Abstract
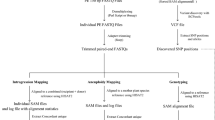
Base editors can achieve targeted genomic base conversion. However, the off-target issue is one of the major concerns in their application. Whole-genome sequencing (WGS) at the individual level can provide direct information on genome-wide specificity, but it is difficult to distinguish true off-target single-nucleotide variants (SNVs) induced by base editors from background variation. Here we describe an unbiased WGS method for evaluating the specificity of base editors in rice. In this protocol, we describe the experimental design and provide details of vector construction, rice transformation and tissue culture, as well as a comprehensive WGS data analysis pipeline for overcoming two related core problems in various plant species: high background mutation rates and the heterogeneity of examined populations. Using this protocol, researchers can straightforwardly and accurately assess the genome-wide specificity of base editors and other genome editing tools in 12–15 weeks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All the sequence data have been deposited in NCBI BioProject under accession code PRJNA522656, in which CBE refers to BE3 and HF1-CBE to HF1-BE3.
Code availability
All the code used in this protocol is available on GitHub at: https://github.com/ReiGao/GWSBE. The code in this protocol has been peer-reviewed.
References
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Nishida, K. et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, 6305 (2016).
Kim, K. et al. Highly efficient RNA-guided base editing in mouse embryos. Nat. Biotechnol. 35, 435–437 (2017).
Liang, P. et al. Effective gene editing by high-fidelity base editor 2 in mouse zygotes. Protein Cell 8, 601–611 (2017).
Liu, Z. Q. et al. Highly efficient RNA-guided base editing in rabbit. Nat. Commun. 9, 2717 (2018).
Tanaka, S. et al. In vivo targeted single-nucleotide editing in zebrafish. Sci. Rep. 8, 11423 (2018).
Zhang, Y. H. et al. Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nat. Commun. 8, 118 (2017).
Zong, Y. et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 35, 438–440 (2017).
Li, C. et al. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome. Biol. 19, 59 (2018).
Zong, Y. et al. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 36, 950–953 (2018).
Komor, A. C., Badran, A. H. & Liu, D. R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168, 20–36 (2017).
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
Jiao, R. & Gao, C. Anything impossible with CRISPR/Cas9? Sci. China Life. Sci. 60, 445–446 (2017).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Tsai, S. Q. & Joung, J. K. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat. Rev. Genet. 17, 300–312 (2016).
Tycko, J., Myer, V. E. & Hsu, P. D. Methods for optimizing CRISPR-Cas9 genome editing specificity. Mol. Cell. 63, 355–370 (2016).
Zhang, F. Development of CRISPR-Cas systems for genome editing and beyond. Q. Rev. Biophys. 52, E6 (2019).
Jin, S. et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292–295 (2019).
Zuo, E. W. et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292 (2019).
Lee, H. K., Smith, H. E., Liu, C., Willi, M. & Hennighausen, L. Cytosine base editor 4 but not adenine base editor generates off-target mutations in mouse embryos. Commun. Biol. 3, 19 (2020).
Doman, J. L., Raguram, A., Newby, G. A. & Liu, D. R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 3, 620-628 (2020).
Gabriel, R. et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 29, 816–823 (2011).
Wang, X. L. et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat. Biotechnol. 33, 175–178 (2015).
Frock, R. L. et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 33, 179–186 (2015).
Kim, D. et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–243 (2015).
Kim, D., Kim, S., Kim, S., Park, J. & Kim, J. S. Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Res. 26, 406–415 (2016).
Cameron, P. et al. Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat. Methods 14, 600–606 (2017).
Tsai, S. Q. et al. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat. Methods 14, 607–614 (2017).
Crosetto, N. et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods 10, 361–365 (2013).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Wienert, B. et al. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science 364, 286–289 (2019).
Li, J. et al. Whole genome sequencing reveals rare off-target mutations and considerable inherent genetic or/and somaclonal variations in CRISPR/Cas9-edited cotton plants. Plant Biotechnol. J. 17, 858–868 (2019).
Willi, M., Smith, H. E., Wang, C., Liu, C. & Hennighausen, L. Mutation frequency is not increased in CRISPR-Cas9-edited mice. Nat. Methods 15, 756–758 (2018).
Iyer, V. et al. No unexpected CRISPR-Cas9 off-target activity revealed by trio sequencing of gene-edited mice. PLoS Genet 14, e1007503 (2018).
Tang, X. et al. A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome. Biol. 19, 84 (2018).
Landrum, M. J. et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44, D862–D868 (2016).
Jackson, S. A. Rice: The first crop genome. Rice 9, 14 (2016).
Shukla, V. K. et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459, 437–441 (2009).
Townsend, J. A. et al. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459, 442–445 (2009).
Li, T., Liu, B., Spalding, M. H., Weeks, D. P. & Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392 (2012).
Wang, Y. P. et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951 (2014).
Wood, A. J. et al. Targeted genome editing across species using ZFNs and TALENs. Science 333, 307–307 (2011).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Yang, L. H. et al. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat. Commun. 5, 5507 (2014).
Feng, Z. Y. et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl Acad. Sci. USA 111, 4632–4637 (2014).
Feng, C. et al. High-efficiency genome editing using a dmc1 promoter-controlled CRISPR/Cas9 system in maize. Plant Biotechnol. J. 16, 1848–1857 (2018).
Nekrasov, V. et al. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7, 482 (2017).
Liang, P. et al. Genome-wide profiling of adenine base editor specificity by EndoV-seq. Nat. Commun. 10, 67 (2019).
Kim, D., Kim, D. E., Lee, G., Cho, S. I. & Kim, J. S. Genome-wide target specificity of CRISPR RNA-guided adenine base editors. