Abstract
Droplet digital PCR (ddPCR) is a sensitive and reproducible technology widely used for quantitation of several viruses. The aim of this study was to evaluate the 2019-nCoV CDC ddPCR Triplex Probe Assay (BioRad) performance, comparing the direct quantitation of SARS-CoV-2 on nasopharyngeal swab with the procedure applied to the extracted RNA. Moreover, two widely used swab types were compared (UTM 3 mL and ESwab 1 mL, COPAN). A total of 50 nasopharyngeal swabs (n = 25 UTM 3 mL and n = 25 ESwab 1 mL) from SARS-CoV-2 patients, collected during the pandemic at IRCCS Sacro Cuore Don Calabria Hospital (Veneto Region, North-East Italy), were used for our purpose. After heat inactivation, an aliquot of swab medium was used for the direct quantitation. Then, we compared the direct method with the quantitation performed on the RNA purified from nasopharyngeal swab by automated extraction. We observed that the direct approach achieved generally equal RNA copies compared to the extracted RNA. The results with the direct quantitation were more accurate on ESwab with a sensitivity of 93.33% [95% CI, 68.05 to 99.83] and specificity of 100.00% for both N1 and N2. On the other hand, on UTM we observed a higher rate of discordant results for N1 and N2. The human internal amplification control (RPP30) showed 100% of both sensitivity and specificity independent of swabs and approaches. In conclusion, we described a direct quantitation of SARS-CoV-2 in nasopharyngeal swab. Our approach resulted in an efficient quantitation, without automated RNA extraction and purification. However, special care needs to be taken on the potential bias due to the conservation of samples and to the heating treatment, as we used thawed and heat inactivated material. Further studies on a larger cohort of samples are warranted to evaluate the clinical value of this direct approach.
Similar content being viewed by others
Introduction
The recent outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) gives rise to a global public health threat (https://covid19.who.int/). SAR-CoV-2 is an enveloped, non-segmented, positive sense RNA virus that is included in the sub-family Coronavirinae, subgenus Sarbecovirus1. As of today, reverse transcriptase real-time PCR (RT-PCR) technology is used as molecular diagnosis for the SARS-CoV-2 and various protocols have been developed and used in the clinical laboratories worldwide2,3. The majority of protocols includes the RNA extraction and purification process before RT-PCR as a necessary step for the measurement of viral RNA load, as it isolates the genomic RNA from the viral capsid and removes PCR-inhibitors from the original material4. Unfortunately, different extraction kits can provide different amounts of both RNA and inhibitors, hampering the agreement on viral loads and increasing the variability of the data4,5. During the SARS-CoV-2 pandemic, it was observed that the relatively low viral load in the throat of patients and the sensitivity limitation of RT-PCR might produce false negatives in the diagnosis6. In this context, the droplet digital PCR (ddPCR) might be more appropriate for quantitation of viral loads, as previously reported7. The ddPCR allows precise quantitation of nucleic acid copies without the need of a calibration curve and with higher resistance to the amplification inhibitors, compared to the quantitative real-time PCR8; some recent studies reported the usage of ddPCR for the quantitation of SARS-CoV-29,10,11,12,13,14,15,16. However, all the described ddPCR procedures included a RNA extraction/purification step, leading to potential amplification errors, due to variable and suboptimal nucleic acid yields17,18. To the best of our knowledge, there is only one study reporting the direct quantitation (meaning without RNA extraction) of SARS-CoV-2 by ddPCR, targeting gene E19. Concerning the SARS-CoV-2 target genes, previous evidences found that SARS-CoV nucleocapsid (N) region is the optimal target with the highest detection sensitivity20,21. Thus, in order to provide new insights on the direct quantitation of SARS-CoV-2 viral loads from swab-derived material, we evaluated the N region (https://www.fda.gov/media/134922/download). In particular, we evaluated the direct ddPCR on two most commonly used nasopharyngeal swabs, the UTM (Universal Transport Medium) 3 mL and the ESwab (Collection and Transport medium) 1 mL (COPAN). In order to evaluate the potential benefits of our ddPCR approach, we compared the ddPCR performance of the direct quantitation with the ddPCR applied to the extracted RNA in both swab types.
