Abstract
Genotypic tropism testing (GTT) for co-receptor usage is a recommended tool for clinical practice before administration of the CCR5-antagonist maraviroc. For some isolates, phenotypic tropism testing (PTT) revealed discordant results with GTT. In this study, we performed a comparative study between GTT and PTT in HIV-1C from East Africa (HIV-1CEA) and compared the data with HIV-1B and 01_AE and described the maraviroc susceptibility in the CCR5-tropic strains. Patient-derived HIV-1 envgp120 region was cloned into a modified pNL4-3 plasmid expressing the luciferase gene. rPhenotyping dissected single clones from 31 HIV-1CEA infected patients and four strains with known phenotype. Additionally, 68 clones from 18 patients (HIV-1B: 5, 01_AE: 7, HIV-1CEA: 6) were used to determine the PTT in GHOST cell line. The respective V3-sequences were used for GTT. R5-tropic strains from HIV-1CEA (n = 20) and non-C (n = 12) were tested for maraviroc sensitivity in TZMbl cell line. The GTT falsely called a higher proportion of X4-tropic strains in HIV-1CET compared to PTT by both rPhenotyping and the GHOST-cell assay. When multiple clones were tested in a subset of patients’ samples, both dual-tropic and R5-tropic strains were identified for HIV-1C. Relatively higher EC50 values were observed in HIV-1C strains than the non-C strains (p = 0.002).
Similar content being viewed by others
Introduction
Predicted genotypic co-receptor tropism testing (GTT) is based on the analysis of human immunodeficiency virus type 1 (HIV-1) envelop (env) V3-loop sequences and is a comparatively inexpensive, rapid and accessible alternative approach to phenotypic tropism testing (PTT) of the HIV-1 tropism in routine clinical practice1. V3-loop sequences can be derived with clonal (i.e., virus sequences are cloned) or population-based methods (i.e., bulk population sequencing of the entire viral quasispecies), and predicted co-receptor tropism can be determined in silico using bioinformatics tools. Numerous such algorithms are available to predict the genotypic tropism of HIV-1 based on the V3-loop sequence. The first and simplest algorithm determined the tropism based on the 11/25 rule, which affirms a tropism based on detecting a charge at amino acid positions 11 and/or 25 of the 35 amino acids of the V3-loop2. However, this algorithm has shown a limited sensitivity for the co-receptor tropism prediction in actual clinical samples (Reviewed in3). Currently, the most widely used GTT tools are WebPSSM4 and Geno2Pheno (G2P)5, which assess the entire V3-loop sequence and assign the viral tropism by a more complex algorithm. However, all the machine-learning GTT tools have been developed primarily for HIV-1B and are now applied also with newly available V3-sequences of non-B subtype HIV-1. More recently a GTT tool called PhenoSeq was developed, which claims to be reliably predictive for the tropism of HIV-1 subtypes A, B, C, D, 01_AE and 02_AG1.
Several guidelines have recommended pre-therapy GTT for patients initiating therapy with the CCR5-antagonist maraviroc (MVC). Thereby therapy is based on the exclusion of all patients having X4-tropic viruses6. Earlier studies have shown that the genotypic co-receptor tropism algorithms were highly sensitive for predicting the treatment outcome for patients receiving MVC7. However, the primary caveat of GTT seem to lie in the subtypes-specific differences affecting the tropism prediction. MVC is a potentially promising new treatment modality in non-B settings, mainly low- and middle-income countries (LMICs). Therefore, optimal use and a high predictability of GTT on non-B subtypes is required to treat the patient with MVC.
Several recent studies from countries, where HIV-1 subtype HIV-1C is dominating, indicated an increase in predicted X4-tropic strains over time8,9. Most of these genotypic and phenotypic tropism correlation studies were performed on HIV-1C sequences from Southern Africa or India. Therefore, training sets and genotypic prediction for HIV-1C were solely based on sequences from those two regions. In contrast, a recent study showed a significant disagreement when these genotypic tools were applied to HIV-1C sequences from Ethiopia9, revealing that data on GTT and PTT for East African HIV-1C (HIV-1CEA) strains are largely lacking. Given the higher heterogeneity among the East African strains10, which can significantly affect the sequence-based tropism prediction1,4,11, we hypothesized in this study that the current GTT tools for subtype C overestimate the X4-tropism in HIV-1CEA.
