hik43. Individual disruption mutants displayed autoaggregation in a static culture. In contrast, unlike in the wild type, high salinity did not induce biofilm formation in Δhik36, Δhik43 and Δrre6. The expression levels of exopolysaccharide (EPS) production genes were higher in Δhik36 and Δhik43, compared with the wild type, but lower in Δrre6, suggesting that the TCS regulated EPS production in Synechocystis. Rre6 interacted physically with the motor protein PilT2, that is a component of the type IV pilus system. This interaction was enhanced in a phosphomimic version of Rre6. Taken together, Hik36–Hik43–Rre6 function as an upstream component of the pili-related signal transduction cascade and control the prevention of cell adhesion and biofilm formation.
Similar content being viewed by others
Introduction
In liquid medium, bacteria generally grow as planktonic single cells. However, when they encounter environmental stress, bacteria can undergo multicellular autoaggregation. This will cause the cells to sediment and settle on the wall or bottom of the culture vessel1. Extracellular cell-surface structures like pili2, EPS, and extracellular DNA3 contribute to the adherence of the cells to solid materials and to the recruitment of other cells, or to the formation of aggregates in solution, resulting in clumps of cells on solid surfaces1,4. A biofilm is a multicellular bacterial community embedded in a self-produced extracellular polymeric matrix, which protects the cells from biotic and abiotic stress5,6. Bacterial cells can switch between a planktonic state and a biofilm as they adapt to various environmental cues.
In Escherichia coli, the life cycle is controlled by the chemosensory system that regulates flagellar chemotaxis7. This complex system consists of numerous components involved in chemical perception, signaling pathways and the rotation of flagella. The model cyanobacterium, Synechocystis sp. PCC6803 (hereafter Synechocystis) does not possess flagella. Instead, Synechocystis possesses pili that mediate cell motility required for adherence, biofilm formation and locomotion8. This system resembles the flagellar signal transduction pathway and has been extensively studied in Pseudomonas aeruginosa9. Light is another factor regulating autoaggregation in cyanobacteria. Thermosynechococcus vulcanus shows light color-dependent cell aggregation as a strategy for light avoidance10,11. Substrains of Synechocystis that display positive phototaxis (PCC-P) and negative phototaxis (PCC-N) use their type IV pilus (TFP) structure for twitching motility12,13,14,15.
When Synechocystis experiences osmotic upshock or high salinity stress, the initial response consists of controlling intracellular ion homeostasis by way of ion uptake systems, followed by the synthesis of osmolytes16,17,18. However, prolonged salinity stress leads to EPS production and biofilm formation which increases stress tolerance19,20. The details of the molecular mechanisms of this response are not yet understood.
Two-component regulatory systems (TCSs) are common in microbes where they function as stimulus–response mechanisms that sense changes in the environment21,22. TCSs consist of a sensor histidine kinase (Hik) and a response regulator (Rre). A conserved histidine residue in the histidine phosphotransfer (HPt) domain in Hik is autophosphorylated by the catalytic and ATP binding (CA) domain. The phosphoryl group is then transferred to an aspartate residue in the receiver (REC) domain of Rre. This triggers a change in the enzymatic activity, in the affinity of DNA binding and so on. In bacteria, the TCS consisting of the histidine kinase CheA and the response regulator CheY controls bacterial chemotaxis, where phosphorylated CheY interacts with the flagellar motor switch complex23,24. In the genome of Synechocystis, forty-four genes are annotated as hik and forty-two genes are annotated as rre25,26. Synechocystis has three classes of CheA-CheY-like chemotaxis-related gene clusters, the tax1/pixG cluster, the tax2/sll1291 cluster and the tax3/pilG cluster2,13,24,27,28. The tax1/pixG cluster is involved in phototaxis and the tax3/pilG cluster is crucial for motility, pilus biogenesis and genetic transformation competency13,27,28,29. The tax3/pilG cluster contains slr1041 (taxP3, pilG, rre6) and slr1042 (taxY3, pilH, rre7)2,13,24,27,28. Interestingly, in the tax3/pilG cluster the cheA homolog is split into two genes, slr0073 (pilL-N, hik36) and slr0322 (taxAY3, pilL-C, hik43). Based on the fact that mutants in the tax3/pilG cluster showed phenotypes related to phototactic cell movement and on the overall similarity in gene arrangement pattern to CheA-CheY, it had been proposed that Hik36 and Hik43 form a single functional Hik that together with Rre7 functions as a TCS28.
