Abstract
Mammary epithelial cells (MECs) affect milk production capacity during lactation and are critical for the maintenance of tissue homeostasis. Our previous studies have revealed that the expression of miR-152 was increased significantly in MECs of cows with high milk production. In the present study, bioinformatics analysis identified ACAA2 and HSD17B12 as the potential targets of miR-152, which were further validated by dual-luciferase repoter assay. In addition, the expressions of miR-152 was shown to be negatively correlated with levels of mRNA and protein of ACAA2, HSD17B12 genes by qPCR and western bot analysis. Furthermore, transfection with miR-152 significantly up-regulated triglyceride production, promoted proliferation and inhibited apoptosis in MECs. Furthermore, overexpression of ACAA2 and HSD17B12 could inhibit triglyceride production, cells proliferation and induce apoptosis; but sh234-ACAA2-181/sh234-HSD17B12-474 could reverse the trend. These findings suggested that miR-152 could significantly influence triglyceride production and suppress apoptosis, possibly via the expression of target genes ACAA2 and HSD17B12.
Similar content being viewed by others
Introduction
Breast fatty gland tissues not only help to support the basic mammary structure, but also serve as the communication bridge between mammary epithelia and their local and systemic environment throughout the development of breast1, which are mainly consisted of a large number of mammary epithelial cell clusters. Cow has four mammary glands in each breast which are responsible for the synthesis and secretion of milk proteins2. More importantly, the growth and development of mammary epithelial cells affect milk production capacity during lactation and new mammary epithelial cells formation is critical for maintenance of tissue homeostasis3.
MicroRNA (miRNA) as an mRNA suppresser regulates protein quantity and controls a variety of cellular and physiological processes by post-transcriptional processing, including cell proliferation, differentiation, apoptosis and tumor metastasis4,5,6,7,8,9. Recent studies have shown that miRNAs are involved in fat formation and lipolysis10,11,12,13,14. Certain miRNAs have been reported to either promote or inhibit triglyceride production and to suppressapoptosis of MECs at growing stages. For instance, miR-224 overexpression could decrease triglyceride synthesis and promote apoptosis12. Bta-miR-29b promotes triglyceride production and suppress apoptosis11. In addition, miR-33a, miR-21, miR-23a, miR-877 have also been reported to affect triglyceride production and apoptosis of MECs10. Nonetheless, the effects of miR-152 expression on MECs have not been well studied.
The mature sequence of miR-152, a member of the miR-148/152 family (miR-148a, miR-148b, and miR-152), is relatively conservative15. Previous studies on miR-152 are mostly focused on tumor. It has been reported that miR-152 might be involved in the carcinogenesis of ovarian cancer through deregulation of cell proliferation and might be a novel biomarker for early detection or therapeutic purpose16. MiR-152 affects cell cycle progression in non-small cell lung cancer and liver cancer by targeting WNT-1 protein17. There are also evidences suggest that miR-152 is associated with epigenetics. MiR-152 and miR-185 could co- target DNMT1 (DNA Methyltransferase 1) in ovarian cancer cells18,19. In recent years, increased studies indicate that miR-152 is involved in lipid metabolism, for example, miR-152 regulates DNMT1, which in turn influences lactation-related genes in dairy cow mammary epithelial cells20. However, miR-152 has not been comprehensively investigated in MECs, especially the involvement of miR-152 in the mammary gland at the molecular level.
MiRNA could control gene expression either via target mRNAs degradation or translation inhibition by binding to the 3′-untranslated regions (3-UTR)19,21,22. Bioinformatics analysis suggest that ACAA2 (acetyl-CoA acyltransferase 2) and HSD17B12 (hydroxysteroid 17-beta dehydrogenase 12) are both potential target genes of miR-152.
Acetyl-CoA acyltransferase 2 (ACAA2) gene encodes an enzyme of the thiolase family, which is involved in mitochondrial fatty acid elongation and degradation by catalyzing the last step of the respective β-oxidation pathway23. Meanwhile, BNIP3, a unique pro-apoptotic protein, belonging to the BH3-only subset of the Bcl-2 family, was found to have a linkage with ACAA2. Cell apoptosis from overexpression of BNIP3 or hypoxia treatment could be abolished by ACAA2 expression24. The Hydroxysteroid (17b) dehydrogenase type 12 (HSD17B12) gene belongs to the hydroxysteroid (17) dehydrogenase superfamily. HSD17B12 encodes a multifunctional enzyme, which is involved in the prolongation of very long chain fatty acid (VLCFA), especially in the conversion of palmitic to archadonic (AA) acid and in the synthesis of arachidonic acid (AA)25,26,27.