Therefore, the present study aims to phenotypically verify the co-receptor usage in HIV-1CEA and compare it using the current versions of several genotypic tools. Further, we sought to study Maraviroc susceptibility among the phenotypically determined R5-tropic stains.
Results
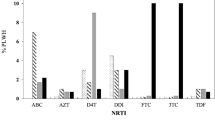
Clonal PTT (cPTT) using the virus produced using the individual clones in GHOST cell lines were performed with 68 individual clones which were infectious from 180 clones tested, obtained from 18 patients samples infected with HIV-1B (n = 5), HIV-1C (n = 6) and 01_AE (n = 7) subtypes. The clonal GTT by sequencing individual clones (cGTT) falsely identified a higher proportion of X4-tropism in HIV-1C compared to phenotypic tropism testing by cPTT (Fig. 1A). Presence of both dual tropic and R5-tropic strains were observed in HIV-1C when multiple clones from the same subset of patients’ samples were tested (Fig. 1). In some patients’ samples (Pt#2, Pt#7) there are no changes in the V3 sequences, but one clone showed R5 tropic and other as dual-tropic. Virus generated from the plasmids pMJ4 (R5-tropic) and pNL43 (X4-tropic) showed R5-tropic and X4-tropic respectively.
The replicative phenotyping (rPhenotyping, population PTT, cPTT) by pooling several clones was performed with 31 patients’ samples infected with HIV-1C and four HIV-1C QC-samples using a co-culture of transfected 293 T cells with the SXR5 reporter cell. cPTT was also performed with six QC viruses with known tropism (97ZA009, 97ZA003, 93IN101, 94KE105, DEMC08NG001, and DEMC09ZA001.S1). All QC viruses identified showed 100% concordance with cGTT and population GTT (pGTT) performed by bulk population sequncing by Sanger Sequencing method. In contrast, discordances were observed between different tools for cGTT/pGTT and cPTT or rPhenotyping. In concordance with cGTT, pGTT also identified more X4-tropic viruses than rPhenotyping (Fig. 2).
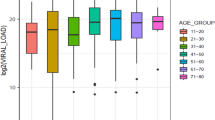
Next, a maraviroc susceptibility assay was performed on phenotypically determined R5-tropic viruses that identified a wide range of susceptibility in HIV-1 C viruses. When comparing with HIV-1B or 01_AE, the HIV-1CEA strains showed reduced susceptibility (p = 0.002) for maraviroc (Fig. 3).
Discussion
In this study using several bioinformatics tools for GTT and two PTT methods, we were able to demonstrate a higher false assignment to X4-tropism by GTT for HIV-1C viruses from East Africa. This indicates the need to train the machine-learning GTT tools with these strains. The maraviroc susceptibility assay further identified a wide range of drug susceptibilities in HIV-1C East African strains ex vivo and a statistically significantly reduced susceptibility compared to non-C viruses.
In our recent study, we showed that, despite strong monophyletic clustering, HIV-1C strains from East Africa possess a significantly higher diversity compared to HIV-1C strains from other geographical locations such as India or South Africa10. Our study also indicated an increase in predicted X4-tropic strains over the decade10. A more recent genotypic study further confirmed the findings in a large number of samples9. A study from India also indicated a marginal temporal increase in X4-tropism prevalence8. However, there is a high chance for false calls of X4-tropism in HIV-1C depending upon the machine-learning GTT tools use in the case of marginal increase12. A recent study comparing GTT and PTT, demonstrated the over-estimation of the X4-tropism in 01_AE and 02_AG isolates11. Also a study by Kalu et al. from Ethiopia showed the disagreements between the GTTs9. Earlier phenotypic studies had reported hardly any (6%)13 or no14 X4-tropic strains for Ethiopia. Our study now confirmed the over-estimation of the predicted X4-tropism by GTT in HIV-1C from East Africa which is in line with our original hypothesis. Our study also indicated that, despite the absence of any changes in the V3-loops, some strains are dual tropic. This is further supported by a recent study, in which the inclusion of the complete gp120 sequence improved the genotypic prediction for HIV-1A and C viruses15. Overall, these studies emphasize the need for improving the present algorithms for GTT, e.g. by including larger segments of gp120 in the tropism prediction.
