Abstract
Mycobacterium avium subsp. paratuberculosis (MAP) is the etiological agent of Johne’s disease or paratuberculosis, a chronic infection affecting domestic ruminants worldwide. Despite sporadic reports of MAP occurrence in non-ruminants, information on the risk factors predisposing for infection is still scarce and evidence of transmission paths linking the livestock-wildlife-environment interfaces also remains lacking. In this study, we predicted that environmental, host-related, land use and human driven disturbance factors would modulate carnivore exposure to MAP. To test these hypotheses, we performed a retrospective survey, based on microbiological and molecular methods, in mainland Portugal including five sympatric species from the Herpestidae, Canidae, Viverridae, and Mustelidae families (n = 202) and examined 16 variables as putative predictors of MAP occurrence. Molecular evidence of MAP using IS900 as proxy was demonstrated in 7.43% (95%CI: 4.55–11.9) of surveyed carnivores, the highest proportions being registered for red fox (Vulpes vulpes) (10%; 95%CI: 4.0–23) and Egyptian mongoose (Herpestes ichneumon) (6.0%; 95%CI: 3.2–11). We demonstrate that important species of the Mediterranean carnivore guild, such as stone marten (Martes foina) and common genet (Genetta genetta), may also be exposed to MAP, being this the first time that occurrence in genet is reported. The high proportion of DNA-positive specimens, concurrent with the apparent lack of gastro-enteric lesions and molecular confirmation of IS900 in feces, argue for the presence of subclinical carriers that occasionally shed bacteria, potentially aiding as source of infection to susceptible species and possibly contributing for environmental contamination. Achievement of MAP isolation would prove beyond any doubt that MAP is present in this wildlife population. Ecological modelling results suggested that the probability of MAP infection using IS900 as proxy in mongoose is positively associated with higher altitude and temperature stability, as well as with lower annual rainfall. Density of livestock farms was found not to be a significant predictor, which may indicate that the livestock-wildlife interface is probably not important as an infection route for mongoose.
Similar content being viewed by others
Introduction
Mycobacterium avium subspecies paratuberculosis (MAP) is a member of the Mycobacterium avium complex (MAC) that also includes M. avium subsp. avium, M. avium subsp. silvaticum, and M. avium subsp. hominissuis. This group of fastidious, phenotypically diverse, bacteria has specific pathogenicity and host range features, worldwide distribution and presumed zoonotic potential1,2,3.
MAP is an infectious enteric pathogen that causes paratuberculosis or Johne’s disease, a chronic infection of the gastrointestinal tract that affects ruminants worldwide4. According to the OIE Worldwide Animal Health Information Database (WAHID interface), 58 out of 241 countries reported MAP infection during 2017. While not all countries report infection, paratuberculosis is a communicable disease that is widespread and represents a major problem for animal health, causing substantial economic losses that differ according to the production system, herd size, or herd management type4. Different figures have been advanced for both dairy and beef cattle and these may vary from country to country (reviewed in Garcia and Shalloo, 20155). In the USA alone, costs have been estimated from $200 to $1,500 million6,7,8. Financial losses are related with decreased milk productivity, reduced feed conversion efficiency and slaughter value, limitations on animal trade and transactions of animal products, and increased costs related with premature culling and veterinary care5.
Ruminants appear to be the preferred or natural host for MAP, primarily domestic ruminants such as cattle, sheep and goat, but the disease in wild ruminants is also well documented, including red deer (Cervus elaphus), roe-deer (Capreolus capreolus), fallow deer (Dama dama), white-tailed deer (Odocoileus virginianus), alpine ibex (Capra ibex) and riverine buffalo (Bubalus bubalis)9,10,11,12,13,14.
In non-ruminant animals, MAP was first detected in wild rabbits (Oryctolagus cuniculus) in Scotland15,16. After this finding, the research for the presence of this pathogen was extended to other wildlife species, firstly in areas with previous history of paratuberculosis in livestock, which allowed the detection of MAP in a very broad host range, including brown bear (Ursus arctos), raccoon (Procyon lotor), opossum (Didelphis virginiana), coyote (Canis latrans), red fox (Vulpes vulpes), stoat (Mustela erminea), weasel (Mustela nivalis), wood mouse (Apodemus sylvaticus), European badger (Meles meles), hare (Lepus europaeus) and jackdaw (Corvus monedula)14,17,18,19,20,21.
Disease progression and clinical signs are best characterized in domestic ruminants, where the pathogen typically enters the host by the oral route and then, eventually, will cause a progressively chronic inflammatory state with the presence of granulomatous lesions in the small and large intestines and in the mesenteric lymph nodes, leading to progressive weight loss, diarrhea and decreased milk production22. MAP excretion occurs mainly in feces but, in advanced stages of infection, the pathogen may also be found in colostrum and milk, saliva, uterine fluid and semen, and cause fetal infections in utero22,23,24,25. As such, the main route of intra- and interspecies transmission is considered fecal-oral by direct ingestion of MAP-contaminated feces, infected colostrum or milk or, indirectly, via MAP-contaminated water and/or feed (livestock feed, vegetation and/or infected prey)26,27.
In Portugal, paratuberculosis was first reported in cattle from Lisboa region by Petisca and colleagues, in 196128. Since then, geographically-limited studies, circumscribed to the North, Alentejo region and Lisbon, have tried to assess its occurrence, mainly in small ruminants29,30,31,32,33. Reports refer mainly to serological or molecular evidences rather than to clinical or anatomo-histological data29,30,31,32,33,34, suggesting that probably this disease is under-diagnosed. These studies, spanning the 1994-2007 period, report varying antibody prevalence rates at the animal-level that can reach as much as 7% in cattle and, roughly, 10–18% and 10% in sheep and goat, respectively, depending on the region and screening method (Fig. 1). MAP infection in slaughtered domestic pigs was also documented35. More recently, regional studies with small sampling sizes reported MAP occurrence in wildlife, specifically wild boar, red deer and also carnivores36,37,38 (Fig. 1). Whether MAP is self-maintained in these wild populations or arise from animal contact with external sources of infection is yet undetermined. Interaction of these wild species with contaminated faeces from infected ruminants, feeding and scavenging on infected prey, or sympatry with yet unidentified reservoir species, possibly contribute to the dynamic maintenance of MAP in the wild, in Portugal.
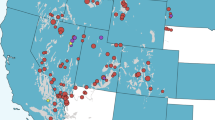
(a) Regional prevalence of MAP in mainland Portugal, showing districts as sampling unit at the administrative level. MAP distribution and prevalence intervals per district, in ovine, caprine and bovine subpopulations are evidenced in grey scale, dots and dashes, respectively, as indicated. Data represented was compiled from previous studies31,32,33. n – number of animals serologically tested; n1 - ovine; n2 - caprine and n3 – bovine; (b) MAP distribution in the wild: carnivore species (grey shade; data from this study), rabbits (dots77) and carnivores, wild boar and red deer from Central East/Castelo Branco district (horizontal lines36,37,38). n1 - number of animals tested in this study; n2 - number of animals tested in previous studies. The proportion of exposed carnivores determined in the current study is shown as intervals.
Control of paratuberculosis can only be achieved through identification of MAP host range and transmission paths linking the livestock-wildlife interface to adjust surveillance and ascertain biosecurity measures. As such, the main objectives of this work were to investigate the ecological spread and spill-over of MAP into non-paradigmatic hosts, contributing to the disclosure of wild species that potentially may be exposed to this pathogen in mainland Portugal, fine-tuning the proportion of exposed animals and the spatiotemporal distribution of MAP in sympatric carnivores. For this purpose, widely distributed animal specimens from the Mustelidae, Herpestidae, Viverridae and Canidae families, sampled during a 10-year period across the whole territory of the mainland, were opportunistically surveyed for the presence of this bacterium using a polyphasic diagnostic approach based on microbiological and molecular methods. We also aimed at identifying environmental (climate and orography), bio-ecological (food resources, population and individual factors), and disturbance factors (human and cattle) that correlate with MAP infection in carnivores which might help to understand this pathogen’s ecology and serve as predictors for ascertaining populations at risk.
Methods
Biological specimen collection and demographic characteristics
The presence of MAP was opportunistically investigated in widely distributed free-ranging animals from four families of the Carnivora order (n = 202): Egyptian mongoose (Herpestes ichneumon, Herpestidae family, n = 149), red fox (Vulpes vulpes, Canidae; n = 40), common genet (Genetta genetta, Viverridae; n = 5), stone marten (Martes foina, Mustelidae; n = 4) and European badger (Meles meles, Mustelidae; n = 4), collected between 2005 and 2014 in mainland Portugal (Supplementary Table 1; Supplementary Fig. 1). The surveyed animals were either found dead on the road (n = 28) or hunted during legal hunting actions aiming the control of predator densities (n = 174). Corpses were donated for scientific purposes by road maintenance companies, hunters, game reserve owners or hunting associations. No animals were sacrificed for the purposes of this specific study. None of the authors were responsible for the death of any animals nor were any samples used in the study collected by the authors. All applicable institutional and/or national guidelines for the care and use of animals were followed.