We have previously shown that addition of high amounts of salt to Synechocystis cultures results in decreased polyamine content, which leads to biofilm formation20. To further dissect the signal transduction pathway leading to biofilm formation, we investigated whether the TCS genes in the tax3/pilG cluster mediate autoaggregation and biofilm formation in Synechocystis. Our analysis of protein–protein interactions and of the phenotype of TCS mutants indicated that TCS play an important role in the prevention of autoaggregation under normal condition and in the promotion of cell adhesion and biofilm formation under salt stress.
Results
Hik36 and Hik43 interact with Rre6 but not with Rre7
The Synechocystis tax3/pilG cluster contains two hik genes, hik36 (pilL-N) and hik43 (taxAY3, pilL-C), and two rre genes rre6 (taxP3, pilG) and rre7(taxY3, pilH). While hik36 and hik43 are at distant positions in the genome, rre6 and rre7 are located next to each other27,28 (Fig. 1A and Supplemental Fig. S1). To identify protein–protein interactions between these potential components of TCS, i.e. between the histidine kinases and the response regulators, we performed a bacterial two-hybrid (BACTH) assay with Hik36, Hik43, Rre6 and Rre730. Escherichia coli expressing combinations of Hik36–Hik43, Hik43–Rre6, and Hik36–Rre6 resulted in blue colonies, indicating direct interactions of the proteins. In contrast, no interactions were detected between Hik36–Rre7, Hik43–Rre7 and Rre6–Rre7 (Fig. 1B). These results indicated that Rre6 formed a TCS with Hik36 and Hik43 (Fig. 1C). To identify which domains of Hik43 were responsible for the interaction with Hik36 and Rre6, we generated two Hik43 variants. Hik43ΔC, lacking the CA domain, the CheW interaction domain (CWI), and the receiver domain (REC), and Hik43ΔN, lacking the homodimeric domain (DIM) (Fig. 1A). Hik43ΔC was able to interact with both Hik36 and Rre6. However, deletion of the DIM domain of Hik43 (Hik43ΔN) resulted in white colonies, indicating that the N-terminal domain was required for interaction with Hik36 and Rre6.
Protein–protein interactions between Hik36, Hik43 and Rre6. (A) Diagram showing conserved regions of histidine kinases, Hik36 and Hik43, and response regulators, Rre6 and Rre7. Motifs were predicted according to SMART (https://smart.embl-heidelberg.de). The scale bar represents 100 amino acids. HPT, histidine phosphotransfer domain; DIM, homo-dimeric domain; CA, catalytic and ATP binding domain; CWI, CheW interaction domain; REC, receiver domain. (B) Bacterial two-hybrid assay. Colonies were grown on solid medium containing 0.5 mM IPTG and 80 ng/mL X-Gal for 48 h. A blue colony indicates a positive interaction between the proteins. Escherichia coli containing pKT25-zip and pUT18C-zip were used as positive control (PC), and cells containing pKNT25 and pUT18C were used as negative control (NC). (C) Diagram summarizing the protein–protein interactions determined in (B).
Deletion of hik36, hik43 or rre6 enhanced autoaggregation and reduced salt stress-induced biofilm formation
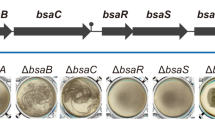
Hik36–Hik43 and Rre7 are known to form at TCS28, but a TCS pair consisting of Hik36–Hik43 and Rre6 has not been studied to date (Fig. 1). To evaluate a potential physiological connection of Hik36–Hik43 and Rre6, individual mutants (Δhik36, Δhik43 and Δrre6) were generated in Synechocystis by insertion of a kanamycin or a spectinomycin resistance gene cassette into the genome sequence (Fig. 2). Reintroduction of the coding sequence of hik36 and hik43 into the native locus in Δhik36 or Δhik43, respectively, failed, presumably due to the low transformation efficiency of the mutants28. In contrast, we were able to reintroduce rre6 into Δrre6, which reflected the normal transformation competency of Δrre628. The complemented strain was designated rre6C. The morphology of wild-type and mutant cells was observed by field emission-scanning electron microscopy (FE-SEM). All mutants were similar to the wild type with respect to size and cell shape (Fig. 3A). We noticed that the disruption mutants spontaneously aggregated and sedimented at the bottom of test tubes when they were left without agitation in the light, a process termed autoaggregation1. Therefore, the sedimentation rates of Δhik36, Δhik43, Δrre6 and rre6C and the wild type were evaluated in static cultures (Fig. 3B). Sedimentation occurred rapidly in Δhik36, Δhik43 and Δrre6 cultures, which appeared transparent after 4 h (OD730 = less than 0.5). In contrast, the wild type and the complemented strain rre6C did not sediment even after 24 h (OD730 = more than 1.5 at 24 h). These results suggested that presence of Rre6 and therefore of a TCS consisting of Hik36–Hik43–Rre6 prevented cell aggregation in the wild-type and rre6C cultures.
