Abstract
We conducted a genome-wide association study (GWAS) of anorexia nervosa (AN) using a stringently defined phenotype. Analysis of phenotypic variability led to the identification of a specific genetic risk factor that approached genome-wide significance (rs929626 in EBF1 (Early B-Cell Factor 1); P = 2.04 × 10−7; OR = 0.7; 95% confidence interval (CI) = 0.61–0.8) with independent replication (P = 0.04), suggesting a variant-mediated dysregulation of leptin signaling may play a role in AN. Multiple SNPs in LD with the variant support the nominal association. This demonstrates that although the clinical and etiologic heterogeneity of AN is universally recognized, further careful sub-typing of cases may provide more precise genomic signals. In this study, through a refinement of the phenotype spectrum of AN, we present a replicable GWAS signal that is nominally associated with AN, highlighting a potentially important candidate locus for further investigation.
Similar content being viewed by others
Introduction
Anorexia nervosa (AN) is a complex and often chronic eating disorder characterized by inability to maintain a normal healthy body weight and a persistent fear of weight gain, resulting in extreme emaciation and even death in some cases1. Previous genetic and epidemiological studies have indicated a multifactorial etiology, where both genetic and environmental factors contribute to disease risk2,3,4,5,6,7.
As sample sizes have increased, genome-wide association studies (GWASs) of AN have begun to identify risk variants8,9,10. To further elucidate the genetic architecture of AN, we performed a GWAS using data from our previously published study8 consisting of 1,033 AN cases by excluding 212 patients with AN who experienced diagnostic crossover during the course of their illness. Specifically, we excluded patients who migrated from or to binge-eating disorder (BED) or bulimia nervosa (BN) as assessed with the Structured Interview for Anorexic and Bulimic Disorders11). Although a previous study indicated women with BN were rarely to cross over to AN12, we observed ~43% of AN/BN crossover cases falls into this category in our cohort, suggestive of a confounding factor. We hypothesized that this reduction in phenotypic heterogeneity, despite the fact that AN and BN may share some genetic risk factors13, would enhance gene discovery.
Results
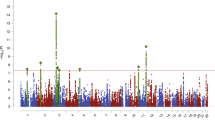
Our discovery cohort included a total of 692 female AN cases of non-Hispanic European (NHE) descent. Cases were included if they were diagnosed with restricting type and binge eating/purging type of AN as defined by DSM-IV. Both types are characterized by below-normal weight and restricted food intake. Individuals diagnosed as restricting type do not experience binge-eating episodes and do not engage in purging, such as vomiting or use of laxatives. Standard quality controls measures were applied, specifically, excluding potential cryptic relatedness and checking for population stratification (details described elsewhere8). The average age of onset of the case subjects was 16.3 years with a standard deviation (SD) of 3 years (Interquartile Range; IQR = 16(14–18)). The control group included 3,570 female matched healthy adolescents of NHE ancestry that had an average age of 18.3 years at the time of data analysis (SD = 5.7; IQR = 19(13–23)) (Supplementary Table 1). Associations were assessed with 507,999 SNPs genotyped on either Illumina HumanHap550 or Human610-Quad BeadChips in an additive model using logistic regression analyses with principal components adjustment, based on the principal component analysis of cases and controls (Supplementary Figure 1), resulting in significantly low level of genomic control inflation factor of 1.03 (Supplementary Figure 2). The analysis yielded one SNP (rs929626) with a P value of 2.04 × 10−7 and 4 other SNPs with marginally larger P values that are in strong linkage disequilibrium (r 2 > 0.8); these SNPs were selected for further analysis (Supplementary Figure 3; Supplementary Table 2).
Using imputation analysis based on data from the 1000 Genomes Project (Phase I integrated variant set, v2, March 2012), we subsequently tested associations with SNPs (imputed info > 0.5, minor allele frequency (MAF) > 0.05) located in a 200-kb window centered on the SNP rs929626. We observed association with a series of markers around this region, of which 34 SNPs supported suggestive associations (P < 1.0 × 10−6) with both imputed and genotyped SNPs, which were in high LD with AN (Fig. 1; Supplementary Table 3). This suggests that the single markers demonstrating nominal association in the GWAS are likely to be true positives.
Region of genome-wide nominal association at 5q33.3. Regional plot of the EBF1-associated interval for the imputation analysis. Foreground shows scatter plot of the −log10 P values plotted against physical position of human reference hg19. Background shows estimated recombination rates plotted to reflect the local LD structure. The color of the dots represents the strength of LD between the top SNP (rs929626) and its proxies (red, r 2 ≥ 0.8; orange, 0.8 > r 2 ≥ 0.6; green, 0.6 > r 2 ≥ 0.4; blue and navy, r 2 < 0.4). Genes, position of exons, and direction of transcription from UCSC genome browser (http://genome.ucsc.edu) are noted.