The present study aims to identify the role of miR-152 in mammary epithelial cells. Our results suggested that miR-152 could regulate fatty acid metabolism by directly targeting ACAA2, HSD17B12, which in turn inhibited cellular apoptosis, promoted cell proliferation and enhanced triglyceride production. Further study of miR-152 in lipid metabolism and apoptosis in MECs might establish this microRNA as a novel biomarker for marker assisted selection of Holstein dairy cows, especially in the selection of butterfat rate.
Results
ACAA2 and HSD17B12 were the direct target genes of miR-152 which were verified in MECs
Many target genes of miR-152 were predicted by high-throughput sequencing. Among all the potential genes, ACAA2 and HSD17B12 were selected for further validation due to their potential roles in fatty acid and triglyceride formation using DAVID system (Fig. 1A). Both ACAA2 and HSD17B12 genes had one miR-152 binding sites in 3′UTR region. (Fig. 1B). Co-transfection results showed that luciferase activities of MECs transfected with ACAA2-WT or HSD17B12-WT vector significantly decreased compared with MECs transfected with ACAA2-si/mut vector and HSD17B12/mut vector (Fig. 1C). These results further confirmed that ACAA2 and HSD17B12 were target genes of miR-152.
Target sites of miR-152 and luciferase assay. (A) The pathways of ACAA2 and HSD17B12 were analyzed using the DAVID system. (B) Binding sites of miR-152 on the 3′UTR of ACAA2 and HSD17B12. (C) Luciferase activities were detected in MECs co-transfected with miR-152 and pmiR-RB-REPORT-ACAA2-mut/WT/NC vector or pmiR-RB-REPORT- HSD17B12-mut/WT/NC vector. (**p < 0.01).
ACAA2 and HSD17B12 were down-regulated by miR-152
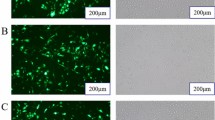
To test whether miR-152 down-regulates the expression of ACAA2 and HSD17B12, miR-152 mimics and miR-152 inhibitor were transfected into MECs and the expressions of GFP protein can be observed in cells transfected with miR-152 mimics, miR-152 inhibitor and miRNA-ShNC (Fig. 2), suggesting that the transfection was successful. The expressions of ACAA2 and HSD17B12 at mRNA and protein level were examined (Fig. 3B,C and D) (Fig. S1.1). The results showed that the expression levels of miR-152 were remarkably increased in MECs transfected with miR-152 mimics as compared with that transfected with miR-152 inhibitor and miRNA-shNC (Fig. 3A). Moreover, transfection of miR-152 mimics significantly decreased the expressions of ACAA2 and HSD17B12 at both mRNA and protein levels, indicating that miR-152 down-regulated the expression of both ACAA2 and HSD17B12 genes.
MEC transfection efficiency. Green fluorescence could be observed under a fluorescence microscope 24 h after the transfection. The expression rate of green fluorescence in mammary epithelial cells of dairy cow cells was 60%. (A) Cells transfected with miR-152 mimics; (B) Cells transfected with miR-152 inhibitor; (C) Cells transfected with miR-shNC.
The expressions of miR-152b and target genes. (A) qPCR analysis of the expressions of miR-152 in MECs transfected with miR-152 mimics, miR-152 inhibitor and miR-shNC. (B) Western blot analysis of the protein levels of ACAA2 and HSD17B12 genes in MECs transfected with miR-152 mimics, miR-152 inhibitor and miR-shNC. (C) qPCR analysis for the mRNA levels of ACAA2 and HSD17B12 genes in MECs transfected with miR-152 mimics, miR-152 inhibitor and miR-shNC. (D) Relative levels of ACAA2 and HSD17B12 protein in MECs transfected with miR-152 mimics, miR-152 inhibitor and miR-shNC. (**p < 0.01, *p < 0.05).
MiR-152 promoted cell proliferation and inhibited apoptosis
The apoptosis rate of MECs transfected with the miR-152 mimics, miR-152 inhibitor and miRNA-shNC were examined using flow cytometry. The results showed that the apoptosis rates were 14.59%, 21.99% and 18.1% in MECs transfected with the mimics, inhibitor and shNC of miR-152, respectively (Fig. 4A), suggesting that overexpression of miR-152 inhibited the apoptosis of MECs.
miR-152 effect on cell apoptosis, cell proliferation and triglycerides production in MECs. (A) Apoptosis ratio of MECs transfected with miR-152 mimics, miR-152 inhibitor and miR-shNC. (B) MECs proliferation determined by the MTT assay in MECs transfected with miR-152 mimics, miR-152 inhibitor and miR-shNC. (C) Triglycerides level in MECs transfected with miR-152 mimics, miR-152 inhibitor and miR-shNC. (*p < 0.05).
The cellular proliferation rate was determined by an MTT assay after the cells were transfected with miR-152 mimics, miR-152 inhibitor, miRNA-ShNC and cultured for 0, 12, 24, 36, 48 or 72 h. As shown in Fig. 4A, miR-152 mimics induced a significant decrease on the proliferation rate of MECs (p < 0.01) (Fig. 4B).
Regulation of triglyceride production by miR-152
Triglyceride production was upregulated in MECs transfected with miR-152 mimics, and no significant differences were observed in MECs transfected with inhibitor, shNC and blank, respectively (Fig. 4C) (p < 0.05).
ACAA2 and HSD17B12 effect on cell apoptosis, proliferation and triglyceride production
PBI-CMV3-ACAA2/HSD17B12 and sh234-ACAA2-181/sh234-HSD17B12-474 plasmids were respectively transfected into cells using lipofectamine TM 2000. The apoptosis rates were detected as follows: PBI-CMV3-ACAA2 (7.49%), PBI-CMV3 (4.89%), sh234-ACAA2-181 (7.08%), sh234 (6.62%), PBI-CMV3-HSD17B12 (8.9%), and sh234-HSD17B12-474 (2.58%) respectively. The results showed that overexpression of ACAA2 and HSD17B12 could induce cells apoptosis; but sh234-ACAA2-181/sh234-HSD17B12-474 could reverse the effect.
After MTT detection, cells proliferation ability was better in group of sh234-ACAA2-181/sh234-HSD17B12-474 transfection than cells transfected PBI-CMV3-ACAA2/HSD17B12 plasmids.
In addition, overexpression of ACAA2 and HSD17B12 could inhibit triglyceride production. But sh234-ACAA2-181/sh234-HSD17B12-474 could improve the expression (Figs 5 and 6).
ACAA2 gene effect on cell apoptosis, proliferation and triglyceride production. (A) PBI-CMV3-ACAA2 and sh234-ACAA2-181 plasmids were respectively transfected into cells. Then qPCR analysis of the expressions of ACAA2. (B) Proteins of ACAA2 in cells after transfection. (C) MECs proliferation were analyzed after PBI-CMV3-ACAA2 and sh234-ACAA2-181 plasmids treatment. (D) triglyceride production in MECs. (E) Apoptosis rate was detected in MECs transfected with PBI-CMV3-ACAA2 and sh234-ACAA2-181.
HSD17B12 gene effect on cell apoptosis, proliferation and triglyceride production. (A) HSD17B12 mRNA detected in MECs that transfected PBI-CMV3-HSD17B12 and sh234-HSD17B12-474 plasmids. (B) Proteins of HSD17B12 were analyzed by western blot in MECs. (C) MTT was used to assay the effect of HSD17B12 on MECs proliferation ability. (D) Influence of HSD17B12 on triglyceride was assessed. (E) Apoptosis rate were analyzed after plasmids transfection.
Discussion
Triglyceride, the main form of energy storage in the body, is stored in adipose tissue which is involved in a range of processes such as energy balance and fatty acids metabolism. Triglycerides are ester molecules derived from glycerol and three fatty acids28,29,30,31. Apoptosis is a process of programmed cell death that occurs in multicellular organisms32,33, which involve coordinated regulation of a broad range of genes.
Here we identified the regulatory roles of miR-152 on ACAA2 and HSD17B12 in triglyceride production and apoptosis of MECs. Our results suggested that miR-152 represses the expression of ACAA2 and HSD17B12 through a direct interaction with the 3′UTR regions of the ACAA2 and HSD17B12. When RNA is bound by miRNAs, miRNA-induced silencing complex is thought to inhibit protein production either through blocking translation or by reducing messenger RNA stability34. Our results also indicated that miR-152 could suppress the expression of ACAA2 and HSD17B12 by reducing the stability of mRNA.
Furthermore, in our study, overexpression of ACAA2 and HSD17B12 could inhibit triglyceride production, cells proliferation and induce apoptosis; but sh234-ACAA2-181/sh234-HSD17B12-474 could reverse the trend. Gene pathway analysis suggests that upregulation of ACAA2 and HSD17B12 could induce fatty acid elongation. In mammals, HSD17B12 is involved in lipid metabolism35, such as in the synthesis of arachidonic acid. HSD17B12 is also essential for normal neuronal development during embryogenesis36. ACAA2 encodes an enzyme of the thiolase family that is involved in fatty acid elongation and degradation by catalyzing the last step of the respective β-oxidation pathway. The increased energy needs for gluconeogenesis and triglyceride synthesis during lactation are met primarily by increased fatty acid oxidation. Therefore, ACAA2 enzyme plays an important role in the supply of energy and carbon substrates for lactation and may thus affect milk production traits23. In addition, the viability and apoptosis of MECs is also a key factor affecting lipid metabolism. Several recent studies show that tumor cell growth was inhibited and apoptosis was increased upon silencing of HSD17B12 37,38. ACAA2 has also been shown to be a functional BNIP3 binding partner, which provides a possible link between fatty acid metabolism and cell apoptosis. ACAA2 counteracts the apoptosis induced by BNIP3. BNIP3 is a unique pro-apoptotic protein which belongs to the BH3-only subset of the Bcl-2 family and localizes on mitochondrial membrane in HepG2 cells24.
It has been reported that simultaneous inhibition of miR-148a and miR-152 could significantly protect MCF-7 cells from 4-OHT induced cell viability reduction and inhibit cell apoptosis39. MiR-152-3p might also act as a tumor suppressor in human breast cancer cells via negatively regulating PIK3CA expression to inhibit the activation of AKT and RPS6, leading to the suppression of HCC1806 cells proliferation40. MiR-152 inhibits tumor cell growth by directly targeting RTKN in hepatocellular carcinoma41. In addition, miR-152 could enhance the viability and multiplication capacity of DCMECs20. Similarly, our results showed that miR-152 could inhibit apoptosis and promote cell proliferation in MECs. In summary, ACAA2 and HSD17B12, two important genes involved in lipid metabolism, were targeted and regulated by miR-152. In addition, miR-152 could influence mammary epithelial cells apoptosis and triglyceride formation. Therefore, miR-152 could be considered as an important indicator for evaluation of cow’s milk fat quality and marker assisted cattle breeding.
Materials and Methods
The experimental design and procedures were performed in accordance with the approved Guidelines for Animal Experiments of Jilin University, China and were approved by the Animal Care and Use Committee of Jilin University, China (Approval ID: SYXK (Ji) 2008-0010/0011).
Experimental Reagents
The primers were synthesized by Shanghai Sangon Biotech Company in China. The RNA Extraction Kit, cDNA reverse transcription Kit and the SYBR Green were from Takara (Takara Biological Company, Japan). ACAA2 antibody and HSD17B12 antibody were from abcam (abacm Reagent Company, USA). The Dual luciferase assay kit was from Promega (Promega Company, USA).
Target Prediction and KEGG Orthology Analysis
Based on the sequences of the miRNAs, target genes were predicted by deep sequencing and screening. The KEGG of all small RNA target genes was analyzed using DAVID system (https://david.ncifcrf.gov/home.jsp). The predicted target genes were subsequently submitted to KOBAS for KEGG Orthology analysis (http://kobas.cbi.pku.edu.cn/home.do) using KEGG database.
Plasmid Construction
Vectors of miR-152 mimics, miR-152 inhibitor and miR-shNC were purchased from GenePharma Company in China. The sequences of ACAA2 and HSD17B12 were amplified with PCR instrument with Not I and Xho I restriction sites engineered at primer ends. Amplified DNA fragments were cloned into pmiR-RB-REPORT vector to construct recombinant vectors pmiR-RB-REPORT- ACAA2-mut/WT/si and pmiR-RB-REPORT- HSD17B12-mut/WT/si. To investigate the association between target genes with apoptosis, cell proliferation and triglyceride production, PBI-CMV3-ACAA2/HSD17B12 and sh234-ACAA2-181/sh234-HSD17B12-474 plasmids were respectively constructed, and then transfected into cells using lipofectamine TM 2000 according to the manufacturer’s instructions.
Cells culture and Transfection
6 heads of healthy cows were selected, then mammary epithelial cells was isolated from dairy cows with high-fat production. Mammary epithelial cells was isolated and cultured from the Laboratory of animal genetics in Jilin University. Progesterone was added into the basal medium. After determination of cell viability and concentration, cells were seeded in six-well culture plates (Corning Inc., Corning, NY) the day before transfection at a density of approximately 1 × 106 per well with DMEM/F12 (GIBCO, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS; PAA, Pasching, Austria) and incubated at 37 °C in a 5% CO2 incubator with humidified atmosphere.
For luciferase activity detection, 150 µL Opti-Minimal Essential Medium (MEM) serum-free medium (GIBCO, Grand Island, NY, USA) was mixed with 5 µL lipofectamine TM 2000 (Invitrogen, USA) and 1.25 µL 20 µmol of the miR-152 mimics and 500 ng pmiR-RB-REPORT vectors. To validate the miR-152 effects on the target genes and triglyceride production, 150 µL Opti-MEM serum-free medium was mixed with 5 µL of lipofectamine TM 2000 and 1.25 µL 20 µmol of the miR-152 mimics, inhibitor and miR-shNC. To investigate the association between target genes with apoptosis and triglyceride production, PBI-CMV3-ACAA2/HSD17B12 and sh234-ACAA2-181/sh234-HSD17B12-474 plasmids were respectively transfected into cells using lipofectamine TM 2000 according to the manufacturer’s instructions.
Luciferase Reporter Assay
MECs were maintained in DMEM/F12 (GIBCO) supplemented with 10% fetal bovine serum (FBS, GIBCO). The cells were transiently co-transfected with 0.5 μg of reporter plasmids (pmiR-RB-REPORT-ACAA2-mut/WT/si or pmiR-RB-REPORT-HSD17B12-mut/WT/si) and miR-152 mimics. The activity of luciferase was detected by using the SpectraMax M5 Microplate Reader.
Real-time PCR Analysis
qRT-PCR was utilized to measure the expression levels of miRNAs and mRNAs. The reverse transcription primers and fluorescence labeled primers for quantitative analysis of miR-152 and target genes were designed using Primer 6.0 as shown in Table 1. MECs transfected with miR-152 mimics, miR-152 inhibitor and miRNA-ShNC were harvested at 48 h post-transfection and total RNA from cultured cells was extracted by using TRIzol reagent (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. The cDNA was synthesized with the Reverse Transcription Kit. The expression levels of miRNA and mRNA were assessed with qRT-PCR using SYBR Green I (Takara, Dalian, China) on an Eppendorf AG-5341 instrument. Three biological repeats and three technical replicates were measured. The expression levels of miRNA and mRNA were defined by the following formula:
Here, 2−ΔΔCt refers to the relative expression ratio and relative expression levels were calculated using the 2 −ΔΔCt method. The expression levels of U6 small nuclear RNA was used as housekeeping gene of miR-152, and GAPDH was used as reference genes of target genes individually.
Western Blot Analysis
Cultured cells were lysed in RIPA buffer (Boster, Wuhan, China) with 1% Phenylmethanesulfonyl fluoride (Beyotime, China) following the manufacturer’s instruction. Protein was loaded and seperated by SDS-PAGE gel and transferred onto PVDF membrane (Bio-Red Laboratories Inc, USA). Then, the polyvinylidene difluoride (PVDF) membrane were incubated with anti-ACAA2 (1:1000, Abcam, Cambridge, MA, USA), anti-HSD17B12 (1:1000, Abcam) or anti-actin (1:1000, Abcam) at 4 °C overnight. After washing 3 times by Tris Buffered Saline with Tween 20 (TBST), the PVDF membrane was incubated with goat anti-rabbit IgG (1:3000, Abcam) for 1 h at room temperature. Finally, protein bandings were obtained by enhanced chemiluminescence (ECL) Western Blotting Substrate (Invitrogen USA). And the signal intensities were captured by a Tanon 5200 chemiluminescence/fluorescence image analysis system. β-actin was used as the endogenous control.
Cell Apoptosis Analysis by Flow Cytometery
After transfection for 36 h, cell apoptosis was analyzed by flow cytometry. The Annexin V-FITC Apoptosis Detection Kit (KeyGEN, Jiangsu, China) was used to stain the cells. Cells were harvested and wahsed twice with 1× PBS at a final concentration of 106 cells per ml. Then, 500 μl cell suspension, 5 μl Annexin V-FITC conjugate and 10 μl propidium iodide solution were added into a test tube sequentially. The tubes were incubated at room temperature for 10 min at dark. Cells were subsequently analyzed by flow cytometry (BD, USA) to verify the effect of miR-152 and target genes on cell apoptosis.