Generation of Synechocystis mutants. Schematic representation of disruption or reintroduction of hik36, hik43 and rre6 (left) and corresponding PCR products on an agarose gel (right). The antibiotic resistance cassettes (Spmr: Spectinomycin, Kmr: Kanamycin) are shown as boxes. The size of expected PCR products is given on the left (in kb). Primers are represented by arrows.
Phenotypes of Synechocystis mutants. (A) Representative FE-SEM images of wild type (WT), Δhik36, Δhik43 and Δrre6 Synechocystis cells. (B) Cell sedimentation of WT (filled circles), Δhik36 (filled squares), Δhik43 (open squares), Δrre6 (open triangles) and rre6C (filled triangles). Samples were taken at the surface of cultures left without shaking and OD730 was determined. Each value corresponds to mean ± SD (n = 3) (left). Representative images of cell cultures after being left to settle for 24 h (right). (C) Growth of WT, Δhik36, Δhik43, Δrre6, and rre6C in media without (white bars) and with (black bars) 500 mM NaCl for 3 days. Each value corresponds to mean ± SD (n = 5). (D) Biofilm formation of WT, Δhik36, Δhik43 and Δrre6 in media without (white bars) and with (black bars) 500 mM NaCl for 3 days. Each value corresponds to mean ± SD (n = 8–13). Significant differences between WT and each mutant were analyzed by Tukey's test (* p < 0.05).
Since Synechocystis forms biofilms under salt stress conditions20, Δhik36, Δhik43, Δrre6 and the wild type were cultured without or with added NaCl (500 mM). After growth for 30 days, the cell density of Δhik36, Δhik43 and Δrre6 cultures was similar to that of the wild type in both conditions (Fig. 3C). Biofilm formation in the wild type increased during salt stress, but no such increase was seen in Δhik36, Δhik43 and Δrre6 (Fig. 3D). Cells of Δhik36, Δhik43 and Δrre6 aggregated but were unable to attach to solid materials like the glass or plastic walls of the culture vessels. These results indicated that Hik36–Hik43–Rre6 promoted biofilm production during salt stress.
Expression of EPS-producing genes was changed in Δhik43, Δhik36 and Δrre6
EPS help to protect cells against salt stress and some mutants affected in EPS production display increased cell-to-cell aggregation19. We therefore evaluated whether expression of the four EPS production genes, sll0923, sll1581, slr1875, and sll5052 was changed in Δhik43, Δhik36 and Δrre619 (Fig. 4). Expression of all four genes increased in Δhik43. In Δhik36 expression of sll5052 was the same as in the wild type, but expression of the other three genes also increased. The differences in accumulation of sll5052 transcripts in Δhik36 and Δhik43 may be caused by a difference in the function of Hik36 and Hik43. In contrast, expression of all four genes decreased in Δrre6. These results suggest that loss of hik36, hik43 and rre6 affects expression of the EPS production genes (Fig. 3B).
Expression of EPS production genes. mRNA levels of sll0923, sll1581, slr1875 and sll05052 were determined by quantitative RT-PCR analysis using the 2−ΔΔCT quantification method and rnpB as internal control. Expression levels are shown as relative values to those of the WT. Each value corresponds to mean ± SD (n = 3). Significant differences between WT and mutants were analyzed by Dunnett’s test (** p < 0.01, *** p < 0.001).
Rre6 interacted with PilT2
The motility of Synechocystis is controlled by pili, whose biogenesis, assembly, retraction and function requires dozens of genes31,32. Based on the gene assignment of the type IV pili (TFP) biogenesis and signal transduction system in Synechocystis, we hypothesized that Rre6 might interact with two ATPases, PilB and PilT, that are membrane peripheral proteins of the pilus system, localized on the cytoplasmic side of the inner membrane. Synechocystis contains two copies of both PilB (PilB1, PilB2) and PilT (PilT1, PilT2)12,13. A BACTH assay was performed to analyze possible protein–protein interactions (Fig. 5). Of all combinations tested (Rre6–PilB1, Rre6–PilB2, Rre6–PilT1 and Rre6–PilT2), only Rre6–PilT2 resulted in light blue colonies, indicating that these two proteins interacted with each other (Fig. 5). To examine whether the interaction between Rre6 and PilT2 was dependent on phosphorylation of Rre6, we converted the predicted phosphorylated residue D318 in Rre6 to E (phosphomimic: Rre6D318E). E. coli co-expressing Rre6D318E/PilT2 resulted in blue colonies but colonies expressing Rre6/PilT2 remained white (Fig. 5). These data suggest that Hik36–Hik43-mediated phosphorylation of Rre6 enabled interaction of Rre6 with PilT2.