Results
Limit of detection (LoD) and assessment of variability
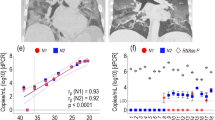
In order to assess LoD and variability of our direct procedure by ddPCR, we performed a tenfold serial dilution of a patient’s sample for each nasopharyngeal swab type. In particular, we used a confirmed SARS-CoV-2 positive sample by our routine molecular diagnostic testing (for the UTM sample the Ct value was 22.06 for gene N1 and 21.62 for gene N2; for the ESwab sample the Ct value was 20.25 for gene N1 and 21.79 for gene N2). We performed the LoD analysis by ddPCR comparing the direct and the extracted RNA approaches. The viral copies were detected up to dilution 10^3 in all replicates for both swab types by the direct approach (data reported in supplementary material). The measurements of the two approaches are reported in Figs. 1 and 2 with the values obtained by the Spearman correlation. For UTM, the measurement from undiluted material was equal by both approaches, while the undiluted Eswab showed lower viral RNA (N1 and N2) signal than the first dilution. Of note, the internal amplification control (IAC) human RPP30 was generally equal, independently of the swab type. Concerning the variability of data, direct quantitation and quantitation from extracted RNA was equally repeatable, independently of the swab type (data reported in supplementary material). Additionally, the coefficient of variation (CV) using direct quantitation generally did not exceed 10% for both swab types and targets, and was in line with the quantitation of the extracted RNA. On the contrary, the undiluted ESwab showed a higher CV (20%) only for target N1.
Comparison of ddPCR direct quantitation and quantitation of extracted RNA on patients’ samples
We investigated a number of clinical samples (total n = 50) comparing the extracted RNA and the direct quantitation. For both approaches, the swab material was used undiluted. Table 1 summarizes the results for each swab and target gene. Supplementary material reports detailed data. Thus, in order to evaluate the clinical value of the approaches, we calculated the sensitivity (SE) and specificity (SP) using the results obtained by our routine RT-PCR diagnostic screening as reference values. Based on the data on the single target gene, the SE and SP of direct approach in UTM were respectively 68.75% [95% CI, 41.34 to 88.98] and 90.00% [95% CI, 55.50 to 99.75] for N1 (with the routine RT-PCR Ct value range of 22.02–35.83, Ct mean 32.77). For N2, the SE and SP were 66.67% [95% CI, 38.38 to 88.18] and 90.00% [95% CI, 55.50 to 99.75] respectively (with the routine RT-PCR Ct value range of 21.62–37.5, Ct mean 32.52). Of note, we observed that the discordant results for N1 and N2 had Ct value ≥ 34 (by the routine RT-PCR).
If we considered a combination of N1 and N2 results, the SE increased to 80% [95% CI, 51.91 to 95.67] and the SP to 93.33% [95% CI, 68.05 to 99.83]. In ESwab direct approach, considering N1 and N2 independently, the SE and SP were respectively 93.33% [95% CI, 68.05 to 99.83] and 100.00% for both N1 (with RT-PCR Ct range of 16.81–38.77, Ct mean 27.36) and N2 (with RT-PCR Ct range of 17.73–37.58, Ct mean 28.42). If we combined N1 and N2, the SE and SP were 100%. On the other hand, using the extracted RNA from the same patient’s samples, the sensitivity (SE) and specificity (SP) in UTM were respectively 93.33% [95% CI, 68.05 to 99.83] and 100% for both N1 and N2. In ESwab the SE was 93.33% [95% CI, 68.05 to 99.83] for N1 and 66.67% [95% CI, 38.38 to 88.18] for N2, while and the SP was 100% for both gene targets. When we combined N1 and N2, the SE was 93.33% [95% CI, 68.05 to 99.83] and SP 100% for both UTM and ESwab. The SE and SP were 100% for RPP30 independently of swab types and approaches. In summary, despite some discordant results, the measurements of RNA copies were generally equal for both viral and human RNA between the direct and the RNA extracted.
Comparison of ddPCR results between UTM and ESwab
We compared the results obtained using ddPCR on UTM swabs with those obtained on ESwab in our cohort (Table 2). No statistically significant differences were detected between the UTM and ESwab in terms of measured copy numbers, for both the direct and extracted RNA approaches, with the exception of N1 quantitation in extracted RNA, in which the UTM method showed a slightly higher signal (p = 0.0105, Wilcoxon test). However, the data need to be confirmed with a larger number of measurements.