Each specimen was assigned to one of four age classes (adults, sub-adults, juveniles and cubs), according to body length and dental growth39,40. Animals were geo-referenced upon collection.
Diversity analyses to assess sampling bias
A diversity analyses to ascertain adequacy of sampling for epidemiological purposes was performed, considering the constraints related to our opportunistic strategy. The relationship between abundance and distribution of samples was established by linear regression; the calculation of diversity indices (Shannon-Wiener, Simpson and Berger-Parker), according to Help and collaborators41, and the determination of non-parametric estimators (chao 1 and chao 2) of species richness, according to Gotelli and Cowell42, were also performed. Mathematical formula used for these calculations are provided in Supplementary Table 2.
Post-mortem examination
Animal carcasses were frozen at −20 °C (most for up to two months, although a few had been stored for one year) until post-mortem examination. The carcasses were thawed and examined. Those macroscopically showing clear signs of putrefaction, or in evident poor preservation condition, were eliminated from the study. A portion of the spleen was removed aseptically to sterile containers, refrozen at −20 °C and later processed for bacteriological culture and nucleic acid extraction as described below. The abdominal cavity of each specimen was opened and the intestines isolated. Solid intestinal content was collected from the rectum of each animal, using a sterile feces collection tube, refrozen at −20 °C, which was subsequently processed for further analysis (Supplementary Table 1).
Fecal (n = 111) and/or spleen (n = 149) samples were tested by microbiological culture and/or molecular analysis in the scope of a master thesis43. No gross lesions suggestive for MAP were detected during necropsy, even though histopathological examinations performed in the scope of another study (unpublished data) evidenced autolysis of a proportion of specimens.
All necropsy procedures and subsequently sample collection followed all applicable institutional guidelines and regulations.
Bacterial culture of feces and spleen
For bacterial culture purposes, 1 g of feces and 2 g of spleen samples removed aseptically from the containers were macerated and homogenized with 20 ml sterile water or sterile saline, respectively. After homogenization, 5 ml were maintained at 4 °C until further processing for DNA extraction, and other 5 ml were decontaminated with 0.9% (v/v) or 0.7% (v/v) of hexadecylpyridinium chloride monohydrate (HPC) (Sigma-Aldrich), respectively, following the methodology described in OIE manual guidelines (2008). After decontamination, the sample was centrifuged and the sediment inoculated (100 μL) into slants containing standard cultivation media: Herrold’s egg yolk medium (HEYM) supplemented with mycobactin J and HEYM without mycobactin (Biogerm, Portugal). MAP positive controls (K10 strain) and physiological saline blanks were seeded in solid media in parallel with each processed batch for culture. After inoculation, the tubes were maintained at 37 °C and observed for bacterial growth every week. The incubation period was at least 6 months. Bacterial culture procedures were carried out in a biosecurity level 2 facility, in accordance with the experimental protocols implemented and approved by INIAV, the national reference laboratory (NRL) for animal health, Portugal.
Microbiological culture in HEYM with and without mycobactin J was attempted for all the fecal samples. Subsequently, based on the positive results from nested real-time IS900 PCR, nine spleen samples from feces-positive specimens were also processed for culture.
DNA extraction from fecal and tissue suspensions
DNA extraction directly from 5 mL of feces or spleen suspensions was performed using the commercial system Invisorb® Spin Tissue Mini Kit (Stratec Molecular) and High Pure PCR Template Preparation Kit (Roche), respectively, following the manufacturer’s instructions. Despite using commercial systems, we optimized nucleic acid extraction by including additional steps for the concentration of samples and the mechanical disruption of cells in order to maximize sensitivity. A bead-beating protocol was used for mechanical disruption, before applying Proteinase K. Zirconium beads were added to sample suspensions in lysis buffer and 6.5 ms−1 cycles for 45 sec, repeated twice, were performed in the FastPrep FP120 Bio101 (Savant Instruments). Disrupted samples were cooled on ice for 15 minutes, 50 µL of proteinase K was added, and the resulting mixture was incubated overnight at 52 °C. The remaining procedure was performed according to the manufacturer’s instructions. The genomic DNA was eluted with 100 µL of elution buffer and stored at −20 °C until further use. Blanks (negative tissue or saline) were included to discard contamination during extraction. A bovine fecal sample known to be MAP-positive by culture was also included in several batches of DNA extraction to confirm that the procedure would adequately extract MAP DNA. All steps were carried out in a laminar flow cabinet and precautions were taken during all stages to avoid nucleic acid carry-over. Obviously, only experiments wherein negative and positive controls performed correctly were considered.
Nested real-time PCR for the detection of IS900 insertion sequence
The detection of MAP in the genomic DNA extracted from biological samples was performed by a novel nested real-time PCR approach targeting IS900, developed at INIAV (LNR) in the scope a PhD thesis44. The genomes of MAP strains contain 15–20 copies of IS90045 making this a good target for molecular diagnosis purposes. This new in-house IS900-PCR includes modifications of a previously validated real-time PCR system optimized for human samples46 in order to increase diagnostic sensitivity in animal samples44,47. These newly designed primers (forward: 5′-TGATCTGGACAATGACGGTTACGGA-3′; reverse: 5′-GGCGTTGAGGTCGATCGCCCACGTGAC-3′) are located within IS900 but are external to the amplicon generated by the methodology described by Sidoti and co-workers46. They target a fragment of 224 bp, located between positions 204 e 427 of IS900 sequence. The second amplification step, amplifying an internal IS900 sequence, maintained the primers and probe described by Sidoti et al.46. The specificity of both primer pairs and hydrolysis probe sequences was assessed against the NCBI nucleotide sequence database. The in silico analysis showed 100% identity between the nucleotide sequences of the primers and probe with the homologous sequences in Mycobacterium avium subsp. paratuberculosis, confirming specificity of this molecular procedure to detect MAP.
The PCR reaction was carried out in a final volume of 25 μl containing 1x NZYTaq Colourless Master mix (NZYTech), 0.5 μM of each primer, DNase-free water and 5 μl of extracted DNA template. The first amplification reaction was performed in C1000 Thermal Cycler (Bio-Rad) at 55 °C (annealing temperature) and the amplified products were stored at 4 °C, until electrophoretic analyses in a 2% agarose gel prepared in TBE (Tris-Borate-EDTA) 1x, or directly used in the second real-time PCR step. The second real-time amplifies a 66 bp sequence within the first amplicon using primers, probe and experimental conditions of Sidoti and collaborators46. This PCR was performed using as template 5 μL of the amplification product from the first PCR. Real-time PCR reactions were performed in CFX96 Real-Time Thermal Cycler (Bio-Rad) and the amplified products were stored at 4 °C until electrophoretic analyses in a 2% agarose gel. Fluorescence measurements were recorded after each annealing step (at 60 °C) and data was analyzed with the software Bio-Rad CFX Manager 3.0 (Bio-Rad) and in relation to standard calibration curves prepared as described below. For all calculations, the baseline and the fluorescence threshold were set automatically.
In all PCR batches, in addition to samples, positive controls (DNA extracted from MAP ATCC 19698T and DNA from a positive bovine fecal sample) and a no-template control (water) were included. Since a nested approach was used, multiple negative controls (uninfected bovine feces extracted as described up above and no-template) interleaved with samples were included for PCR to discard cross-contamination and pseudopositives. No-template and positive controls were included in each single assay (including DNA extraction, molecular detection, and also culture steps). All samples were tested by PCR (standard and real-time) at least three times and considered positive or negative if yielding coherent results in at least two independent amplification experiments. Only experiments wherein negative and positive controls performed correctly were considered. PCR steps (reaction mix preparation and template addition) were carried out in different laminar flow cabinets located in separate rooms, according to routine laboratorial practices at INIAV IP. Precautions were taken during all stages to avoid nucleic acid carry-over. For quality control purposes, six positive and 16 negative samples were re-extracted and re-tested by PCR. Result agreement was 100%. In several occasions, the amplified products from both PCR reactions were analyzed in 2% agarose gels prepared in TBE 1x and amplicons checked for the expected size. This procedure was always employed whenever the threshold cycles (Cq) of the real-time PCR analyses crossed 35, to confirm amplification of fragments with expected size.
An animal was classified as MAP-positive if the bacterium was isolated from at least one biological matrix in selective media and/or if MAP IS900 was detected by means of PCR at least twice (out of three independent tests).
Performance of the IS900 nested real-time PCR
Spiked faecal samples with ten-fold dilutions of a suspension of MAP cells were used to assess detection limits (LOD) of the nested approach44. Faecal samples were pooled from cattle that tested negative for paratuberculosis by traditional culture, immunological and PCR tests, and showed no histopathology consistent with paratuberculosis following slaughter. Six grams of pooled faeces was spiked with ten-fold dilutions of a suspension of MAP cells, either with the MAP K10 (Type C) or MAP 235G (Type S) strains, in a range of 104 to 101 cells per gram of faeces, tested in triplicate. An additional 6 g of faeces without MAP cells was used as a negative control. The number of MAP cells was estimated by microscope count. The tubes were mixed well and stored at −20 °C until required. The genomic DNA of the faecal spiked samples was extracted and tested by nested real-time PCR approach targeting IS900 with the same procedures as the faecal samples44.