Analysis of protein–protein interactions of Rre6 with PilB1/2 and PilT1/2. Bacterial two-hybrid assays. Rre6 and Rre6D318E were expressed as T25-fusion proteins. PilB1, PilB2, PilT1 and PilT2 were expressed as T18-fusion proteins. Colonies were grown on solid medium containing 0.5 mM IPTG and 80 mg/mL X-Gal for 48 h. T25 fragments expressed by pKT were shown as ○-. T18 fragments expressed by pUT18C or pUT18 were shown as ●- or -●, respectively. Escherichia coli containing pKT25-zip and pUT18C-zip, or pKT25-zip and pUT18-zip were used as positive control (PC), and the cells containing pKT25 and pUT18, or pKT25 and pUT18C were used as negative controls (NC).
Discussion
Synechocystis is known to induce biofilm formation as an adaptation to environmental changes19,20. The signal transduction pathway regulating this process remains to be elucidated. This study investigated the role of members of the tax3/pilG cluster, namely two genes encoding response regulators, rre6 and rre7, as well as two genes, hik36 and hik43, that together encode a histidine kinase. We showed that Hik36–Hik43 and Rre6 comprise a TCS, and function in the prevention of autoaggregation in Synechocystis (Fig. 6). Yoshihara et al. predicted that Rre7 was the receiver for Hik36–Hik43 in the phosphotransfer reaction due to the fact that the tax3/pilG cluster has a similar genome arrangement to the corresponding cluster in P. aeruginosa28. Contrary to this prediction, our study showed that Hik36–Hik43 physically interacted with Rre6, but not with Rre7. However, we cannot exclude the possibility that Rre7 might also interact with Hik36–Hik43. Rre6 (388 amino acids) and Rre7 (146 amino acids) share high homology in their receiver domains, but Rre6 has an extended N-terminal sequence which is absent from Rre728. This difference of the N-terminal regions might be responsible for the difference in function between Rre6 and Rre7. Hik36 contains a histidine phosphotransfer domains (HPt), responsible for phosphotransfer to an aspartate on a response regulator28. The interaction between Hik36 and Rre6 in the BACTH assay (Fig. 1) therefore suggested a possible phosphotransfer reaction between them. In contrast, the physiological function of the detected interaction between Hik43 and Rre6 remains to be elucidated.
Model for Hik36–Hik43–Rre6 functions in TFP regulation and biofilm formation in Synechocystis. Hik36–Hik43–Rre6 prevents autoaggregation but promotes biofilm formation under high salinity conditions. According to the model by Bertrand et al.9 and Schuergers and Wilde32, PilB and PilT conduct the regulation of pilus extension and retraction, respectively. The major pilin PilA forms the filament and PilB1 assembles the filament. PilT depolymerizes TFP. Hik36–Hik43 phosphorylates Rre6. The phosphomimic Rre6D318E interacts with PilT2. OM, outer membrane; IM, inner membrane. Rre6 affects the expression of EPS production genes, sll0923, sll1581 and sll0505219.
According to the proposed model of the Chp-chemosensory system in Pseudomonas aeruginosa9, PilB and PilT are peripheral membrane proteins of the TFP system that function as motor proteins with ATPase activity (Fig. 6). PilB drives pilus extension, and PilT conducts pilus retraction. Our study detected interaction between Rre6 and PilT2, but not PilB. A phosphomimic form, Rre6D318E was able to bind to PilT2, indicating that Rre6 is part of the signal transduction pathway regulating TFP function (Fig. 5).
Cell aggregation and sedimentation enhance biofilm formation, and the process of cells shifting from a planktonic state to a biofilm involves multiple steps1. In Synechococcus elongatus PCC 7942, loss of pili promotes cell sedimentation and biofilm development33. In Synechocystis, absence of the major pilin PilA1 has little effect on flocculation and TFP are not absolutely required for biofilm development34. Δhik36, Δhik43 and Δrre6 showed an increased rate of sedimentation compared with the wild type, but unlike the wild type they did not form biofilms under salt stress conditions (Fig. 3). This inconsistency between cell aggregation and biofilm formation has been observed in other bacteria35. The outer membrane-associated ligand binding protein, BipA in Bordetella holmesii prevents autoaggregation but promotes biofilm production35. In Synechocystis, sll1581 deletion mutants show increased autoaggregation but less adhesion to glass surfaces because of a reduction of electrostatic repulsion around the cells36. Synechocystis has pili, S-layer and EPS as cell surface structures, which all affect cell to cell interactions and adhesion to solid surfaces in the culture. The rate of sedimentation of Synechocystis is regulated by electrostatic repulsion, which depends on net charges of the EPS36. The expression of EPS genes in Δhik36, Δhik43 and Δrre6 differed from those of the WT (Fig. 4). Therefore, loss of function of the TCS might lead to a disruption of the normal cell surface net charge. Lack of Sll1581 impairs biofilm formation, and loss of Sll0923 decreases EPS by approximately 50%36. Our data indicate that Rre6 promoted the expression of the EPS production genes, sll0923, sll1581 and sll505219 (Fig. 4). Decrease of the amount of EPS in Δrre6 may account for a loss of repulsion, resulting in autoaggregation. In the case of Δhik36 and Δhik43, production of different amounts of individual EPS components might also result in different net charges of the cell surface.