In our study, a number of the HIV-1C strains from East Africa possessed a reduced drug sensitivity to the CCR5 antagonist maraviroc compared to isolates of the non-C HIV-1 subtypes B and 01_AE. Other studies have indicated that mutations in the V3 loop of gp120 could mainly be responsible for this variability in drug sensitivity to maraviroc16,17,18. However, this difference appears to be strain specific16,19,20. Of note, the various sensitivity to maraviroc in this study could not in all cases assigned to V3 changes, hinting the involvement of mutations in gp120 and gp41 regions other than the V3 loop sequence itself21,22.
In conclusion, our study demonstrates that there is a high need to re-train the machine-learning GTT tropism tools for HIV-1 non-B isolates, necessitating the inclusion of a larger number of relevant non-B strains for the accurate tropism prediction. As current GTT highly overestimates the X4-tropism e.g. in HIV-1C strains from East Africa, where a higher heterogeneity in the V3-loop is common, it may falsely limit the use of drugs such as maraviroc that are becoming available in these resource-limited areas of Africa and other places. Moreover, as the introduction of CCR5 antagonists such as maraviroc is becoming a therapeutic option there, additional studies are needed that virologically monitor the clinical response to maraviroc, particularly of HIV-C viruses.
Materials and Methods
Cell lines, viruses, and plasmids
TZM-bl, GHOST (3) CXCR4+, and GHOST (3) CCR5+ Cells (Hi-5) cells were obtained from NIH AIDS Reagent Program, NIH, US. 293 T cells were purchased from ATCC, US. SXR5 cells are property of the laboratory of TK. TZM-bl, SXR5, and 293 T cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (Sigma, US) supplemented with 10% Fetal Bovine Serum and 2mM L-glutamine. GHOST cell lines were maintained in high glucose DMEM supplemented with 10% fetal bovine serum. To examine the performance of the replicative phenotypic tropism test (rPTT), we used five viruses (QC panel herein) obtained from either the NIH AIDS Reagent Program, NIH, US (97ZA009, 97ZA003, 93IN101 and 94KE105) or the EQAPOL Viral Diversity program, Duke University, US (DEMC08NG001 and DEMC09ZA001.S1). Additionally, plasmids pMJ4 and pNL4-3 obtained from NIH AIDS Reagent Program, NIH, US.
Clinical specimens, Ethical Considerations and data availability
Stored patients’ plasma samples included in this study were from two different cohorts: (1) Swedish InfCare Cohort (n = 35)23 and (2) Ethiopian cohort (n = 25)10. All samples had been subjected to near full-length sequencing as described by us recently24. Only pure subtypes were selected for the study. Ethical permissions were obtained from the respective sites. Swedish samples: Regional Ethics Committee Stockholm (Dnr: 2006/1367-31/4 and 2014/928-31/2). Ethiopian samples: the Ethiopian Science and Technology Agency (Ref. No. RPHE/126-83/08), and the Drug Administration and Control, Authority of Ethiopia (Ref. No. 02/6/22/17). All the data is available along with the manuscript. All methods were performed in accordance with approved institutional guidelines. The patient identity was anonymised and delinked prior to analysis.