The performance of this novel PCR assay was tested44 using a set of 75 ruminant faecal samples, showing a diagnostic sensitivity of 96.6% and a kappa coefficient of 0.60 (moderate strength of agreement) when compared with culture gold standard44. The sensitivity of this nested approach was shown to be superior to that of the single-step real-time PCR46 when this was applied as a single approach to the same samples (96.6% versus 74.6%, respectively)44. For MAP Type C, positive amplification results were detected at an infection rate of 101 cells per gram of faeces, while for Type S the detection limit was 102 cells per gram44,47, thus showing that this procedure is superior in its ability to detect MAP.
Nineteen reference, clinical and environmental strains of Mycobacterium avium complex (MAC) (M. avium subsp. hominissuis, M. avium subsp. avium, M. avium subsp. paratuberculosis, M. scrofulaceum), non-MAC mycobacteria (M. bovis BCG, M. tuberculosis) and non-mycobacterial (gram positive and gram-negative) clinical species maintained at INIAV were used44. Only MAP strains yielded amplification of IS900 with the amplification system herein described (100% specificity and sensitivity), confirming that this novel PCR methodology is fully specific to detect MAP.
The analytical sensitivity, specificity and kappa coefficient were computed using the previously mentioned panel of strains and samples and the publicly available clinical research calculators on VassarStats48 website (http://vassarstats.net).
The details of these methodologies and the validation work are fully described elsewhere44.
Sequence analyses of IS900 nested real-time PCR amplicons
Three 224 bp amplicons from red fox, mongoose and genet, generated from the first PCR round with the newly established IS900 primers were excised from the agarose gel, purified using QIAquick gel extraction kit (Qiagen) and commercially sequenced (GATC Biotech and Eurofins). The obtained sequences were aligned using Basic Local Alignment Search Tool (BLASTn) in order to find regions of local similarity with sequences from the database. Eleven IS900 sequences from 11 MAP strains genomes (Accession numbers AJ250023.1, AJ250018.1, AJ250016.1, AJ250017.1, AJ250015.1, AJ250021.1, AJ250022.1, S74401.1, AF416985.1, X16293.1 and HM015765.1) and two IS900-like sequences (from Mycobacterium sp. 2333, accession number AF455252.1; Mycobacterium porcinum, accession number EU126150.1) were retrieved from NCBI nucleotide database and aligned with the amplicon sequences obtained in this work using BioEdit Sequence Alignment Editor v7.0.5.3.
Real-time PCR for the detection of MAP F57 sequence
Confirmatory testing of IS900 positive samples was attempted by a specific real-time PCR assay for the single-copy MAP F57 sequence, following the procedure, primers and probe described by Sidoti and collaborators46. Real-time PCR assays aiming the amplification of a 80 bp fragment were performed using QuantiFast Pathogen PCR + IC Kit (Qiagen). This kit includes an Internal Amplification Control (IAC), as well as specific reagents and primers/probes required for its amplification. The optimized setup with a final 25 μl volume per reaction, contained 1x QuantiFast Pathogen Master Mix, 1x IAC assay, 1x Internal control DNA, 0.4 μM of each primer, 0.2 μM of probe, DNase-free water and 5 μl of DNA template. All PCR reactions were also performed in CFX96 Real-Time Thermal Cycler (Bio-Rad) according to the following cycling conditions: 95 °C for 10 min, followed by 45 two-step cycles of 95 °C for 15 s and 60 °C for 1 min, with data acquisition at this step. The software Bio-Rad CFX Manager 3.0 (Bio-Rad) was used for data analysis. The baseline and the fluorescence threshold were set automatically. In all PCR batches, in addition to samples, positive control (DNA extracted from MAP ATCC 19698T) and a no-template control (water) were included. The products of amplification reactions were stored at 4 °C until electrophoretic analyses in a 2% agarose gel.
Statistical analyses of the mongoose’s results
Chi-square test and Mann-Whitney test (α = 0.05) were performed whenever appropriate in IBM SPSS Statistics 22.0; and graphics were produced in Excel and GraphPad Prism 6.0. Significant statistical differences were established at p < 0.05. The confidence intervals (95% CI) were calculated online on the website: VassarStats: Website for Statistical Computation48. Based on the known ecology of mongoose40,49,50,51,52 and MAP occurrence in ruminants, we tested the influence of variables that could directly or indirectly influence MAP infection (using IS900 detection as proxy) in mongoose populations, the only carnivore species with enough samples to allow a modeling procedure. Thus, we considered five categories of variables, representing different working hypotheses: H1) productivity/food resources availability (Normalized Difference Vegetation Index-NDVI, wild rabbit and red-legged partridge abundances); H2) disturbance (human population density and road network density); H3) life-history traits [(gender, age and morphometry (e.g. body size), mongoose abundance]; H4) Climate and orography [season, average annual temperature, annual rainfall, and annual temperature range- data were calculated by interpolations of observed data, representing average values for the period 1960–199053; altimetry, river network density]; and H5) Cattle (density of cattle farms). Each variable was represented by mean values of the 2 × 2 km grid cell, considering the home range of the Egyptian mongoose54. Climate data comprised variables gathered from BioClim (2015)55 at 30 arc-second resolution53. The Normalized Difference Vegetation Index (NDVI) was used as a proxy of primary productivity56. NDVI values for each record was calculated from satellite images supplied by Moderate Resolution Imaging Spectroradiometer57 at a spatial resolution of 250 m. The elevation data (altimetry) was computed using the ASTER Global Digital Elevation Model platform58. Hydrographic data (river network) was gathered from the Sistema Nacional de Informação de Recursos Hídricos59. The human population density was derived from the European Commission (2015) and road network from IGP (2015)60. Latitude and longitude were used as a measure of geographic position of the collected samples. Abundances of Egyptian mongoose, European rabbit (Oryctolagus cuniculus) and red-legged partridge (Alectoris rufa) were estimated based on annual hunting yields published by the National Authority for Nature Conservation (Instituto da Conservação da Natureza e Florestas/ICNF). Cattle breeding farms density per municipality, i.e. farms/km2, (provided by the National Veterinary Authority, Direção Geral de Alimentação e Veterinária/DGAV) was also included as variable given the occurrence of MAP in livestock and its potential spill-over to wildlife.
Body size was estimated as the first component of a principal component analysis (PCA) performed to enable a single estimate based on the covariance matrix of various measures. Calculations included combining weight and six biometric measurements as described by Bandeira et al.40 [snout-tail length (terminal hairs not included), right hind leg length, right hind foot length, shoulder height, neck perimeter and head width)] of each mongoose specimen. Variables were tested for normality with Kolmogorov-Smirnov Test (with correction of Lilliefors for the significance level)61. The categorical co-variates “Gender” and “Season” were composed by two and four categories, respectively. For “Gender” the defined reference category was “Female” and for “Season” we selected “Winter”.
Regarding the modelling procedure, first we tested for data spatial autocorrelation, to account for the degree of geographical dependency of data, using the Moran’s I index with the R’s “ape” package62. Multicolinearity was tested by estimating the Spearmans correlation rank (r)61 and the statistical significance between variables included in two groups of variables: (1) wildlife and cattle abundance (Egyptian mongoose, wild rabbit and partridge abundances and cattle farms density); and (2) climatic and disturbance factors (human density, road network, river network, altimetry, average annual temperature, annual temperature range, annual rainfall and NDVI). When the correlation coefficients (r) between two variables were equal or superior to 0.7, we excluded those that presented a lower correlation with the dependent variable (i.e. MAP IS900 detection)63.
All the remaining continuous candidate variables were standardized prior to the model building procedure to facilitate results interpretation and modelling procedures. Due to the low detection rate of IS900, we used a Logistic Regression in Rare Events data approach (LRRE) to test which variables might be driving/constraining detection in Egyptian mongooses. This approach is adequate to study binary events in situations where non-detections are hundreds or thousands of times more frequent than detections64, as it controls for the underestimation of occurrence that might take place when using simple logistic regression approaches in such situations64. Modelling procedures were implemented using the “logistf” package65 in R 3.1.2.
We implemented a two-folded analytical procedure. First, we used only the variables associated to each working hypothesis (see above), and built several model for each hypothesis, corresponding to all combination of variables associated to the hypothesis. Then we used an Information Criteria approach (Akaike Information Criteria, corrected for small samples – AICc)66 to select the best models for each hypothesis. All produced models were ranked according with their ΔAICc (corresponding to the difference between the model’s AICc and the smallest AICc value). Those models with a ΔAICc < 2 were selected as best models66.