A domain search in Rre6 using SMART (https://smart.embl-heidelberg.de/) and Pfam (https://pfam.xfam.org/) found that Rre6 has only one REC domain without DNA binding motif and enzymatic domain. This is similar to the response regulator PilG from Pseudomonas aeruginosa, a homolog of Rre6, which also contains only one REC domain. PilG pairs with the histidine kinase ChpA in a TCS that stimulates the membrane bound-adenylated cyclase, CyaB37,38. Synechocystis has two adenylate cyclase genes, cya1 and cya2. Mutational analysis revealed that cya1is essential for cell motility, and application of cyclic AMP to the cells confirmed that cyclic AMP is involved in regulation of cell motility39. Understanding the connection of Rre6 with cyclic AMP may be a clue to understanding the signaling pathway regulating the expression of the EPS production genes sll0923, sll1581 and sll5052.
The pili of Synechocystis can be classified into two types, thick pili and thin pili12. The pilus structure of Δrre6 (ΔpilG) is the same as that of the wild type. Δhik36 (ΔpilL-N) has longer and more numerous thick pili, and Δhik43 (ΔpilL-C) has almost no thick pili, but both mutants possess normal thin pili13,28. The data indicated that these mutations affect pilus structure differently although Hik36, Hik43 and Rre6 are part of the same TCS (Fig. 6). A comprehensive study of E. coli TCS revealed that one Hik can form a pair with multiple Rre and that one Rre can form a pair with multiple Hik40. It is therefore possible that other Hik or Rre could also participate in the phospho-relay of Hik36, Hik43 and Rre6, at least with respect to pilus formation. Deletion of PilT1 resulted in a drastic increase in the number of thick pili, whereas deletion of PilT2 had no effect on the number and shape of pili12,13. Deletion of either Rre6 or PilT2 resulted in the same phenotype and neither was involved in regulation of pilus structure. This was also consistent with the fact that Rre6D318E interacted with PilT2 (Fig. 5).
Our study indicates that Hik36–Hik43–Rre6 form a TCS, and that the phosphomimic form, Rre6D318E can physically interact with PilT2. Hik36–Hik43–Rre6 function as an upstream component of the pili-related signal transduction cascade and control the prevention of cell adhesion and biofilm formation. Further studies will be required to work out the details of in vivo protein–protein interactions between Rre6 and Hik36–Hik43 and to fully understand the molecular switch involved in cell sedimentation and cell motility in response to complex environmental changes, for example changes in light and salinity.
Experimental procedures
Cells and growth conditions
Synechocystis sp. PCC 6803 GT-strain was grown at 29 °C in BG11 medium with shaking and continuous light (50 µE m−2 s−1)41. Growth was monitored by measuring the OD730 of the culture.
Crystal violet assay
Crystal violet assay was performed as previously described20. Briefly, cells were cultured in liquid BG11 medium with or without NaCl in a 96-well polystyrene microtiter plate (Merck, https://www.merckgroup.com/en) at 29 °C. The cells attached to the microtiter plate wells were stained with crystal violet. The bound crystal violet was then extracted with 70% ethanol and the absorbance was measured at 590 nm using a plate reader.
Detection by field emission scanning electron microscopy (FE-SEM)
Cells were gently suspended in PBS and fixed in 1.25% glutaraldehyde for 12 h at 4 °C. After dehydration through an ethanol series (from 0 to 100%) and t-butyl alcohol, cells were transferred to polycarbonate membranes and lyophilized in a vacuum evaporator. The membranes were sputter-coated with osmium (POC-3 osmium plasma coater, MEIWAFOSIS Co., https://www.meiwafosis.com/) and examined under a FE-SEM (S-4800; Hitachi, https://www.hitachi.com/).