Cell Proliferation Assays
To determine the effects of miR-152 and target genes on MECs proliferation, miR-152, ACAA2, HSD17B12 were overexpressed or silenced in MECs using the cell transfection methods individually. Briefly, cells (2 × 104 per well) were seeded in 96-well plates. After transfection, proliferation was examined in the surviving fractions at 0, 12, 24, 36, 48 and 72 hours using the MTT assay (cellchipbj, Beijing, China). The absorbance was recorded at 450 nm using a micro-plate spectrophotometer (ACTGene, USA).
Triglyceride Detection
Triglyceride was extracted from cells transfected with miR-152 mimics, miR-152 inhibitor, miRNA-ShNC, PBI-CMV3-ACAA2/HSD17B12 and sh234-ACAA2-181/sh234-HSD17B12-474 following the manufacturer’s instructions (Sigma, USA). Then the absorbance of the samples was detected with a SpectraMax M5 Microplate Reader (MD, USA).
References
Pavlovich, A. L., Manivannan, S. & Nelson, C. M. Adipose stroma induces branching morphogenesis of engineered epithelial tubules. Tissue Eng Part A 16, 3719–3726, https://doi.org/10.1089/ten.TEA.2009.0836 (2010).
Wiseman, B. S. & Werb, Z. Stromal effects on mammary gland development and breast cancer. Science 296, 1046–1049, https://doi.org/10.1126/science.1067431 (2002).
Kass, L., Erler, J. T., Dembo, M. & Weaver, V. M. Mammary epithelial cell: influence of extracellular matrix composition and organization during development and tumorigenesis. Int J Biochem Cell Biol 39, 1987–1994, https://doi.org/10.1016/j.biocel.2007.06.025 (2007).
Qu, K. et al. Circulating miRNA-21-5p as a diagnostic biomarker for pancreatic cancer: evidence from comprehensive miRNA expression profiling analysis and clinical validation. Sci Rep 7, 1692, https://doi.org/10.1038/s41598-017-01904-z (2017).
Singh, J. et al. Role of differentially expressed microRNA-139-5p in the regulation of phenotypic internal anal sphincter smooth muscle tone. Sci Rep 7, 1477, https://doi.org/10.1038/s41598-017-01550-5 (2017).
Ioannidis, J. & Donadeu, F. X. Changes in circulating microRNA levels can be identified as early as day 8 of pregnancy in cattle. PLoS One 12, e0174892, https://doi.org/10.1371/journal.pone.0174892 (2017).
Ul Hussain, M. Micro-RNAs (miRNAs): genomic organisation, biogenesis and mode of action. Cell Tissue Res 349, 405–413, https://doi.org/10.1007/s00441-012-1438-0 (2012).
Wang, H. et al. miR-26b promoter analysis reveals regulatory mechanisms by lipid-related transcription factors in goat mammary epithelial cells. J Dairy Sci 100, 5837–5849, https://doi.org/10.3168/jds.2016-12440 (2017).
Li, N. et al. MiR-106b and miR-93 regulate cell progression by suppression of PTEN via PI3K/Akt pathway in breast cancer. Cell Death Dis 8, e2796, https://doi.org/10.1038/cddis.2017.119 (2017).
Shen, B. et al. Deep Sequencing and Screening of Differentially Expressed MicroRNAs Related to Milk Fat Metabolism in Bovine Primary Mammary Epithelial Cells. Int J Mol Sci 17, 200, https://doi.org/10.3390/ijms17020200 (2016).
Yang, Y. et al. miR-29b Targets LPL and TDG Genes and Regulates Apoptosis and Triglyceride Production in MECs. DNA Cell Biol 35, 758–765, https://doi.org/10.1089/dna.2016.3443 (2016).
Shen, B. et al. miR-224 Affects Mammary Epithelial Cell Apoptosis and Triglyceride Production by Downregulating ACADM and ALDH2 Genes. DNA Cell Biol 36, 26–33, https://doi.org/10.1089/dna.2016.3540 (2017).
Guo, Y., Zhang, X., Huang, W. & Miao, X. Identification and characterization of differentially expressed miRNAs in subcutaneous adipose between Wagyu and Holstein cattle. Sci Rep 7, 44026, https://doi.org/10.1038/srep44026 (2017).
Guan, L. et al. bta-miR-23a involves in adipogenesis of progenitor cells derived from fetal bovine skeletal muscle. Sci Rep 7, 43716, https://doi.org/10.1038/srep43716 (2017).
Yang, J. S. et al. Serum miR-152, miR-148a, miR-148b, and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumour Biol 36, 3035–3042, https://doi.org/10.1007/s13277-014-2938-1 (2015).