In a second stage, we compared the best model(s) for each hypothesis, using the AICc approach, to test what hypothesis was more supported by our data. As more than one model fulfilled the best model criterion (i.e. ΔAICc < 2), we implemented a model averaging procedure to estimate variables coefficients. Variables with coefficients 95% confidence intervals that did not include zero, and with a significant relation with the dependent variable, were those considered in the discussion (as only for those we could assess if their influence in MAP detection was significantly positive or negative). Odds-ratio for each variable included in the best model(s) were also estimated. Model selection procedures were implemented in R package “MuMIn”67. Best model validation was tested by calculation the Receiver Operating Characteristic (ROC) Curve, and the Area Under the Curve (AUC)68, using the R package “pROC”69.
Results
Demography and diversity analyses of the samples under study
Two-hundred and two carnivores belonging to five carnivore species (Egyptian mongoose, red fox, common genet, stone marten, and European badger) were surveyed in this study. Egyptian mongoose and red fox were the most represented species (73.8% and 19.8%, respectively). The majority of surveyed individuals were adult (n = 168; 83%). Regarding geographical distribution, the districts of mainland Portugal (first level administrative divisions; n = 18) were considered the sampling unit. Five districts (Braga, Porto, Bragança, Viana do Castelo and Setúbal) were not represented in our sampling effort. Most individuals originated from Beja (n = 68), while Vila Real, in the inner North, was the district with more animal species analysed (four species) (Fig. 1 and Supplementary Fig. 1).
A diversity analysis was performed to assess if our eventually biased sampling would enable proper inferences on MAP occurrence and the underlying epidemiological situation. A positive relationship between the abundance of animals from a particular species and their distribution across districts was registered by linear regression (r2 = 0,6152), meaning that the two species from which more individuals were sampled (mongooses and foxes) are indeed the species that are distributed in a higher number of geographic locations. The diversity indices Shannon-Wiener, Simpson and Berger-Parker represent a coherent system for diversity estimates and allow the appraisal of the balance between the number of individuals and the number of species under analysis in a given community (district)70. Since only one animal species was sampled from the districts of Leiria and Faro, this analysis was not carried out for those regions. Regarding the estimated diversity indices, the higher values were attained for the districts of Vila Real and Aveiro, which thus can be considered the most diverse and balanced sampled communities (Supplementary Table 3); in contrast, Beja and Lisboa were the most unbalanced. The same diversity indices were re-calculated for clustered geographical areas, the statistical terrestrial units (NUTS). The North region, which includes Vila Real, is, according to this criteria, the most balanced community amongst those of our study (Supplementary Table 3).
Non-parametric estimators of species richness chao 1, which examines species abundance data, and chao 2, that considers species incidence data, were also calculated, evidencing completeness of the sampling method of 100% and 91%, respectively (Supplementary Table 3).
Detection of MAP-positive animals using IS900 as proxy
MAP bacteria were not isolated from −20 °C preserved tissues in none of the selective media used, but MAP IS900 was detected by nested real-time PCR in specimens from four out of the five surveyed species, the exception being the badger (Table 1, Supplementary Table 4). The samples from which amplification of IS900 was accomplished (n = 15) were also tested for F57 by real-time PCR. Amplification was only obtained for the positive control (supplementary Figure). The internal amplification control (IAC) performed correctly for all samples (30 < Ct < 33). Eletrophoretic analyses of the PCR reaction products evidenced an amplicon corresponding to the IAC in each sample but no F57 amplicon (with the exception of the positive control). MAP’ IS900 identity of three IS900 amplicons generated by the first step of the nested approach was confirmed by sequencing using first amplification primers. Alignment with eleven IS900 sequences from 11 MAP strains genomes deposited in Genbank and two IS900-like sequences from Mycobacterium sp. and Mycobacterium porcinum evidenced 100% similarity of the red fox, mongoose and common genet IS900 fragments with genuine IS900 from MAP, in parallel with the lack of the point mutations that IS900-like exhibit, allowing distinction of IS900 from IS900-like (Supplementary Fig. 2) and thereby confirming the specificity of the carnivore amplicons as IS900 and the ability of this novel PCR to detect MAP using as a proxy IS900.
According to established positivity criterion (see methods), 15 out of the 202 animals tested were considered MAP IS900-positive [7.43% (95% CI: 4.55–11.89%]: nine Egyptian mongoose, four red fox, one stone marten and one common genet collected in 2005 (n = 1), 2009 (n = 1), 2010 (n = 2) and 2011 (n = 10). There were DNA positive animals among the road-killed (n = 4) as well as the hunted (n = 11). Most DNA-positive specimens were adult (n = 14; 93%) (Table 1, Supplementary Table 4).
In the most represented species, the percentage of IS900-positive animals was 6% (95% CI: 3.2–11%) for Egyptian mongoose and 10% (95% CI: 4.0–23%) for red fox.
MAP IS900 was detected in both feces and spleen samples (5.4% and 6% of total samples, respectively). DNA detection in the spleen of positive specimens was confirmed for a subset of mongooses and foxes (n = 9, 60%). The presence of DNA in feces was confirmed in all four positive species (n = 6, 40% of positive specimens), including 75% of DNA-positive foxes and 11% of DNA-positive mongooses. Statistical analysis by the Chi-square test between feces and spleen samples evidenced no significant difference in the probability of detecting MAP IS900in any of the biological matrices tested (p > 0.05).
The distribution of Cq values among fecal and spleen samples ranged from 7.74 to 37.41 and from 10.28 to 44.08, respectively. Amplification reactions yielding Cq above 40 were considered a positive result if amplification of the 66 bp fragment was confirmed by electrophoresis. The mean Cq values were 23.0 and 27.3, respectively (Supplementary Fig. 3). The mean Cq values of the two matrix types under analysis were compared by the Mann-Whitney test and no significant differences were obtained (i.e. p > 0.05). The mean Cq values obtained for biological samples from the most represented hosts (red fox and Egyptian mongoose) were also compared by the Mann-Whitney test and, again, no significant differences were found (i.e. p > 0.05; Supplementary Fig. 3).
Regarding the geographic location, IS900-positive animals were detected in eight out of the 13 (61.5%) districts analyzed (Fig. 1 and Supplementary Table 4). None of the animals sampled from the central region of the mainland (Santarém, Coimbra, Guarda, Castelo Branco and Leiria districts) tested positive, while Beja, from where most animals were sampled, was the district with more IS900-positive specimens (n = 5; 7.35%). Faro and Aveiro presented the highest proportion of DNA-positive animals (40.0% and 66.7%, respectively) (Fig. 1 and Supplementary Table 4).
Unraveling significant predictors for MAP using IS900 detection in mongooses as proxy
The influence of productivity/food resources availability, disturbance, life-history traits, climate and orography, and density of cattle farms, which potentially could influence MAP detection using IS900 as proxy was investigated through modeling strategies on PCR positive and negative mongooses, whose representative sampling (n = 149), in comparison with the remaining surveyed species, would enable robust modeling analyses. Five specimens were excluded as gender information was not available.
The components retained from principal component analysis (PCA) of body size (based on weight, snout-tail length, neck perimeter) had loadings higher than 0.70. PCA of body size for each specimen was explained by a proportion of variance of 85.6%, with an eigenvalue of 2.568. No significant spatial autocorrelation of the collected samples was detected (Moran’ I = −0.006; P = 0.956), indicating a negligible influence of spatial dimension on our data structure. Multicollinearity testing indicated that several variables among those selected as potential predictors were highly correlated: wild rabbit and red partridge abundance: r = 0.69, P < 0.001; human density and road network: r = 0.84, P < 0.001; average annual temperature and annual rainfall: r = 0.92, P < 0.001; altimetry and average annual temperature: r = 0.79, P < 0.001; altimetry and annual rainfall: r = 0.91, P < 0.05. Thus, partridge abundance, human density and average annual temperature were excluded from the analyses based on lower rho correlation estimates from Spearmans correlation rank test with the dependent variable. Thus, we tested all combinations of the following candidate predictors: H1 - NDVI and Abundance of wild rabbits; H2 - Road network; H3 - Gender, Age, Body size and abundance of Egyptian mongooses; H4 - Season, Annual temperature range, Annual rainfall, Altimetry, and River network; H5 - density of cattle farms. Although several models were identified as the best models for each working hypothesis (see Table 2), those related with the fourth hypothesis, i.e. Climate and orography hypothesis, were the ones that reached lower AICc values and a ΔAIC < 2), thus representing the most parsimonious and explanatory hypothesis to describe the probability of IS900 detection in mongoose (Table 2). Those models were composed by a combination of four variables (Tables 2 and 3). Model averaging procedures of the two best models indicated that annual temperature range, annual rainfall and altimetry as the variables with most significant relative importance (with p < 0.05) and with coefficients 95% confidence intervals that did not cross zero, enabling to determine the type of effect (Table 3). Annual rainfall and annual temperature range exerted a negative influence on MAP infection probability in mongooses (with an odds-ratio of 0.034 and 0.146, respectively), while altimetry had a positive effect (with an odds-ratio of 21.575) (Table 3). The AUC value derived from the ROC curve reached 0.855, revealing good accuracy of the average model to predict MAP infection in carnivores71.