Discussion
As of today, few studies reported the use of ddPCR for a more sensitive SARS-CoV-2 detection compared to RT-PCR12,14,15,16. To our knowledge, this is the first report of direct quantitation of SARS-CoV-2 RNA performed on a consistent number of clinical samples and comparing two different nasopharyngeal swabs. Indeed, the RNA extraction from nasopharyngeal swabs of patients affected by SARS-CoV-2 might slacken the diagnostic process due principally to the shortage of reagents for the RNA extraction. To overcome this issue, direct protocols from swab samples before conducting molecular diagnostics have been assessed and reported19,22,23. In our study, we analysed results obtained by ddPCR using an assay based on the Center for Disease Control and Prevention (CDC) recommended targets, directly on material derived from two different nasopharyngeal swab types (UTM and ESwab). The results were compared with those obtained from the RNA extracted from the same swabs. In particular, the data obtained from the human IAC (RPP30), showed that the direct quantitation approach achieved generally equal RNA copies compared to those from the extracted RNA, independently of the swab. On the other hand, for the viral load (N1 and N2 genes), the ddPCR measurements showed that the direct quantitation was generally equal to that obtained from the RNA extracts, but when we performed the limit of detection on ESwab, we observed that the undiluted material might be underestimated. One possible explanation for the different results between UTM and ESwab, could be due to the fact that the varying volume of swab media might denote different amount of both inhibitors and viral capsid proteins influencing the direct quantitation17. In order to overcome this issue, we could hypothesize that the introduction to our procedure of a supplementary pre-treatment using proteinase K19 could be helpful. Moreover, we used two different viral targets (N1 and N2), but future study targeting additional viral regions could be valuable in order to minimize this potential bias and to increase the chance of amplification17. In our work, other potential bias of ddPCR quantities should be taken into consideration as we used thawed material and we chose to pre-heat samples for viral inactivation instead of using chemical treatment. Indeed we avoided to use the most commonly used guanidinium to circumvent possible cause of inhibition of the amplification24,25. However, the heating step was used only for the direct quantitation and it could be another possible cause of underestimation of the amplification as reported recently26.
To conclude, with this work we have demonstrated that our procedure allows the direct quantitation of SARS-CoV-2 RNA. Our ddPCR procedure is simple and direct, avoiding the possible limitations due to the lack of commercial kits for the extraction. The strategy that we proposed does not require great changes of the workflow for laboratories performing the CDC assay. Concerning the performance of direct quantitation on different swab-derived material, although the data obtained on UTM showed generally equal measurement to the RNA extracted, a low SE was found on our cohort. The results from ESwab were more accurate in terms of SE and SP on the cohort samples, but with potential higher amount of inhibitors. Thus, a larger number of specimens and data from other laboratories are needed to evaluate the clinical value of the direct procedure. Further investigations will be necessary focusing on the assessment of the performance of the direct RNA quantitation i) on fresh swab-derived material and ii) using additional viral targets.
Methods
Setting of the study
A total of 50 anonymized samples were used. All the samples were previously screened by RT-PCR with our routine diagnostic testing based on the CDC protocol (N1 and N2 genes) (https://www.fda.gov/media/134922/download). N = 30 samples were positive and N = 20 were negative to SARS-CoV-2. We analysed samples collected using two different nasopharyngeal swab types: N = 15 positive and N = 10 negative in UTM 3 mL (COPAN) and N = 15 positive and N = 10 negative in ESwab 1 mL (COPAN) (Fig. 3). For the ddPCR analysis performed in the preset study, we used aliquots of samples stored at -80 °C. The aliquots were thawed and used for both the automated extraction of RNA and the direct quantitation. All the procedures were performed in BLS2 laboratories, according to the biosafety guidelines for handling and processing specimens associated with Coronavirus Disease 2019 (COVID-19) (https://www.cdc.gov/coronavirus/2019-nCoV/lab/lab-biosafety-guidelines.html#guidance).
Automated RNA extraction
RNA was isolated from 200 μl of nasopharyngeal swab medium by the Nextractor NX-48 robot, using the NX-48S Viral NA Kit (Genolution Inc.), according to the manufacturer’s instructions. Samples were eluted in 50μL of elution buffer. The isolated RNA was immediately used by ddPCR.
Heat inactivation for direct quantitation
For the direct ddPCR, 20 μl of thawed medium from UTM and ESwab were added in a 96-well plate and incubated at 56 °C for 10 min27,28,29 (https://www.who.int/csr/sars/survival_2003_05_04/en/), followed by 4 °C for 5 min and then immediately used by ddPCR.