Zhou, X. et al. Altered expression of miR-152 and miR-148a in ovarian cancer is related to cell proliferation. Oncol Rep 27, 447–454, https://doi.org/10.3892/or.2011.1482 (2012).
Nie, L. et al. Progesterone-Induced miR-152 Inhibits the Proliferation of Endometrial Epithelial Cells by Downregulating WNT-1. Reprod Sci, 1933719116689595, https://doi.org/10.1177/1933719116689595 (2017).
Ji, W. et al. MicroRNA-152 targets DNA methyltransferase 1 in NiS-transformed cells via a feedback mechanism. Carcinogenesis 34, 446–453, https://doi.org/10.1093/carcin/bgs343 (2013).
Xiang, Y. et al. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene 33, 378–386, https://doi.org/10.1038/onc.2012.575 (2014).
Wang, J. et al. MicroRNA-152 regulates DNA methyltransferase 1 and is involved in the development and lactation of mammary glands in dairy cows. PLoS One 9, e101358, https://doi.org/10.1371/journal.pone.0101358 (2014).
Lopez, M. F. et al. Opposing activities of oncogenic MIR17HG and tumor suppressive MIR100HG clusters and their gene targets regulate replicative senescence in human adult stem cells. NPJ Aging Mech Dis 3, 7, https://doi.org/10.1038/s41514-017-0006-y (2017).
Celia-Terrassa, T. et al. Normal and cancerous mammary stem cells evade interferon-induced constraint through the miR-199a-LCOR axis. Nat Cell Biol 19, 711–723, https://doi.org/10.1038/ncb3533 (2017).
Miltiadou, D. et al. Variants in the 3′ untranslated region of the ovine acetyl-coenzyme A acyltransferase 2 gene are associated with dairy traits and exhibit differential allelic expression. J Dairy Sci 100, 6285–6297, https://doi.org/10.3168/jds.2016-12326 (2017).
Cao, W. et al. Acetyl-Coenzyme A acyltransferase 2 attenuates the apoptotic effects of BNIP3 in two human cell lines. Biochim Biophys Acta 1780, 873–880, https://doi.org/10.1016/j.bbagen.2008.02.007 (2008).
Visus, C. et al. Identification of Hydroxysteroid (17beta) dehydrogenase type 12 (HSD17B12) as a CD8+ T-cell-defined human tumor antigen of human carcinomas. Cancer Immunol Immunother 60, 919–929, https://doi.org/10.1007/s00262-011-1001-y (2011).
Bellemare, V., Phaneuf, D. & Luu-The, V. Target deletion of the bifunctional type 12 17beta-hydroxysteroid dehydrogenase in mice results in reduction of androgen and estrogen levels in heterozygotes and embryonic lethality in homozygotes. Horm Mol Biol Clin Investig 2, 311–318, https://doi.org/10.1515/HMBCI.2010.036 (2010).
Kemilainen, H. et al. The Hydroxysteroid (17beta) Dehydrogenase Family Gene HSD17B12 Is Involved in the Prostaglandin Synthesis Pathway, the Ovarian Function, and Regulation of Fertility. Endocrinology 157, 3719–3730, https://doi.org/10.1210/en.2016-1252 (2016).
Wang, Q., Wang, G., Qiu, Z., He, X. & Liu, C. Elevated Serum Triglycerides in the Prognostic Assessment of Acute Pancreatitis: A Systematic Review and Meta-Analysis of Observational Studies. J Clin Gastroenterol 51, 586–593, https://doi.org/10.1097/MCG.0000000000000846 (2017).
Rodriguez-Carrio, J. et al. High triglycerides and low high-density lipoprotein cholesterol lipid profile in rheumatoid arthritis: A potential link among inflammation, oxidative status, and dysfunctional high-density lipoprotein. J Clin Lipidol, https://doi.org/10.1016/j.jacl.2017.05.009 (2017).
Mustafa, G. et al. The relationship between erythrocyte membrane fatty acid levels and cardiac autonomic function in obese children. Rev Port Cardiol 36, 499–508, https://doi.org/10.1016/j.repc.2016.10.013 (2017).
Khabbush, A. et al. Neuronal decanoic acid oxidation is markedly lower than that of octanoic acid: A mechanistic insight into the medium-chain triglyceride ketogenic diet. Epilepsia, https://doi.org/10.1111/epi.13833 (2017).