Cloning and recombinant virus production
HIV RNA was extracted by using QIAamp Viral RNA Mini Kit (Qiagen, US) from 140uL of patients’ plasma and supernatant of QC viruses. cDNA was converted using the SuperScript® IV RT enzyme (Invitrogen, Life Technologies, MA, USA) with Oligo (dT)18 primer (Thermo Scientific). The first round PCR was performed using the high fidelity KAPA HiFiHotStart Ready Mix (2×) (KAPA Biosystem, MA, USA) with primers: 5550 F, 5′-AGARGAYAGATGGAACAAGCCCCAG-3′ (HXB2 co-ordinates: 5550 → 5574) and 9555 R, 5′-TCTACCTAGAGAGACCCAGTACA-3′ (HXB2 co-ordinates: 9555 → 9533). The second round PCR was performed to amplify and clone the complete gp120 using 6433 F, 5′-CYACCAACGCGTGTGTACCCACAGA-3′ (HXB2 co-ordinates: 6433 → 6457) and 8329 R (5′-CCCTGCCGGCCTCTATTYAYTATAGAAA-3′) (HXB2 co-ordinates: 8356 → 8329). The primers contained the NgoMIV and MluI restriction sites respectively. Plasmids pNL4-3 and pMJ4 were also amplified with 6435F and 8329R primers. The resultant fragments were gel purified using QIAquickGel Extraction Kit (Qiagen, USA) and cloned into a pMN-K7-Luc-IRESs-NefΔgp120 plasmid25 following digestion with NgoMIV and MluI-HF (New England Biolabs, US) and ligation with T4 DNA ligase (New England Biolabs, US). The recombinant viruses were produced transfecting the plasmids using FuGENE® HD Transfection Reagent (Promega, US) in 293 T cell line. A total of 10 colonies from each of the 18 patients (180 colonies) were screened for molecular clones by restriction digestion with NgoMIV and MluI-HF followed by sequencing.
Genotypic tropism testing
Genotypic tropism testing (GTT) were performed amplifying the V3 region of HIV-1 envelope gene using the plasma viral RNA, followed by population-based Sanger sequencing as described by us previously (pGTT)26. Individual clones also sequenced to perform the clonal GTT (cGTT).
Phenotypic tropism testing
Phenotypic tropism testing was performed using two methods: (1) Tropism testing in GHOST cell-lines27 of the viruses generated from individual clones (cPTT) and (2) replicative phenotypic tropism test rPhenotyping (pPTT) whereas described previously25. In cPTT co-receptor tropism was determined by measuring Renilla luciferase activity (relative light units [RLU]) using Bright-Glo™ Luciferase Assay System (Promega, US). We consider a 10-fold shift in mean RLU of infected cells over non-infected. The pNL4-3 and pMJ4 were used as positive control for X4- and R5-tropic strains respectively. In addition to luciferase expression, in a subset of clones’ green fluorescent protein (GFP) expression was also captured using confocal microscopy (Olympus Fluoview v2.0b). The rPhenotyping was performed with 31 patients’ samples infected with HIV-1C and four QC-samples. The ligated mixture was transformed into TOP10 bacteria (Life Technologies) and inoculated directly in the LB broth supplemented with ampicillin to retain the viral diversity. The tropism was inferred by using serial dilutions of the CCR5 antagonist TAK-779 (obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, Bethesda, MD, USA) or the CXCR4 inhibitor AMD3100 (Sigma-Aldrich, St. Louis, MO, USA). rPhenotyping was compared with pGTT while cPTT were compared with cGTT.
Ex vivo Maraviroc Drug Sensitivity Assay
Drug sensitivity assay to Maraviroc (Selzentry) (obtained through NIH AIDS Reagent Program, NIH, US) of recombinant viruses was measured by determining the extent to which the antiretroviral drugs inhibited viral replication in TZM-bl cell. Briefly, serial dilutions spanning 10 µM to 0.000001 µM were added in triplicate in 96-well plates in complete DMEM media containing TZM-bl followed by infection with reference virus (R5 tropic, pMN-NL43-MJ4env) or the corresponding patient derived R5-tropic viruses from HIV-1C (n = 20) and non-C (HIV-1B and 01_AE; n = 12), at a multiplicity of infection (MOI) of 0.05 IU/cells into the 96 well plate in the presence of 10 µg/ml concentration of DEAE. Virus replication was quantified by measuring Renilla luciferase activity (relative light units [RLU]) using Bright-Glo™ Luciferase Assay System (Promega, US). Drug concentrations required to inhibit virus replication by 50% (EC50) were calculated using nonlinear regression analysis (GraphPad Prism, version 5.01; GraphPad Software, La Jolla, CA).