Discussion
The high apparent prevalence rates that have been found in livestock populations over the years, together with two recent reports of regional distribution in wild boar and red deer36,37, suggest that MAP occurrence in mainland Portugal is enzootic (Fig. 1). Thirteen out of 18 administrative regions from mainland Portugal were represented in our sampling effort. Despite the constraints related to an opportunistic strategy, species diversity, abundance and richness estimates, calculated with different algorithms/indicators at the district and NUTS levels, suggest that the mesocarnivore communities sampled here were, in general, balanced, both in composition and structure, and that our strategy is appropriate to appraise MAP occurrence and ecological interactions. Although sample sizes were intrinsically limited for low-density Mustelidae and Viverridae species, we show that important terrestrial species of the Mediterranean carnivore guild, such as stone marten and genets, may be exposed to MAP, while we provide the first report of MAP IS900 detection in common genet. Our survey is also the first to report IS900 in feces from wildlife in Portugal as this matrix was not tested in any of the previous surveys, although fecal shedding was reported in bovines from the North of Portugal72.
We found epidemiologically relevant proportions of MAP IS900-positive animals among extensively sampled wild carnivores from the Canidae and Herpestidae families, particularly red fox (10%) and Egyptian mongoose (6%). Detection of positive wild carnivores from Northeastern Portugal down to Alentejo (Beja) and further south, in the Algarve, represents important new data, since all the information available so far on MAP incidence was limited to free-ranging animals from the Centre and Northeast. This is particularly important in terms of conservation because Guadiana Valley region (in Beja district) is a conservation priority area for some Portuguese menaced predators, such as the Iberian lynx (Lynx pardinus), Spanish Imperial eagle (Aquila adalberti), and the European wildcat (Felis silvestris)73,74,75, which might be susceptible to MAP. Iberian lagomorphs such as wild rabbit (Oryctolagus cuniculus) and Iberian hare (Lepus granatensis) also occur in this region. Potential exposure to MAP is also of relevance since these two species are already very fragilized in mainland Portugal due to rabbit hemorrhagic disease (rabbit) and myxomatosis (rabbit and hare) epizootics in recent years.
A previous report described MAP isolation or MAP DNA detection in 27% of 74 surveyed wild carnivores from the central region of mainland Portugal38. Although our study design included almost three times more animals than those surveyed by Matos and collaborators (2014)38, the mean proportion of exposition to MAP using IS900 as proxy we estimated is much lower, around 7.4% (95% CI: 4.5–11.9), with considerably fewer positive mongooses [6% (95% CI: 3.2–11%), relative to the 46.7% reported by Matos et al. (2014)]38. Regarding red fox, our estimates are also underneath those of Matos et al. (2014) [10% (95% CI: 4.0–23%) versus 14% determined by Matos et al. (2014)]38. Also in contrast, none of the animals that we sampled from the central region of the country tested positive for MAP IS900. Differences in estimated proportions by both studies might be related to several factors, namely lower number of matrices tested per specimen in the current work, prolonged sample storage diminishing detection probability and the fact that our sampling array spans a longer time period (ten versus four years) and a wider geographical area. Furthermore, it is possible that sites tested by Matos et al.38 might be located near livestock herds with higher MAP prevalence than herds near our sampling sites, resulting in spillover to wildlife. However, we lack data to test this idea. Furthermore, since MAP bacterial isolation is hampered in the present work, we cannot discard the hypothesis of having detected a non-culturable (myco)bacteria present in small amounts in mongoose and in other carnivore species that has an IS900 sequence undistinguishable from the one present in MAP.
Intersecting data from the studies published so far in Portugal suggests that MAP may have a wider natural host range than previously supposed and that enzootic occurrence in wildlife may be a recent phenomenon, occurring in parallel in different geographical areas and possibly related with different infection sources. However, we also cannot discard the hypothesis of MAP expansion from “traditional” hotspots, since we detected IS900-positive animals both in areas formerly recognized as positive but also in novel ranges.
MAP occurrence among sympatric wild populations is expected to be influenced by ecological interactions dependent on behavior, social and spatial structure, and also by physiological characteristics of the hosts. The carnivore species that we explore in this study as susceptible are considered habitat and trophic generalist76. The surveyed species also explore the environment in different ways and may, therefore, interact with domestic and wild species differently. While common genets and stone martens are arboreal and solitary, with the first predating more on rodents, foxes and mongooses are cursorial predators that use the landscape mainly in two dimensions and, thus, have higher opportunities to contact with domestic animals in rural settings. In spite of being considered as a solitary animal, the observation of mongoose family groups is frequent77, indicating a certain tendency for sociability. Besides social behavior, some of these predators, like genets and mongooses, use a promiscuous scent marking behavior, characterized by defecation of several individuals in latrines78. This behavior may increase the chances for intra- and inter-specific infection with MAP or other mycobacteria due to intermittent shedding in feces and contamination of the environment, namely water and pasture, thus perpetuating the infection cycle. Furthermore, fox and mongoose’ diets include small vertebrates, such as free-ranging wild rabbit in which MAP was previously reported, both in Portugal79 and Scotland15,16. Therefore, in addition to contamination via social interaction and/or the environment, predator infection from ingestion of contaminated prey may also be a plausible infection source.
Besides the effects of host ecological features mentioned above, numerous environmental (e.g. climate), other host biological characteristics (e.g. age, level and source of exposure), and bacterial factors (e.g. infective dose and strain genotype), are hypothesized to affect the natural resilience and burden of MAP outside the host, amplifying (or decreasing) the infection risk and routes of transmission. Although based on a single species models, our results clearly suggest that probability of MAP IS900 detection in mongoose was determined by region’s altitude, climate stability and dryness.
The likelihood of mongoose exposure to MAP or to a non-culturable bacterium carrying IS900 was positively associated with higher altitude, narrower annual temperature range (i.e. climate stability) and lower annual rainfall, an overlapping combination that is mostly encountered in the North of Portugal, with the exception of the north western region, where mongooses are currently absent52. In this region, 90% of the land is above 400 m, and the highest areas of Portugal are located. The dominant northern lithology in the upper inland is granitoids and a flysh-type series of schists and graywackes80, wherein pH is more acidic and topsoil iron, zinc and nickel, are more abundant81. Also in this northern region, the temperature range and solar irradiation are less pronounced, rainfall is intermediate, and hydrographic network is denser. Although we did not test for MAP environmental presence in these regions, we speculate that the combination of these environmental variables may favour MAP survival off-host. Some experimental studies have shown that MAP survival correlates with temperature and pH, with the bacterium being moderately susceptible to long-term desiccation, exposure to sunlight, and sensitive to soils with an alkaline pH or low iron content82. MAP survival for long periods of time in other extreme conditions such as alkaline soils is, on the other hand, underreported. In contrast with the south, the north is also characterized by higher human density and transformation by human action, more fragmented habitats and lower densities of prey target species. Although the number of livestock farms is also higher in the north, we did not detect a significant effect of this factor, which may indicate that this is probably not an important MAP infection route for mongoose. Our knowledge about MAP distribution and genotypes, the guild of susceptible species across regions, and prevalence rates remains limited owing to several factors, including MAP isolation and genotyping difficulties that continue to challenge microbiologists. The results assembled in this work were partially restricted by the lack of isolation of MAP and the possibility of DNA extraction from pure cultures, which would have been essential to definitely confirm IS900-positive animals as being MAP-infected and to complete downstream application of molecular typing techniques. Bacterial isolation achievement would enable the identification of infection sources and of transmission directionality. We thus cannot discard the possibility of having detected a non-culturable bacterium present in small amounts in mongoose and in other carnivore species that has an IS900 sequence undistinguishable from the one present in MAP. Adding to the extraordinary challenge of culturing MAP and its intrinsic extremely slow growth rate, the necessity of aggressive chemical decontamination of samples to eliminate rapidly growing microorganisms often impair bacilli growth up to detectable levels83. On the other hand, opportunistic testing of hunter-harvested animal specimens habitually implies using autolysed samples or samples poorly preserved by donors in domestic freezers (<−20 °C), further complicating the recovery of injured bacteria. Indeed, the conditions related to the management of samples (e.g. long term storage, freeze–thawing) and sub-optimal preservation most probably hampered culture success in this work. MAP does not survive well over-time at −20 °C. To retain viable bacteria for long-term storage, samples would have needed to be stored at −80 °C. The absence of microbiological isolation in selective media has often been reported by others47,83,84,85 and, while expected in opportunistic studies, it has been the driver underlying several reports informing on more sensitive methods. For diagnosis purposes, we were able to overcome these limitations by using a more sensitive technique, such as nested real-time PCR approach directed towards IS900. This procedure has been successfully used to detect MAP DNA in feces44 and in milk and bulk milk samples47. The improvement and validation of this molecular detection system was also performed together with enhancement of the nucleic acid extraction method. Additional steps for the concentration of faecal samples and the mechanical (bead beating) disruption of the cells were included in order to maximize sensitivity.