Site-directed mutagenesis of Rre6
The 318th codon (GAC) of rre6, encoding aspartate (D318) was changed to GAA, encoding glutamate (D318E) as a phosphomimic or to GCC, encoding alanine (D318A) as a mimic of the dephosphorylated form using PCR. To generate rre6D318E, two fragments were generated using primers rre6_XbaI_Fw and rre6_D318E_Rv, or rre6_D318E_Fw and rre6_XbaI_Rv and then fused by PCR using primers rre6_XbaI_Fw and rre6_XbaI_Rv. To generate rre6D318A,two fragments were generated using rre6_XbaI_Fw and rre6_D318A_Rv, or rre6_D318A_Fw and rre6_XbaI_Rv and then fused by PCR using rre6_XbaI_Fw and rre6_XbaI_Rv. Each rre6 variant was subcloned into the XbaI site of pUC19 and the mutations were confirmed by DNA sequencing. All primer sequences are listed in Supplemental Table S1.
Inactivation and reintroduction of genes in Synechocystis sp. PCC 6803
A kanamycin resistance gene (Kmr) amplified with pUC19_Kmr_Fw and pUC19_Kmr_Rv or a spectinomycin resistance (Spmr) gene amplified with pUC19_Spmr_Fw and pUC19_Spmr_Rv was cloned into pUC19, respectively. The resultant plasmids were designated pUC19_Spmr and pUC19_Spmr. To disrupt hik36 (Δhik36), hik43 (Δhik43) or rre6 (Δrre6) in Synechocystis, the coding sequence from start to stop codon of each individual gene was replaced by the Kmr or Spmr gene cassette in this way: The upstream region of hik36, amplified using hik36_U_Fw and hik36_U_Rv, and the downstream region of hik36, amplified using hik36_D_Fw and hik36_D_Rv, were inserted into EcoRV site or HpaI site of pUC19_Spmr. The upstream region of hik43 amplified using hik43_U_Fw and hik43_U_Rv, and the downstream region of hik43 amplified using, hik43_D_Fw and hik43_D_Rv were inserted into EcoRV site or HpaI site of pUC19_Spmr. The upstream region of rre6 amplified using rre6_U_Fw and rre6_U_Rv, and the downstream region of rre6 amplified using rre6_D_Fw and rre6_D_Rv were inserted into EcoRV site or NcoI site of pUC19_Kmr. These plasmids were introduced into Synechocystis to disrupt hik36 (Δhik36), hik43 (Δhik43) or rre6 (Δrre6). After segregation in BG11 medium containing kanamycin (25 µg/mL) or spectinomycin (20 µg/mL), the gene disruptions were confirmed by PCR using hik36_U_Fw and hik36_D_Rv, hik43_U_Fw and hik43_D_Rv, or rre6_U_Fw and rre6_D_Rv with genomic DNA as template.
For reintroduction of rre6 into the native locus, the upstream and the coding sequence of rre6 amplified using rre6_U_Fw and rre6_U_Rv2, was introduced into EcoRV site of pUC19_Spmr. The downstream sequences of rre6 amplified using rre6_D_Fw2 and rre6_D_Rv2 was introduced into HpaI site of the same plasmid. Reintroduction of rre6 with Spmr gene was performed by integration into the Kmr gene in Δrre6. Upon segregation in BG11 medium containing spectinomycin (20 µg/mL), the correct integration was confirmed by PCR using rre6_U_Fw and rre6_D_Rv. The resultant strains were named rre6C. The primer sequences used in this experiment are listed in Supplemental Table S1.
Bacterial two-hybrid assay
A bacterial two-hybrid assay was performed according to the Euromedex manual (Euromedex, https://web.euromedex.com/). For the analysis of interactions among TCSs, hik36 was amplified using hik36_pKNT25_Fw and hik36_pKNT25_Rv and introduced into the BamHI-KpnI sites of pKNT25. The hik36 fragment, digested by BamHI and KpnI was also introduced into the BamHI-KpnI sites of pUT18C. The hik43, rre6 and rre7 were amplified using a pair of primers listed in Supplemental Table S1 and introduced into the BamHI-KpnI sites of pUT18C. The hik43 fragment and rre6 fragment digested by BamHI and KpnI was further introduced into BamHI-KpnI site of pKNT25. The 614 amino acids of C-terminal deletion mutant of hik43 (hik43ΔC) was amplified using hik43_pUT18C_Fw and hik43ΔC_pUT18C_Rv, and the 360 amino acids of N-terminal deletion mutant of hik43 (hik43ΔN) was amplified using hik43ΔN_pUT18C_Fw and hik43_pUT18C_Rv, and introduced into BamHI-KpnI site of pUT18C. The primer sequences used in this experiment are listed in Supplemental Table S1. For the analysis of interactions between Rre6 and pilin related proteins pilB1, pilB2, pilT1, and pilT2, rre6 and rre6D318E were amplified using rre6_pKT25_Fw and rre6_pKT25_Rv, and introduced into SmaI site of pKT25, respectively. The pilB1, pilB2, pilT1, and pilT2 sequences were amplified using a pair of corresponding primers listed in Table S1 and the PCR products were introduced into the SalI site of pUT18 or pUT18C, respectively. Escherichia coli BTH101 cells were co-transformed with each pK and pUT construct. The precultured transformed cells were diluted to an OD600 = 0.005 and 5 µL of them were spotted onto solid Luria–Bertani (LB) medium containing 0.5 mM IPTG and 80 mg/mL X-Gal.