Di Stefano, S. et al. Apoptotic cell death and genetic control in graft coronary artery disease in heart transplant. J Cardiovasc Surg (Torino) 44, 577–582 (2003).
Perez, A. R. et al. Death of adrenocortical cells during murine acute T. cruzi infection is not associated with TNF-R1 signaling but mostly with the type II pathway of Fas-mediated apoptosis. Brain Behav Immun, https://doi.org/10.1016/j.bbi.2017.05.017 (2017).
Osella, M., Bosia, C., Cora, D. & Caselle, M. The role of incoherent microRNA-mediated feedforward loops in noise buffering. PLoS Comput Biol 7, e1001101, https://doi.org/10.1371/journal.pcbi.1001101 (2011).
Lima, D. et al. Cloning and expression analysis of the 17beta hydroxysteroid dehydrogenase type 12 (HSD17B12) in the neogastropod Nucella lapillus. J Steroid Biochem Mol Biol 134, 8–14, https://doi.org/10.1016/j.jsbmb.2012.10.005 (2013).
Rantakari, P. et al. Hydroxysteroid (17{beta}) dehydrogenase 12 is essential for mouse organogenesis and embryonic survival. Endocrinology 151, 1893–1901, https://doi.org/10.1210/en.2009-0929 (2010).
Szajnik, M. et al. 17beta Hydroxysteroid dehydrogenase type 12 (HSD17B12) is a marker of poor prognosis in ovarian carcinoma. Gynecol Oncol 127, 587–594, https://doi.org/10.1016/j.ygyno.2012.08.010 (2012).
Plourde, M. et al. Analysis of 17beta-hydroxysteroid dehydrogenase types 5, 7, and 12 genetic sequence variants in breast cancer cases from French Canadian Families with high risk of breast and ovarian cancer. J Steroid Biochem Mol Biol 116, 134–153, https://doi.org/10.1016/j.jsbmb.2009.05.005 (2009).
Chen, M. J., Cheng, Y. M., Chen, C. C., Chen, Y. C. & Shen, C. J. MiR-148a and miR-152 reduce tamoxifen resistance in ER+ breast cancer via downregulating ALCAM. Biochem Biophys Res Commun 483, 840–846, https://doi.org/10.1016/j.bbrc.2017.01.012 (2017).
Ge, S., Wang, D., Kong, Q., Gao, W. & Sun, J. Function of MiR-152 As a Tumor Suppressor in Human Breast Cancer By Targeting PIK3CA. Oncol Res, https://doi.org/10.3727/096504017X14878536973557 (2017).
Zhou, J. et al. MicroRNA-152 inhibits tumor cell growth by directly targeting RTKN in hepatocellular carcinoma. Oncol Rep 37, 1227–1234, https://doi.org/10.3892/or.2016.5290 (2017).
Acknowledgements
This work was supported by the National Major Special Project on New Varieties Cultivation for Transgenis Organisms (2016ZX08009003-006), the National High Technology Research and Development Program (863 Program, no. 2013AA102505), the National Natural Science Foundation of China (no. 31372278 and no. 31672389), the Jilin Scientific and Technological Development Program (No. 20170519014JH), and the Jilin province industrial technology research and development program (2016C032).
Author information
Authors and Affiliations
Contributions
In our current research, Yuwei Yang and Xibi Fang were mainly to complete the experiments and writing works. Zhihui Zhao and Boxing Sun were proposed experimental design and ideas. And then Haibin Yu and Ping Jiang were given help during the experiment process; Runjun Yang was assisting in data analysis.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, Y., Fang, X., Yang, R. et al. MiR-152 Regulates Apoptosis and Triglyceride Production in MECs via Targeting ACAA2 and HSD17B12 Genes. Sci Rep 8, 417 (2018). https://doi.org/10.1038/s41598-017-18804-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-18804-x
This article is cited by
-
Cullin-associated and neddylation-dissociated protein 1 (CAND1) alleviates NAFLD by reducing ubiquitinated degradation of ACAA2
Nature Communications (2023)
-
ACAA2 is a novel molecular indicator for cancers with neuroendocrine phenotype
British Journal of Cancer (2023)
-
Impact of 17β-HSD12, the 3-ketoacyl-CoA reductase of long-chain fatty acid synthesis, on breast cancer cell proliferation and migration
Cellular and Molecular Life Sciences (2020)
-
Chi-miR-3031 regulates beta-casein via the PI3K/AKT-mTOR signaling pathway in goat mammary epithelial cells (GMECs)
BMC Veterinary Research (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.