We further explored this finding using the meta-analysis results from 15 previously reported AN cohorts10. Interestingly, two SNPs were also nominally significant (rs929626 with P = 0.037 and rs17543752 with P = 0.05) in the same direction as in the GWAS (Table 1 ). Meta-analysis results in a P value of 1.52 × 10−7.
We next used the ENCODE project14 data to predict possible functional effect of the SNPs identified in this study. The top SNP, rs929626, and other significant markers located in the 6th intron of the EBF1 gene (Early B-Cell Factor 1), as well as two SNPs (rs113252656 and rs1081071) flanking the top SNP rs929626 at r 2 > 0.5 function as binding sites for EBF1 itself (HaploReg v4.1; ref. 15). This suggests that these genetic variants may modulate the expression of EBF1. Indeed, we observed a positive correlation with the rs929626 C allele carriers compared with TT homozygotes on the EBF1 expression level in nine independent subjects (the FPKM value for TT homozygotes (3 subjects) versus C allele carriers (6 individuals) is 5.0 versus 6.4) with both whole genome sequencing data of blood and corresponding RNA-Seq data of heart right ventricle selected from the Pediatric Cardiac Genomics Consortium cohort (dbGaP Study Accession: phs000571.v3.p2). By using the Genotype-Tissue Expression Portal database (http://www.gtexportal.org), we also observed nominally significant expression quantitative trait loci (eQTLs) association (P = 0.0024, tested in 97 samples) in the putamen for rs929626 in the same direction. A few comorbid psychiatric disorders have been linked with the function of the putamen, such as anxiety, obsessive-compulsive disorder and attention deficit-hyperactivity disorder16,17,18. Taken together, these suggest the minor allele C carriers have relatively higher EBF1 expression.
Discussion
EBF1 encodes a transcription factor that originally thought to function as necessary for the development of the immune system19, but it has since been shown to regulate the development of both osteoblast and adipocyte lineages20,21,22. Two EBF1 variants, rs11953630-T and rs9313772-T, showed significant association at genome-wide level (P < 5 × 10−10) in a study testing blood pressure in European whites23, 24. In addition, rs17056278-C was also identified as a metabolic risk allele, interacting with psychosocial stress to contribute to increased hip circumference (P = 3 × 10−8)25. However none of these is in LD with any markers in our identified locus. In animal studies, Ebf1−/− mice showed increased adipose tissue within marrow, whereas peripheral white adipose tissue was severely reduced. Circulating levels of leptin, a hormone released by adipocytes and one of the major players in food intake regulation, were also decreased in Ebf1−/− mice compared with controls26. This concurs with the reported generalized loss of accumulation of subcutaneous and visceral adipose accompanied by significant increases in yellow marrow in AN patients27, 28. Also notable is the finding that circulating levels of leptin are very low in AN patients29, 30 and a decline in levels of circulating leptin can lead to changes in brain activity in areas involved in regulatory, emotional, and cognitive control of appetite5.
Understanding the genetics of AN is currently a major within-field initiative, in parallel to other neuropsychiatric/neurodevelopmental disorders such as schizophrenia, bipolar disorder, and autism spectrum disorders. Although the clinical and etiologic heterogeneity is universally recognized, in practice, many studies still failed to account for sample heterogeneity. In this study, by focusing on individuals with AN who have not crossed over to BN or BED, we have identified a marginally replicating GWAS signal that approached genome-wide significance. One limitation of our study is that all participants may not yet have experienced the full course of their eating disorder (The average duration of follow-up was 8.6 years with a SD of 7.0 years in the discovery cohort, while the average crossover time was 2.8 years with a SD of 2.6 years for the excluded AN patients), and a portion of the sample may develop BN or BED at later stages of illness. This would represent a conservative bias and underscores the importance of further investigation of this locus in the future focusing on individuals with lifetime AN who have never crossed over to other eating disorder presentations.
Methods
Discovery data set and quality control
We conducted a GWAS using data from our previously published study8 consisting of 1,033 AN cases by excluding 212 patients with AN who experienced diagnostic crossover during the course of their illness (i.e. migrated from or to binge-eating disorder (BED) or bulimia nervosa (BN) as assessed with the Structured Interview for Anorexic and Bulimic Disorders11) plus 100 patients without such information. A total of 692 female AN cases and 3,570 female matched controls that were carefully selected from Center for Applied Genomics (CAG) database were included in the analysis after Standard quality controls, namely, excluding potential cryptic relatedness and checking for population stratification by using the PLINK software31 version 1.90a. The Research Ethics Board of CHOP and other participating centers approved the study. Informed consent was obtained from all adult participants and from a parent or legal guardian in the case of children and all work followed was in accordance with an IRB-approved protocol.
Association tests
For the genome-wide association analysis for SNPs, we utilized the PLINK software31 version 1.90a, through Cochran–Armitage trend test.