Quantitative RT-PCR
Total RNA was extracted with TRI Reagent (Merck, https://www.merckgroup.com/en) following the manufacturer’s protocol. cDNA was synthesized using ReverTra Ace quantitative PCR reverse transcription master mix with genomic DNA Remover (Toyobo, https://www.toyobo-global.com/). Gene expression levels were analyzed by quantitative RT-PCR using the 2−ΔΔCT quantification method and rnpB as internal control. The primer sequences used in this experiment are listed in Table S1.
Measurement of autoaggregation
Cells were cultured in BG11 medium until they reached OD730 = 0.5–1.0, and then adjusted to OD730 = 3.0 in 10 mL of BG11 medium in test tubes. Cells were left in the growth chamber at 29 °C in continuous light. At every time point, 50 µl of culture were taken from the surface of the liquid.
Abbreviations
- BACTH:
-
Bacterial two-hybrid
- CA:
-
Catalytic and ATP binding
- CWI:
-
CheW interaction
- DIM:
-
Homodimeric
- EPS:
-
Exopolysaccharides
- FE-SEM:
-
Field emission-scanning electron microscopy
- Hik:
-
Histidine kinase
- HPT:
-
Histidine phosphotransfer
- REC:
-
Receiver
- RT-PCR:
-
Real time-polymerase chain reaction
- TCS:
-
Two-component regulatory system
- TFP:
-
Type IV pilus
References
Trunk, T., Khalil, S. H. & Leo, J. C. Bacterial autoaggregation. AIMS Microbiol. 4, 140–164 (2018).
Craig, L., Pique, M. E. & Tainer, J. A. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378 (2004).
Flemming, H. C. et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575 (2016).
Michael Dunne Jr, W. Bacterial adhesion: seen any good biofilms lately?. Clin. Microbiol. Rev. 15, 155–166 (2002).
Toole, G. O., Kaplan, H. B. & Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79 (2000).
Pereira, S. et al. Complexity of cyanobacterial exopolysaccharides: composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol. Rev. 33, 917–941 (2009).
He, K. & Bauer, C. E. Chemosensory signaling systems that control bacterial survival. Trends Microbiol. 22, 389–398 (2014).
Chen, Z., Li, X., Tan, X., Zhang, Y. & Wang, B. Recent advances in biological functions of thick pili in the cyanobacterium Synechocystis sp. PCC 6803. Front. Plant Sci. 11, 1–10 (2020).
Bertrand, J. J., West, J. T. & Engel, J. N. Genetic analysis of the regulation of type IV pilus function by the Chp chemosensory system of Pseudomonas aeruginosa. J. Bacteriol. 192, 994–1010 (2010).
Enomoto, G., Narikawa, R. & Ikeuchi, M. Three cyanobacteriochromes work together to form a light color-sensitive input system for c-di-GMP signaling of cell aggregation. Proc. Natl. Acad. Sci. USA 112, 8082–8087 (2015).
Enomoto, G., Okuda, Y. & Ikeuchi, M. Tlr1612 is the major repressor of cell aggregation in the light-color-dependent c-di-GMP signaling network of Thermosynechococcus vulcanus. Sci. Rep. 8, 1–10 (2018).
Bhaya, D., Bianco, N. R. & Bryant, D. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 37, 941–951 (2000).
Yoshihara, S. et al. Mutational analysis of genes involved in pilus structure, motility and transformation competency in the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 42, 63–73 (2001).
Narikawa, R. et al. Novel photosensory two-component system (PixA-NixB-NixC) involved in the regulation of positive and negative phototaxis of cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 52, 2214–2224 (2011).
Savakis, P. et al. Light-induced alteration of c-di-GMP level controls motility of Synechocystis sp. PCC 6803. Mol. Microbiol. 85, 239–251 (2012).
Hagemann, M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol. Rev. 35, 87–123 (2011).
Nanatani, K. et al. Comparative analysis of kdp and ktr mutants reveals distinct roles of the potassium transporters in the model cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 197, 676–687 (2015).
Pade, N. & Hagemann, M. Salt acclimation of cyanobacteria and their application in biotechnology. Life 5, 25–49 (2015).
Jittawuttipoka, T. et al. Multidisciplinary evidences that Synechocystis PCC6803 exopolysaccharides operate in cell sedimentation and protection against salt and metal stresses. PLoS ONE 8, e55564 (2013).