References
Cashin, K. et al. Reliable genotypic tropism tests for the major HIV-1 subtypes. Sci Rep 5, 8543, https://doi.org/10.1038/srep08543 (2015).
Fouchier, R. A. et al. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol 66, 3183–3187 (1992).
Low, A. J., Swenson, L. C. & Harrigan, P. R. HIV coreceptor phenotyping in the clinical setting. AIDS Rev 10, 143–151 (2008).
Jensen, M. A., Coetzer, M., van‘t Wout, A. B., Morris, L. & Mullins, J. I. A reliable phenotype predictor for human immunodeficiency virus type 1 subtype C based on envelope V3 sequences. J Virol 80, 4698–4704, https://doi.org/10.1128/jvi.80.10.4698-4704.2006 (2006).
Lengauer, T., Sander, O., Sierra, S., Thielen, A. & Kaiser, R. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol 25, 1407–1410, https://doi.org/10.1038/nbt1371 (2007).
Vandekerckhove, L. P. et al. European guidelines on the clinical management of HIV-1 tropism testing. Lancet Infect Dis 11, 394–407, https://doi.org/10.1016/s1473-3099(10)70319-4 (2011).
Panos, G. & Watson, D. C. Effect of HIV-1 subtype and tropism on treatment with chemokine coreceptor entry inhibitors; overview of viral entry inhibition. Crit Rev Microbiol 41, 473–487, https://doi.org/10.3109/1040841x.2013.867829 (2015).
Gupta, S., Neogi, U., Srinivasa, H., Banerjea, A. C. & Shet, A. HIV-1 coreceptor tropism in India: increasing proportion of X4-tropism in subtype C strains over two decades. J Acquir Immune Defic Syndr 65, 397–404, https://doi.org/10.1097/qai.0000000000000046 (2014).
Kalu, A. W. et al. Monophylogenetic HIV-1C epidemic in Ethiopia is dominated by CCR5-tropic viruses-an analysis of a prospective country-wide cohort. BMC Infect Dis 17, 37, https://doi.org/10.1186/s12879-016-2163-1 (2017).
Amogne, W. et al. Phylogenetic Analysis of Ethiopian HIV-1 Subtype C Near Full-Length Genomes Reveals High Intrasubtype Diversity and a Strong Geographical Cluster. AIDS Res Hum Retroviruses 32, 471–474, https://doi.org/10.1089/aid.2015.0380 (2016).
Mulinge, M. et al. HIV-1 tropism determination using a phenotypic Env recombinant viral assay highlights overestimation of CXCR4-usage by genotypic prediction algorithms for CRF01_AE and CRF02_AG [corrected]. PLoS One 8, e60566, https://doi.org/10.1371/journal.pone.0060566 (2013).
Gupta, S., Neogi, U., Srinivasa, H. & Shet, A. Performance of genotypic tools for prediction of tropism in HIV-1 subtype C V3 loop sequences. Intervirology 58, 1–5, https://doi.org/10.1159/000369017 (2015).
Abebe, A. et al. HIV-1 subtype C syncytium- and non-syncytium-inducing phenotypes and coreceptor usage among Ethiopian patients with AIDS. AIDS 13, 1305–1311 (1999).
Bjorndal, A., Sonnerborg, A., Tscherning, C., Albert, J. & Fenyo, E. M. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res Hum Retroviruses 15, 647–653, https://doi.org/10.1089/088922299310944 (1999).
Riemenschneider, M. et al. Genotypic Prediction of Co-receptor Tropism of HIV-1 Subtypes A and C. Sci Rep 6, 24883, https://doi.org/10.1038/srep24883 (2016).
Roche, M. et al. A common mechanism of clinical HIV-1 resistance to the CCR5 antagonist maraviroc despite divergent resistance levels and lack of common gp120 resistance mutations. Retrovirology 10, 43, https://doi.org/10.1186/1742-4690-10-43 (2013).