The IS900 is defined as a 1,451 bp multicopy element inserted into 15 to 20 conserved loci in the MAP genome45 depending on the strain and has been the marker of choice for most molecular assays. However, as IS900-like sequences have been demonstrated in other unrelated Mycobacterium species, we conducted F57 PCR46 on positive samples from the first step of the nested approach and also sequenced the amplicons from three IS900-positive samples to further confirm specificity of MAP DNA amplification. Although no F57 amplicon was obtained in carnivore samples, specificity of IS900 sequences amplified from mongoose and red fox was confirmed by nucleotide sequence alignments of amplicons displaying 100% similarity with genuine IS900 from 11 MAP genomes. Our samples also lacked the point mutations that distinguish IS900 and IS900-like, thereby confirming the identity of the amplicons as IS900 and the ability of this novel PCR to specifically detect MAP IS900.
Although F57 is highly specific for MAP detection, it is present only in one copy, and so analytical methodologies based on F57 amplification have reduced lower sensitivity when compared to qPCR for IS900. The odds to detect it in poorly preserved samples or in samples with low quantities of DNA are thus lower. The specificity of the external and internal primers and probe used in this survey for IS900 DNA detection in a nested methodology was originally assessed by homology searches in the NCBI nucleotide sequence database. In silico analysis showed 100% identity with the homologous sequences in MAP genomes. In parallel with sequencing results, specificity concerns were also ruled out by the performance parameters we obtained when validating the method with a panel of mycobacterial and non-mycobacterial species, thus confirming specificity and accuracy of selected primers to avoid cross-reactive elements, such as IS900-like.
While the mean Cq values of the nested approach were 23.0 and 27.3 in feces and spleen of positive carnivores, respectively, lower values (but not statistical significant) possibly reflect a higher MAP burden in the sample, although better DNA extraction efficiency or better sample preservation may also jointly contribute to lower quantification cycles. Under these low Cq values and according to current understanding of the pathogenesis of the disease, pathology and culture would be expected to be positive. The proportion of detection in parenchymal organs was also anticipated to be lower relative to intestine/feces since systemic infection is considered a late stage event, at least in ruminants. When systemic infection occurs, the disease is advanced, and pathological lesions are assumed but we did not see any, reinforcing the idea that more studies are needed to fully apprehend the pathophysiology of MAP occurrence in carnivores and for which MAP isolation will be crucial. Although MAP can infect many wildlife species, evidence of clinical disease is more frequent among wild ruminants and lagomorphs14,26, while in predators and scavengers the intestinal and lymphoid tissues, if any, only have minor histopathological changes18,86,87.
MAP has been isolated from clinically healthy domestic and wild carnivores, but also from animals with diffuse microscopic lesions, for example red foxes in Scotland18, and sometimes dogs88, with clinical signs compatible with chronic gastrointestinal disease or lymphadenitis. Still, inflammatory lesion patterns are usually less extensive in carnivores than in ruminants. Pathological findings in red foxes and mongooses associated to MAP presence were reported by Matos et al.38, who described macroscopic lesions in mesenteric lymph nodes. Bacilli isolation and/or DNA detection from several anatomic regions, including the brain of foxes and stone marten, were also reported by the same authors, suggesting disseminated infection. In contrast, we did not find any macroscopic lesions nor pathological evidence of generalized infection, even in specimens collected in the same study region. Thus, our results rather suggest that MAP-exposed carnivores were either sampled during a preclinical phase or may be able to control pathogen multiplication and limit inflammatory responses, lessening tissue damage in an eventual life-long infection. The apparent high proportion of MAP IS900-positive red fox and mongoose found during this study, concurrent with the apparent lack of gastro-enteric lesions and confirmation of IS900 shedding through feces, strongly suggests the presence of subclinical carriers that may occasionally shed bacteria. Although signs of true infection could not be confirmed and since detection of MAP IS900 in feces does not imply the microorganism is viable, at this stage we can only speculate that mongoose and fox may serve as source of infection to other animals and contribute for environmental contamination. MAP contagion through the environment is epidemiologically relevant, especially when dealing with infection-resistant competitor species, which appears to be the case of red fox and mongoose, as these may potentially influence infection dynamics in susceptible hosts, alleviating or aggravating downstream effects such as prevalence rates and density or range of susceptible populations. Establishment of insidious pathogens in already fragile communities may thereafter compromise viability of natural populations.
Concluding Remarks
Paratuberculosis has a major impact in animal health, being associated with important economic losses when species of commercial interest or species under costly conservation programs are affected. Results from this work give strong indications that MAP circulates in wildlife species from specific geographic regions of mainland Portugal but further work is required to obtain an isolate of MAP that proves beyond any doubt that MAP is present in this wildlife population. At this time point, we cannot discard the possibility of having detected a non-culturable microorganism carrying IS900. Although directionality of transmission cannot be inferred at this stage, our data supports the findings from previous reports of MAP isolation in carnivores from Portugal, adding to the increasing body of evidence of wilderness exposure to MAP and possibly to subclinical MAP shedders. Environmental contamination as an indirect transmission path may be likely, which suggests that MAP ecology and resilience outside natural hosts, as well as sustained transmission in carnivores, may be dependent on abiotic factors (e.g. climate). Prolonged survival of MAP in the environment enacts risks to grazing livestock, to paratuberculosis-infected adjacent farms, and also at the livestock-wildlife interface.
Suspicion for the ecological spread of MAP reinforces the need for increased surveillance and action. Quantitative information on the prevalence and concentration of MAP in putative contaminated sources of exposure is required. Although transmission intervals and paths tend to be uncertain for slow-growing and slow-evolving bacteria such as MAP, supplementary sampling efforts should be carried out in the future in order to include more wild specimens from the less represented species and to reach more balanced communities across the Iberian territory. Also, optimization of sample storage (−80 °C) to retain MAP viability and optimization of culture procedures to enable bacterial isolation in future studies should allow quantification of the extent and directionality of MAP transmission between livestock and wild species and amongst the latter. Whole genome sequencing following culture of MAP is to be recommended to confirm transmission between wildlife and livestock. Finally, additional studies are needed to identify landscape-specific risk factors, particularly across environmental gradients. This information is critical to understanding the risk of exposure under a climate change scenario and to decreasing the occurrence of new infections and the prevalence of existing ones.
Data availability
The authors declare that all data enclosed in the manuscript will be made available to the community.
References
Thorel, M. F., Krichevsky, M. & Lévy-Frébault, V. V. Numerical taxonomy of mycobactin-dependent Mycobacteria, emended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov., Mycobacterium avium subsp. paratuberculosis subsp. nov., and Mycobacterium avium subsp. silvaticum. Int. J. Syst. Bacteriol. 40, 254–260 (1990).
Mijs, W. et al. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and M. avium subsp. hominissuis’ for the human/porcine type of M. avium. Int. J. Syst. Evol. Microbiol. 52, 1505–1518 (2002).
Turenne, C. Y., Collins, D. M., Alexander, D. C. & Behr, M. A. Mycobacterium avium subsp. paratuberculosis and M. avium subsp. avium are independently evolved pathogenic clones of a much broader group of M. avium organisms. J. Bacteriol. 190, 2479–2487 (2008).
Whittington et al. Control of paratuberculosis: who, why and how. A review of 48 countries. BMC Vet. Res. 15(1), 198, https://doi.org/10.1186/s12917-019-1943-4 (2019).
Garcia, A. B. & Shalloo, L. The economic impact and control of paratuberculosis in cattle. Dairy Sci. 98(8), 5019–39, https://doi.org/10.3168/jds.2014-9241 (2015).
Jones, R. L. Review of the economic impact of Johne’s disease in the United States. Milner, A. R. & Wood, P. R. (Eds.) Johne’s Disease. Current Trends in Research, Diagnosis and Management, CSIRO, Melbourne, Victoria, Australia, pp. 46–50 (1989).
Ott, S. L., Wells, S. J. & Wagner, B. A. Herd level economic losses associated with Johne’s disease on US dairy operations. Prev. Vet. Med. 40, 179–192 (1999).
Losinger, W. C. Economic impact of reduced milk production associated with Johne’s disease on dairy operations in the USA. J. Dairy Res. 72, 425–32 (2005).
Chiodini, R. J. & Van Kruiningen, H. J. Eastern white-tailed deer as a reservoir of ruminant paratuberculosis. J. Am. Vet. Med. Assoc. 182, 168–169 (1983).
Ferroglio, E., Nebbia, P., Robino, P., Rossi, L. & Rosati, S. Mycobacterium paratuberculosis infection in two free-ranging Alpine Ibex. Rev. sci. tech. Off. int. Epiz. 19, 859–862 (2000).
Pavlik, I. et al. Epidemiology of paratuberculosis in wild ruminants studied by restriction fragment length polymorphism in the Czech Republic during the period 1995-1998. Vet. Microbiol. 77, 231–51 (2000).