One step reverse transcriptase—ddPCR
The ddPCR procedure was performed following the manufacturer’s instructions of the 2019-nCoV CDC ddPCR triplex probe assay (dEXS28563542, Bio-Rad). The PCR reaction mixture was assembled as follows: One-Step supermix 2 × for probe (no dUTP) (Bio-Rad), 20 × Assay (for N1, N2, RPP30 detection), reverse transcriptase 20U/μl, RNase free water 7 μl, and RNA template 5 μl or inactivated swab medium 5 μl, in a final volume of 22 μl. Then, QX200 droplet generator (Bio-Rad) was used to convert 20 μl of each reaction mix into droplets. The Droplet-partitioned samples were transferred to a 96-well plate, sealed and processed in a C1000 touch Thermal Cycler (Bio-Rad) under the following cycling protocol: 25 °C for 3 min, 50 °C for 60 min for reverse transcription, 95 °C for 10 min for enzyme activation, 95 °C for 30 s for denaturation and 55 °C for 60 s for annealing/extension for 40 cycles, 98 °C 10 min for enzyme deactivation followed by infinite 4-degree hold. The amplified samples were then transferred and read in the FAM and HEX channels using the QX200 reader (Bio-Rad). The experiments were performed using a negative control (no template control, NTC) and a positive control (a patient’s sample confirmed positive by RT-PCR with our routine diagnostic testing). The reactions with less than 10,000 droplets and discordant results were repeated. Data were analysed using the QuantaSoft™ v1 AnalysisPro Software (Bio-Rad) and expressed as Log10 (copies/mL).
Limit of detection analysis
For each nasopharyngeal swab, we used a patient’s sample (confirmed positive by RT-PCR with our routine diagnostic testing) to generate tenfold serial dilutions for a total of five points of both RNA extracted and swab-derived material. Each dilution point was analysed by ddPCR in triplicate and the repeatability intra-assay was assessed. Results were expressed as Log10 (copies/mL).
Statistics
The statistical analyses and graphical representations were performed by GraphPad Prism 8. Data are reported as mean ± SD. Spearman’s correlation was performed between measurements, due to the small sample size. Paired non parametric Wilcoxon Test was performed to compare the two approaches, ddPCR on RNA extracted and direct ddPCR on the swab-derived material UTM and ESwab. A p value ≤ 0.05 was considered statistically significant.
Ethical statement
The study (No. 39528/2020 Prog. 2832CESC) was approved by the competent Ethics Committee for Clinical Research of Verona and Rovigo Provinces. Written informed consent was obtained from the patients and all research was performed in accordance with relevant guidelines/regulations.
Data availability
All data generated or analysed during this study are included in this published article (and its supplementary information files).
References
Astuti, I. & Ysrafil, Y. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndr. Clin. Res. Rev. 14(4), 407–412 (2020).
Venter, M. & Richter, K. Towards effective diagnostic assays for COVID-19: a review. J. Clin. Pathol. 73(7), 370–377 (2020).
Corman, V. M. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 25(3), 2000045. https://doi.org/10.2807/1560-7917.ES.2020.25.3.2000045 (2020).
Monleau, M. et al. Evaluation of different RNA extraction methods and storage conditions of dried plasma or blood spots for human immunodeficiency virus type 1 RNA quantification and PCR amplification for drug resistance testing ∇. J. Clin. Microbiol. 47(4), 1107–1118 (2009).
Lim, N. Y. N., Roco, C. A. & Frostegård, Å. Transparent DNA/RNA co-extraction workflow protocol suitable for inhibitor-rich environmental samples that focuses on complete DNA removal for transcriptomic analyses. Front. Microbiol. 7, 1588. https://doi.org/10.3389/fmicb.2016.01588 (2016).
Arevalo-Rodriguez, I. et al. False-negative results of initial RT-PCR assays for COVID-19: a systematic review. medRxiv https://doi.org/10.1101/2020.04.16.20066787 (2020).
Strain, M. C. et al. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS ONE 8(4), e55943. https://doi.org/10.1371/journal.pone.0055943 (2013).
Kuypers, J. & Jerome, K. R. Applications of digital PCR for clinical microbiology. J. Clin. Microbiol. 55(6), 1621–1628 (2017).
Suo, T. et al. ddPCR: a more sensitive and accurate tool for SARS-CoV-2 detection in low viral load specimens. medRxiv 9(1), 1259–1268 (2020).
Dong, L. et al. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. medRxiv https://doi.org/10.1101/2020.03.14.20036129 (2020).
Wang, X. et al. Limits of detection of 6 approved RT–PCR kits for the novel SARS-Coronavirus-2 (SARS-CoV-2). Clin. Chem. 66(7), 977–979 (2020).
Yu, F. et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin. Infect. Dis. 71(15), 793–798 (2020).