Expression studies
The extended locus around associated SNP was then defined by identification of all SNPs showing r 2 > 0.5. Linkage disequilibrium (LD) was defined with the HaploReg v4.1 (ref. 15) based on Phase I of the 1000 Genomes project. Variants showing evidence of LD with associated AN variants were explored for impact on gene function via regulatory function (including eQTLs) by HaploReg v4.1, which both collate data from the Encyclopedia of DNA Elements (ENCODE)14. We also referred to the Genotype-Tissue Expression Portal database (http://www.gtexportal.org) for eQTLs analysis.
Change history
21 August 2017
A correction to this article has been published and is linked from the HTML version of this paper. The error has been fixed in the paper.
References
Klump, K. L., Bulik, C. M., Kaye, W. H., Treasure, J. & Tyson, E. Academy for eating disorders position paper: eating disorders are serious mental illnesses. Int J Eat Disord 42, 97–103, doi:10.1002/eat.20589 (2009).
Zipfel, S., Giel, K. E., Bulik, C. M., Hay, P. & Schmidt, U. Anorexia nervosa: aetiology, assessment, and treatment. Lancet Psychiatry 2, 1099–1111, doi:10.1016/S2215-0366(15)00356-9 (2015).
Hinney, A. & Volckmar, A. L. Genetics of eating disorders. Curr Psychiatry Rep 15, 423, doi:10.1007/s11920-013-0423-y (2013).
Bulik, C. M., Slof-Op’t Landt, M. C., van Furth, E. F. & Sullivan, P. F. The genetics of anorexia nervosa. Annu Rev Nutr 27, 263–275, doi:10.1146/annurev.nutr.27.061406.093713 (2007).
Clarke, T. K., Weiss, A. R. & Berrettini, W. H. The genetics of anorexia nervosa. Clin Pharmacol Ther 91, 181–188, doi:10.1038/clpt.2011.253 (2012).
Bulik, C. M. et al. Prevalence, heritability, and prospective risk factors for anorexia nervosa. Arch Gen Psychiatry 63, 305–312, doi:10.1001/archpsyc.63.3.305 (2006).
Pinheiro, A. P., Root, T. & Bulik, C. M. The Genetics of Anorexia Nervosa: Current Findings and Future Perspectives. Int J Child Adolesc health 2, 153–164 (2009).
Wang, K. et al. A genome-wide association study on common SNPs and rare CNVs in anorexia nervosa. Mol Psychiatry 16, 949–959, doi:10.1038/mp.2010.107 (2011).
Scott-Van Zeeland, A. A. et al. Evidence for the role of EPHX2 gene variants in anorexia nervosa. Mol Psychiatry 19, 724–732, doi:10.1038/mp.2013.91 (2014).
Boraska, V. et al. A genome-wide association study of anorexia nervosa. Mol Psychiatry 19, 1085–1094, doi:10.1038/mp.2013.187 (2014).
Fichter, M. & Quadflieg, N. The structured interview for anorexic and bulimic disorders for DSM-IV and ICD-10 (SIAB-EX): reliability and validity. Eur Psychiatry 16, 38–48, doi:10.1016/S0924-9338(00)00534-4 (2001).
Eddy, K. T. et al. Diagnostic crossover in anorexia nervosa and bulimia nervosa: implications for DSM-V. Am J Psychiatry 165, 245–250, doi:10.1176/appi.ajp.2007.07060951 (2008).
Bulik, C. M. et al. Understanding the relation between anorexia nervosa and bulimia nervosa in a Swedish national twin sample. Biol Psychiatry 67, 71–77, doi:10.1016/j.biopsych.2009.08.010 (2010).
Consortium, T. E. P. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74, doi:10.1038/nature11247 (2012).
Ward, L. D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40, D930–934, doi:10.1093/nar/gkr917 (2012).
Teicher, M. H. et al. Functional deficits in basal ganglia of children with attention-deficit/hyperactivity disorder shown with functional magnetic resonance imaging relaxometry. Nat Med 6, 470–473, doi:10.1038/74737 (2000).
Radua, J. & Mataix-Cols, D. Voxel-wise meta-analysis of grey matter changes in obsessive-compulsive disorder. Br J Psychiatry 195, 393–402, doi:10.1192/bjp.bp.108.055046 (2009).
Radua, J., van den Heuvel, O. A., Surguladze, S. & Mataix-Cols, D. Meta-analytical comparison of voxel-based morphometry studies in obsessive-compulsive disorder vs other anxiety disorders. Arch Gen Psychiatry 67, 701–711, doi:10.1001/archgenpsychiatry.2010.70 (2010).
Lukin, K., Fields, S., Hartley, J. & Hagman, J. Early B cell factor: Regulator of B lineage specification and commitment. Semin Immunol 20, 221–227, doi:10.1016/j.smim.2008.07.004 (2008).