Berro, R. et al. V3 determinants of HIV-1 escape from the CCR5 inhibitors Maraviroc and Vicriviroc. Virology 427, 158–165, https://doi.org/10.1016/j.virol.2012.02.006 (2012).
Garcia-Perez, J. et al. A single-residue change in the HIV-1 V3 loop associated with maraviroc resistance impairs CCR5 binding affinity while increasing replicative capacity. Retrovirology 12, 50, https://doi.org/10.1186/s12977-015-0177-1 (2015).
Berro, R., Sanders, R. W., Lu, M., Klasse, P. J. & Moore, J. P. Two HIV-1 variants resistant to small molecule CCR5 inhibitors differ in how they use CCR5 for entry. PLoS Pathog 5, e1000548, https://doi.org/10.1371/journal.ppat.1000548 (2009).
Wensing, A. M. et al. 2014 Update of the drug resistance mutations in HIV-1. Top Antivir Med 22, 642–650 (2014).
Jiang, X. et al. Characterizing the Diverse Mutational Pathways Associated with R5-Tropic Maraviroc Resistance: HIV-1 That Uses the Drug-Bound CCR5 Coreceptor. J Virol 89, 11457–11472, https://doi.org/10.1128/JVI.01384-15 (2015).
Anastassopoulou, C. G., Ketas, T. J., Sanders, R. W., Klasse, P. J. & Moore, J. P. Effects of sequence changes in the HIV-1 gp41 fusion peptide on CCR5 inhibitor resistance. Virology 428, 86–97, https://doi.org/10.1016/j.virol.2012.03.008 (2012).
Haggblom, A., Svedhem, V., Singh, K., Sonnerborg, A. & Neogi, U. Virological failure in patients with HIV-1 subtype C receiving antiretroviral therapy: an analysis of a prospective national cohort in Sweden. Lancet HIV 3, e166–174, https://doi.org/10.1016/S2352-3018(16)00023-0 (2016).
Aralaguppe, S. G. et al. Multiplexed next-generation sequencing and de novo assembly to obtain near full-length HIV-1 genome from plasma virus. J Virol Methods 236, 98–104, https://doi.org/10.1016/j.jviromet.2016.07.010 (2016).
Edwards, S. et al. A diagnostic HIV-1 tropism system based on sequence relatedness. J Clin Microbiol 53, 597–610, https://doi.org/10.1128/jcm.02762-14 (2015).
Neogi, U. et al. Higher prevalence of predicted X4-tropic strains in perinatally infected older children with HIV-1 subtype C in India. J Acquir Immune Defic Syndr 59, 347–353, https://doi.org/10.1097/QAI.0b013e3182405c7b (2012).
Morner, A. et al. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J Virol 73, 2343–2349 (1999).
Acknowledgements
The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Maraviroc (Cat #11580) and TAK-779 (Cat# 4983), Ghost(3)X4/R5 from Drs Vineet N. Kewal Ramani and Dan R. Littman. Authors would like to that EQAPOL Viral Diversity program, Duke University, US for providing the EQA panel. The study is funded by grants from Swedish Research Council (2016-01675), Stockholm County Council (ALF 20160074) and the Swedish Research Council Establishment grant (2017-01330) in part to UN. UN acknowledges the support received from Jonas Söderquist’s Stipendium for Experimental Virology and Immunology Research-2016.
Author information
Authors and Affiliations
Contributions
U.N. conceives and designed the study. A.B.S., A.H., M.S.R. and S.G.A. performed the experiments. A.B.S., A.H., U.N. and J.B. analysed the data. W.A. maintained the clinical database of Ethiopia and provided the clinical and epidemiological overview. U.N., J.B. and T.K. supervised the overall study. U.N. wrote the first draft of the manuscript reviewed by T.K. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Siddik, A.B., Haas, A., Rahman, M.S. et al. Phenotypic co-receptor tropism and Maraviroc sensitivity in HIV-1 subtype C from East Africa. Sci Rep 8, 2363 (2018). https://doi.org/10.1038/s41598-018-20814-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-20814-2
This article is cited by
-
Immune activation correlates with and predicts CXCR4 co-receptor tropism switch in HIV-1 infection
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.