Liu, X. et al. Analytical comparisons of SARS-COV-2 detection by qRT-PCR and ddPCR with multiple primer/probe sets. Emerg. Microbes Infect. 9(1), 1175–1179 (2020).
Falzone, L. et al. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int. J. Mol. Med. 46(3), 957–964 (2020).
Lv, J. et al. Detection of SARS-CoV-2 RNA residue on object surfaces in nucleic acid testing laboratory using droplet digital PCR. Sci. Total Environ. 742, 140370. https://doi.org/10.1016/j.scitotenv.2020.140370 (2020).
Suo, T. et al. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 9(1), 1259–1268 (2020).
Pavšič, J., Žel, J. & Milavec, M. Digital PCR for direct quantification of viruses without DNA extraction. Anal. Bioanal. Chem. 408(1), 67–75 (2016).
Huggett, J. F., Cowen, S. & Foy, C. A. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin. Chem. 61(1), 79–88 (2015).
Marzinotto, S. et al. A streamlined approach to rapidly detect SARS-CoV-2 infection, avoiding RNA extraction. medRxiv https://doi.org/10.1101/2020.04.06.20054114 (2020).
Huang, J. L. et al. Rapid and sensitive detection of multiple genes from the SARS-coronavirus using quantitative RT-PCR with dual systems. J. Med. Virol. 77(2), 151–158 (2005).
Chu, D. K. W. et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 66(4), 549–555 (2020).
Bruce, E. A. et al. RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swab using Qiagen RNEasy kits or directly via omission of an rna extraction step. bioRxiv https://doi.org/10.1101/2020.03.20.001008 (2020).
Grant, P. R., Turner, M. A., Shin, G. Y., Nastouli, E. & Levett, L. J. Extraction-free COVID-19 (SARS-CoV-2) diagnosis by RT-PCR to increase capacity for national testing programmes during a pandemic. bioRxiv https://doi.org/10.1101/2020.04.06.028316 (2020).
Andreychuk, D. B. et al. Armoured exogenous internal control for real-time PCR diagnosis of avian influenza. Avian Pathol. 48(5), 492–498 (2019).
Arezi, B., McCarthy, M. & Hogrefe, H. Mutant of Moloney murine leukemia virus reverse transcriptase exhibits higher resistance to common RT-qPCR inhibitors. Anal. Biochem. 400(2), 301–303 (2010).
Zou, J. et al. Heat inactivation decreases the qualitative real-time RT-PCR detection rates of clinical samples with high cycle threshold values in COVID-19. Diagn. Microbiol. Infect. Dis. 98(1), 115109. https://doi.org/10.1016/j.diagmicrobio.2020.115109 (2020).
Darnell, M. E. R., Subbarao, K., Feinstone, S. M. & Taylor, D. R. Inactivation of the coronavirus that induces severe acute respiratory syndrome, SARS-CoV. J. Virol. Methods 121(1), 85–91 (2004).
Kariwa, H., Fujii, N. & Takashima, I. Inactivation of SARS coronavirus by means of povidone-iodine, physical conditions and chemical reagents. Dermatology 212(1), 119–123 (2006).
Kampf, G., Voss, A. & Scheithauer, S. Inactivation of coronaviruses by heat. J. Hosp. Infect. 105(2), 348–349 (2020).
Acknowledgments
We thank Giulia La Marca, Martina Leonardi and Claudio Duffini (IRCCS Don Calabria Sacro Cuore Hospital) for their contribution to the study.
Funding
This work was supported by the Italian Ministry of Health “Fondi Ricerca corrente – L1P6″ to IRCCS Sacro Cuore – Don Calabria Hospital”.
Author information
Authors and Affiliations
Contributions
EP and MD conceived and designed the analyses. AM contributed to design the experiments. SS contributed to the collection of samples. EP and MD performed the experiments and analysed the data. EP draft the paper. MD, AM, CP and MF contributed to the revision of the draft. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Deiana, M., Mori, A., Piubelli, C. et al. Assessment of the direct quantitation of SARS-CoV-2 by droplet digital PCR. Sci Rep 10, 18764 (2020). https://doi.org/10.1038/s41598-020-75958-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-75958-x
This article is cited by
-
On-demand ferrofluid droplet formation with non-linear magnetic permeability in the presence of high non-uniform magnetic fields
Scientific Reports (2022)
-
Nested pool testing strategy for the diagnosis of infectious diseases
Scientific Reports (2021)
-
The impact of coronavirus SARS-CoV-2 (COVID-19) in water: potential risks
Environmental Science and Pollution Research (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.