Hesslein, D. G. et al. Ebf1-dependent control of the osteoblast and adipocyte lineages. Bone 44, 537–546, doi:10.1016/j.bone.2008.11.021 (2009).
Akerblad, P., Lind, U., Liberg, D., Bamberg, K. & Sigvardsson, M. Early B-cell factor (O/E-1) is a promoter of adipogenesis and involved in control of genes important for terminal adipocyte differentiation. Mol Cell Biol 22, 8015–8025, doi:10.1128/MCB.22.22.8015-8025.2002 (2002).
Jimenez, M. A., Akerblad, P., Sigvardsson, M. & Rosen, E. D. Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol Cell Biol 27, 743–757, doi:10.1128/MCB.01557-06 (2007).
Ehret, G. B. et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478, 103–109, doi:10.1038/nature10405 (2011).
Wain, L. V. et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet 43, 1005–1011, doi:10.1038/ng.922 (2011).
Singh, A. et al. Gene by stress genome-wide interaction analysis and path analysis identify EBF1 as a cardiovascular and metabolic risk gene. Eur J Hum Genet 23, 854–862, doi:10.1038/ejhg.2014.189 (2015).
Fretz, J. A. et al. Altered metabolism and lipodystrophy in the early B-cell factor 1-deficient mouse. Endocrinology 151, 1611–1621, doi:10.1210/en.2009-0987 (2010).
Bredella, M. A. et al. Increased bone marrow fat in anorexia nervosa. J Clin Endocrinol Metab 94, 2129–2136, doi:10.1210/jc.2008-2532 (2009).
Abella, E. et al. Bone marrow changes in anorexia nervosa are correlated with the amount of weight loss and not with other clinical findings. Am J Clin Pathol 118, 582–588, doi:10.1309/2Y7X-YDXK-006B-XLT2 (2002).
Haluzik, M., Papezova, M., Nedvidkova, J. & Kabrt, J. Serum leptin levels in patients with anorexia nervosa before and after partial refeeding, relationships to serum lipids and biochemical nutritional parameters. Physiol Res 48, 197–202 (1999).
Kilic, M., Taskin, E., Ustundag, B. & Aygun, A. D. The evaluation of serum leptin level and other hormonal parameters in children with severe malnutrition. Clin Biochem 37, 382–387, doi:10.1016/j.clinbiochem.2003.12.010 (2004).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575, doi:10.1086/519795 (2007).
Acknowledgements
We gratefully thank all the patients and their families who were enrolled in this study, as well as all the control subjects who donated blood samples to The Children’s Hospital of Philadelphia (CHOP) for genetic studies. Dong Li is funded in part by 2012–2015 Davis Foundation Postdoctoral Fellowship Program in Eating Disorders Research Award. Bingshan Li was partially supported by Klarman Family Foundation for eating disorders. All genome-wide genotyping for controls was funded by an Institute Development Award to Center for Applied Genomics from CHOP. We thank the Eating Disorders Working Group of the Psychiatric Genomics Consortium (PGC-ED) for providing summary results data for the replication analysis.
Author information
Authors and Affiliations
Author notes
A comprehensive list of consortium members appears at the end of the paper.
An erratum to this article is available at https://doi.org/10.1038/s41598-017-06409-3.
Consortia
Contributions
D.L. and H.H. were leading contributions in the design, analysis and writing; D.L., X.C., Y.L., J.P.B. and P.S contributed to data analysis. J.J.C., L.T., N.R., D.A., Y.R.L. contributed samples and phenotypes. C.E.K., J.L., F.W., J.S., M.L., C.H., Z.W., Y.G., H.Q., F.M., K.T., R.C., B.L., and R.C. provided assistance with samples and data processing. Eating Disorders Working Group of the Psychiatric Genomics Consortium and Price Foundation Collaborative Group provided data for the replication and helped with the discussion; D.L. drafted the manuscript. D.L., J.J.C., E.J.B. and H.H. revised the manuscript. All authors approved final version of manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Change History: A correction to this article has been published and is linked from the HTML version of this paper. The error has been fixed in the paper.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
An erratum to this article is available at https://doi.org/10.1038/s41598-017-06409-3.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, D., Chang, X., Connolly, J.J. et al. A genome-wide association study of anorexia nervosa suggests a risk locus implicated in dysregulated leptin signaling. Sci Rep 7, 3847 (2017). https://doi.org/10.1038/s41598-017-01674-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-01674-8
This article is cited by
-
Stressful life events among individuals with a history of eating disorders: a case-control comparison
BMC Psychiatry (2021)
-
Increased plasma asprosin levels in patients with drug-naive anorexia nervosa
Eating and Weight Disorders - Studies on Anorexia, Bulimia and Obesity (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.