Yadav, D. et al. Pathogenic ‘bison-type’ Mycobacterium avium subspecies paratuberculosis genotype characterized from riverine buffalo (Bubalus bubalis) in north India. Comp. Immunol. Microbiol. Infect. Dis. 31, 373–387 (2008).
Sleeman, J. M. et al. Johne’s disease in a free-ranging white-tailed deer from Virginia and subsequent surveillance for Mycobacterium avium subspecies paratuberculosis. J. Wildl. Dis. 45, 201–206 (2009).
Stevenson, K. et al. Occurrence of Mycobacterium avium subspecies paratuberculosis across host species and European countries with evidence for transmission between wildlife and domestic ruminants. BMC Microbiol. 9, 212 (2009).
Greig, A. et al. Paratuberculosis in wild rabbits (Oryctolagus cuniculus). Vet. Rec. 140, 141–43 (1997).
Greig, A. et al. Epidemiological study of paratuberculosis in wild rabbits in Scotland. J. Clin. Microbiol. 37, 1746–1751 (1999).
Beard, R. M. et al. Evidence of paratuberculosis in fox (Vulpes vulpes) and stoat (Mustela erminea). Vet. Rec. 145, 612–613 (1999).
Beard, P. M. et al. Paratuberculosis infection of nonruminant wildlife in Scotland. J. Clin. Microbiol. 39, 1517–1521 (2001).
Corn, J. L., Manning, E. J. B., Sreevatsan, S. & Fischer, J. R. Isolation of Mycobacterium avium subsp. paratuberculosis from free-ranging birds and mammals on livestock premises. Appl. Environ. Microbiol. 71, 6963–6967 (2005).
Kopecna, M. et al. Detection of Mycobacterium avium subsp. paratuberculosis in two brown bears in the central European carpathians. J. Wildl. Dis. 42, 691–695 (2006).
Anderson, J. L. et al. Mycobacterium avium subsp. paratuberculosis in scavenging mammals in Wisconsin. J. Wildl. Dis. 43, 302–308 (2007).
Arsenault, R. J. et al. From mouth to macrophage: mechanisms of innate immune subversion by Mycobacterium avium subsp. paratuberculosis. Vet. Res. 45, 1–15 (2014).
Herthnek, D., Englund, S., Willemsen, P. T. J. & Bolske, G. Sensitive detection of Mycobacterium avium subsp. paratuberculosis in bovine semen by real-time PCR. J. Appl. Microbiol. 100, 1095–1102 (2006).
Whittington, R. J. & Windsor, P. In utero infection of cattle with Mycobacterium avium subsp. paratuberculosis: a critical review and meta-analysis. Vet. J. 179, 60–69 (2009).
Khol, J. L. et al. Consecutive excretion of Mycobacterium avium subspecies paratuberculosis in semen of a breeding bull compared to the distribution in feces, tissue and blood by IS900 and F57 quantitative real-time PCR and culture examinations. J. Vet. Med. Sci. 72, 1283–1288 (2010).
Daniels, M. J. et al. Do non-ruminant wildlife pose a risk of paratuberculosis to domestic livestock and vice versa in Scotland? J. Wildl. Dis. 39, 10–15 (2003).
Biet, F., Boschiroli, M. A., Thorel, M. F. & Guilloteau, G. Zoonotic aspects of Mycobacterium bovis and Mycobacterium avium-intracellulare Complex (MAC). Vet. Res. 36, 411–436 (2005).
Afonso, A. F. Contributo para o Estudo da paratuberculose nos pequenos ruminantes em Portugal. Colectânea SPOC, 6, 73 [(http://www.ovinosecaprinos.com/bibliografia/afonso95.PDF) (Portuguese)] (1995).
Amado, A., Albuquerque, T. & Ferreira, A.F. Epidemiological study in goats and sheep in the vouzela area of Portugal. In Proceedings of Fourth International Colloquium on Paratuberculosis, pp. 10–11 (1994).
Ferreira, A. et al. Epidemiological study of paratuberculosis in ruminants in alentejo Portugal. In Proceedings of Seventh International Colloquium on Paratuberculosis, pp. 108 (2002).
Mendes, S. et al. Epidemiological studies on paratuberculosis in small ruminants in Portugal. Epidémiol. et Santé Anim. 45, 61–71 (2004).
Coelho, A. C. et al. Seroprevalence of ovine paratuberculosis infection in the northeast of Portugal. Small Rumin. Res. 71, 298–303 (2007).
Vala, H. et al. Paratuberculosis in sheep from serra da estrela region, Portugal. In Proceedings of Nineth International Colloquium on Paratuberculosis, pp. 250–253 (2007).
Coelho, A. C. et al. Estimation of the prevalence of Mycobacterium avium subsp. paratuberculosis by PCR in sheep blood. Small Rumin. Res. 76, 201–206 (2008).
Miranda, C. et al. Mycobacterium avium subsp. paratuberculosis infection in slaughtered domestic pigs for consumption detected by molecular methods. Food Res. Int. 44, 3276–3277 (2011).
Matos, A. C. et al. Renal lesions in deer (Cervus elaphus): involvement of Mycobacterium avium subsp. paratuberculosis. J. Comp. Pathol. 148, 85 (2013).
Matos, A. C. et al. Granulomatous lesions and Mycobacterium avium subsp. paratuberculosis Portuguese wild boars (Sus scrofa). J. Comp. Pathol. 148, 85 (2013).
Matos, A. C. et al. Survey of Mycobacterium avium subspecies paratuberculosis in road-killed wild carnivores in Portugal. J. Zoo Wildl. Med. 45, 775–781 (2014).
Bandeira, V. Bio-ecology of the Egyptian mongoose (Herpestes ichneumon) in Portugal. PhD Thesis in Biology and Ecology of Global Changes. Departamento de Biologia da Universidade de Aveiro e Faculdade de Ciências da Universidade de Lisboa, Portugal (2016).
Bandeira, V., Virgós, E., Barros, T., Cunha, M. V. & Fonseca, C. Geographic variation and sexual dimorphism in body size of the Egyptian mongoose, Herpestes ichneumon in the western limit of its European distribution. Zoolog Anz. 264, 1–10 (2016).
Help, C. H. R., Herman, P. M. J. & Soetaert, K. Indices of Diversity and Evenness. Oceanis 24, 61–87 (1998).
Gotelli, N. J. & Colwell, R. K. Estimating species richness. In Frontiers in measuring biodiversity. pp. 39–54. Magurran, A. E. & McGill, B. J. (Eds). Oxford University Press, New York (2011).
Reis, A. C. On the trail of Mycobacterium avium subsp. paratuberculosis and Mycobacterium bovis in mainland Portugal: a microbiological and molecular survey in wildlife, MSc. Thesis, Faculdade de Ciências, Universidade de Lisboa, Portugal, retrieved from, http://hdl.handle.net/10451/23072 (2015).
Leão, C. Molecular tools in the diagnostic and epidemiology of infections caused by members of Mycobacterium avium complex, PhD thesis, Faculdade de Ciências e Tecnologia, Universidade Nova de Lisboa, Portugal, retrieved from, http://hdl.handle.net/10362/16513 (2015).
Motiwala, A. S., Li, L., Kapur, V. & Sreevatsan, S. Current understanding of the genetic diversity of Mycobacterium avium subsp. paratuberculosis. Microb. Infect. 8, 1406–1418 (2006).
Sidoti, F. et al. Validation and standardization of IS900 and F57 real-time quantitative PCR assays for the specific detection and quantification of Mycobacterium avium subsp. paratuberculosis. Can. J. Microbiol. 57, 347–354 (2011).
Leão, C. et al. Presence of Mycobacterium avium subsp. paratuberculosis DNA in milk used to feed calves in Portugal. J. Dairy Res. 84, 124–27 (2017).
VassarStats: Website for Statistical Computation, http://vassarstats.net/ (accessed on 02-05-2016) (2016).
Ben-Yaacov, R. & Yom-Tov, Y. On the biology of the Egyptian Mongoose, Herpestes ichneumon, in Israel. Z. Säugetierkd 48, 34–45 (1983).
Delibes, M., Aymerich, M. & Cuesta, L. Feeding habits of the Egyptian mongoose or Ichneumon in Spain. Acta Theriol. 29, 205–218 (1984).
Palomares, F. & Delibes, M. Habitat preference of Large grey mongooses Herpestes ichneumon in Spain. Acta Theriol. 35, 1–6 (1990).
Barros, T., Carvalho, J., Pereira, M. J. R., Ferreira, J. P. & Fonseca, C. Following the trail: factors underlying the sudden expansion of the Egyptian mongoose (Herpestes ichneumon) in Portugal. PLoS One 10, e0133768, https://doi.org/10.1371/journal.pone.0133768 (2015).
Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G. & Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978 (2005).