Kera, K. et al. Reduction of spermidine content resulting from inactivation of two arginine decarboxylases increases biofilm formation in Synechocystis sp. strain PCC 6803. J. Bacteriol. 200, e00664-17 (2018).
Wuichet, K. & Zhulin, I. B. Molecular evolution of sensory domains in cyanobacterial chemoreceptors. Trends Microbiol. 11, 200–203 (2003).
Ashby, M. K. & Houmard, J. Cyanobacterial two-component proteins: structure, diversity, distribution, and evolution. Microbiol. Mol. Biol. Rev. 70, 472–509 (2006).
Stock, A., Chen, T., Welsh, D. & Stock, J. CheA protein, a central regulator of bacterial chemotaxis, belongs to a family of proteins that control gene expression in response to changing environmental conditions. Proc. Natl. Acad. Sci. USA 85, 1403–1407 (1988).
Sarkar, M. K., Paul, K. & Blair, D. Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc. Natl. Acad. Sci. USA 107, 9370–9375 (2010).
Marin, K. et al. Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 100, 9061–9066 (2003).
Murata, N. & Suzuki, I. Exploitation of genomic sequences in a systematic analysis to access how cyanobacteria sense environmental stress. J. Exp. Bot. 57, 235–247 (2006).
Bhaya, D., Takahashi, A. & Grossman, A. R. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc. Natl. Acad. Sci. USA 98, 7540–7545 (2001).
Yoshihara, S., Geng, X. & Ikeuchi, M. pilG gene cluster and split pilL genes involved in pilus biogenesis, motility and genetic transformation in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 43, 513–521 (2002).
Yoshihara, S., Katayama, M., Geng, X. & Ikeuchi, M. Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant Cell Physiol. 45, 1729–1737 (2004).
Karimova, G., Ullmann, A. & Ladant, D. Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 3, 73–82 (2001).
Yoshihara, S. & Ikeuchi, M. Phototactic motility in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Photochem. Photobiol. Sci. 3, 512–518 (2004).
Schuergers, N. & Wilde, A. Appendages of the cyanobacterial cell. Life 5, 700–715 (2015).
Conradi, F. D., Zhou, R., Oeser, S. & Schuergers, N. Factors controlling floc formation and structure in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 201, e00344-19 (2019).
Nagar, E. et al. Type 4 pili are dispensable for biofilm development in the cyanobacterium Synechococcus elongatus. Environ. Microbiol. 19, 2862–2872 (2017).
Hiramatsu, Y. et al. BipA is associated with preventing autoagglutination and promoting biofilm formation in Bordetella holmesii. PLoS ONE 11, 1–18 (2016).
Allen, R., Rittmann, B. E.& Ill, R. C. Axenic biofilm formation and aggregation by Synechocystis concentration and require cell surface structures. Appl. Environ. Microbiol. 85, 1–18 (2019).
Fulcher, N. B., Holliday, P. M., Klem, E., Cann, M. J. & Wolfgang, M. C. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol. Microbiol. 76, 889–904 (2010).
Inclan, Y. F. et al. A scaffold protein connects type IV pili with the Chp chemosensory system to mediate activation of virulence signaling in Pseudomonas aeruginosa. Mol. Microbiol. 101, 590–605 (2016).
Terauchi, K. & Ohmori, M. An adenylate cyclase, Cya1, regulates cell motility in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 40, 248–251 (1999).
Yamamoto, K. et al. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 280, 1448–1456 (2005).
Zavřel, T., Očenášová, P. & Červený, J. Phenotypic characterization of Synechocystis sp. PCC 6803 substrains reveals differences in sensitivity to abiotic stress. PLoS ONE 12, 1–21 (2017).
Acknowledgements
We thank Kei Nanatani and Iwane Suzuki for research discussions, Naomi Hoshi-Ohnuma for technical assistance and Anke Reinders for critical reading of the manuscript. We would also like to thank two anonymous reviewers for their helpful comments.
Funding
This work was supported by JSPS KAKENHI Grant Number (16H06558, 18H03762, 19H02880 and 19K22264 to N.U.).
Author information
Authors and Affiliations
Contributions
K.K., Y.Y., T.S., T.N., M.T. and S.T. performed biochemical experiments and isolated the mutants. K.K., Y.Y., T.S., T.N. and N.U. designed the research, K.K. prepared the figures and NU wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kera, K., Yoshizawa, Y., Shigehara, T. et al. Hik36–Hik43 and Rre6 act as a two-component regulatory system to control cell aggregation in Synechocystis sp. PCC6803. Sci Rep 10, 19405 (2020). https://doi.org/10.1038/s41598-020-76264-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-76264-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.