Palomares, F. & Delibes, M. Ecología comparada de la gineta Genetta genetta (L.) y el meloncillo Herpestes ichneumon (L.) (Mammalia, Viverridae) en Doñana (SO de la Península Ibérica). Bol. R. Soc. Esp. Hist. Nat. (Sec Biol) 87, 257–266 (1991).
BioClim. [http://www.worldclim.org/bioclim (accessed 24.06.2015)] (2015).
Pettorelli, N. The Normalized Difference Vegetation Index. 1st ed. Oxford: Oxford University Press, UK (2013).
MODIS, Moderate Resolution Imaging Spectroradiometer, http://modis.gsfc.nasa.gov (accessed 15.06.24) (2015).
ASTER, Global Digital Elevation Model platform, http://gdem.ersdac.jspacesystems.or.jp/ (accessed 24.06.2015) (2015).
SNIRH, Sistema Nacional de Informação de Recursos Hídricos, http://snirh.apambiente.pt/ (accessed 15.06.24) (2015).
IGP, Instituto Geográfico Português, http://www.igeo.pt/ (accessed 24.06.2015) (2015).
Zar, J. H. Biostatistical analysis. 5th Edition, Prentice Hall/ Pearson, Upper Saddle River, xiii, 944 p.
Paradis, E., Claude, J. & Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004).
Hosmer, D. & Lemeshow, S. Applied Logistic Regression. John Wiley & Sons, Inc., New York (2000).
King, G. & Zeng, L. Logistic regression in rare events data. Polit. Anal. 9, 137–163 (2001).
Heinze, G., Ploner, M., Dunkler, D. & Southworth, H. logistf: firth’s bias reduced logistic regression. R package version 1.21 (2013).
Burnham, K.P., Anderson, D.R. Model selection and multimodel inference: a practical information-theoretic approach, 2nd Edition. Springer-Verlag, New York, USA (2002).
Barton, K. MuMIn: Multi-Model Inference. R package version 1.13.4. (2015).
van Erkel, A. R. & Pattynama, P. M. T. Receiver operating characteristic (ROC) analysis: Basic principles and applications in radiology. Eur. J. Radiol. 27, 88–94 (1998).
Robin, X. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77 (2011).
Chandy, S., Gibson, D. J. & Robertson, P. A. Additive partitioning of diversity across hierarchical spatial scales in a forested landscape. J. Appl. Ecol. 43, 792–801 (2006).
Manel, S., Williams, H. C. & Ormerod, S. J. Evaluating presence-absence models in ecology: the need to account for prevalence. J. Appl. Ecol. 38, 921–931 (2001).
Leão, C., Amaro, A., Santos-Sanches, I., Inácio, J. & Botelho, A. Paratuberculosis asymptomatic cattle as spillovers of Mycobacterium avium subsp. paratuberculosis: consequences for disease control. Rev. Port. Ciências Vet. 110, 69–73 (2015).
Sánchez, B., González, L. & Barov, B. Action plan for the Spanish Imperial Eagle Aquila adalberti in the European Union. available at, http://ec.europa.eu/environment/nature/conservation/wildbirds/action_plans/docs/aquila_adalberti.pdf (accessed 26.12.2016) (2008).
Sarmento, P. et al. Status survey of the critically endangered Iberian lynx Lynx pardinus in Portugal. Eur. J. Wildl. Res. 55, 247–253 (2009).
Monterroso, P. Ecological interactions and species coexistence in Iberian mesocarnivore communities. PhD thesis, Universidade do Porto, Porto, Portugal (2013).
Hunter, L & Barrett, P. Carnivores of the World. Princeton University Press, Princeton, USA (2011).
Palomares, F. & Delibes, M. Social organization in the Egyptian mongoose: group size, spatial behaviour and inter-individual contacts in adults. Anim. Behav. 45, 917–925 (1993).
Palomares, F. Faecal marking behaviour by free-ranging common genets Genetta genetta and Egyptian mongooses Herpestes ichneumon in southwestern Spain. Zeitschrift für Säugetierkunde 58, 225–231 (1993).
Maio, E. et al. Paratuberculosis in European wild rabbits from the Iberian Peninsula. Res. Vet. Sci. 91, 212–218 (2011).
Inácio, M., Pereira, V. & Pinto, M. The soil geochemical atlas of Portugal: overview and applications. J. Geochem. Explor. 98, 22–33 (2008).
ESDAC, European Soil Data Centre, http://eusoils.jrc.ec.europa.eu/content/european-soil-database-v20-vector-and-attribute-data (accessed 04.11.2016) (2016).
McKenna, S. L., Keefe, G. P., Tiwari, A., VanLeeuwen, J. & Barkema, H. W. Johne’s disease in Canada part II: disease impacts, risk factors, and control programs for dairy producers. Can. Vet. J. 47, 1089–1099 (2006).
Reddacliff, L. A., Vadali, A. & Whittington, R. J. The effect of decontamination protocols on the numbers of sheep strain Mycobacterium avium subsp. paratuberculosis isolated from tissues and faeces. Vet. Microbiol. 95, 271–282 (2003).
De Juan, L. et al. Comparison of four different culture media for isolation and growth of type II and Type I/III Mycobacterium avium subsp. paratuberculosis strains isolated from cattle and goats. Appl. Environ. Microbiol. 72, 5927–5932 (2006).
Sohal, J. S., Singh, S. V., Singh, A. V. & Singh, P. K. Strain diversity within Mycobacterium avium subspecies paratuberculosis - a review. Indian J. Exp. Biol. 48, 7–16 (2010).
Carta, T., Álvarez, J., Pérez de la Castra, J. M. & Gortázar, C. Wildlife and paratuberculosis: a review. Res. Vet. Sci. 94, 191–197 (2013).
Coelho, A. C., Pinto, M. L., Matos, A., Matos, M. & Pires, M. A. Mycobacterium avium Complex in domestic and wild animals. In Insights from Veterinary Medicine. Payan-Carreira, R (Ed.), InTechOpen (2013).
Glanemann, B. et al. Detection of Mycobacterium avium subspecies paratuberculosis-specific DNA by PCR in intestinal biopsies of dogs. J. Vet. Intern. Med. 22, 1090–1094 (2008).
Acknowledgements
We are grateful to hunters, land owners, the entities managing hunting areas, and their representatives (Jacinto Amaro, Paula Simões, FENCAÇA; ANPC; CNCP), as well as to Estradas de Portugal IP, which generously contributed to animal sampling and carcass donation for scientific purposes. We acknowledge Madalena Monteiro, Paulo Carvalho and Paula Mendonça from INIAV IP, for performing some of the post-mortem examinations in the scope of another unpublished study. We thank the national authority ICNF for data sharing regarding hunting bags. We are also indebted with the late Dr. Pina Fonseca [DGAV (DSSPA)] and DGAV collaborators in general for kindly providing baseline data (cattle farms density) included in the modelling analyses. We thank anonymous reviewers for constructive discussion during the peer review process and for providing suggestions that improved an earlier draft of the manuscript. This work was partially performed in the scope of a Master thesis by co-author Ana C. Reis, entitled “On the trail of Mycobacterium avium subsp. paratuberculosis and Mycobacterium bovis in mainland Portugal: a microbiological and molecular survey in wildlife”. Part of the molecular strategies herein adopted were based on the work included in the PhD thesis of co-author Célia Leão, entitled “Molecular tools in the diagnostic and epidemiology of infections caused by members of Mycobacterium avium complex”. This work was partially funded by Fundação para a Ciência e a Tecnologia (FCT/MEC), Portugal, through strategic funding to cE3c and BioISI Research Units (UID/BIA/00329/2019 and UID/Multi/04046/2019). This work was also supported by Programa Operacional de Competitividade e Internacionalização (POCI) (FEDER component), Programa Operacional Regional de Lisboa, and FCT, Portugal, in the scope of project COLOSSUS (reference POCI-010145-FEDER-029783). VB, LMR and CF were financially supported by the University of Aveiro (Department of Biology), CESAM (UID/AMB/50017) and FCT through national funds, and the co-funding by the FEDER, within the PT2020 Partnership Agreement and Compete 2020. A.C.R. and V.B. were also supported by FCT through doctoral grants (PD/BD/128031/2016 and SFRH/BD/51540/2011, respectively).
Author information
Authors and Affiliations
Contributions
M.V.C. conceived the study. V.B. and C.F. established the strategy for collection of animal samples. A.C.R., M.V.C. and C.L. performed the experimental work. M.V.C. and L.M.R. performed the statistical analyses. M.V.C. and A.C.R. wrote the manuscript. M.V.C. and L.M.R. contributed to conception of ideas within the manuscript. V.B., C.F., C.L. and A.B. gave critical feedback on final draft. All authors contributed and approved the final version for submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cunha, M.V., Rosalino, L.M., Leão, C. et al. Ecological drivers of Mycobacterium avium subsp. paratuberculosis detection in mongoose (Herpestes ichneumon) using IS900 as proxy. Sci Rep 10, 860 (2020). https://doi.org/10.1038/s41598-020-57679-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-57679-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.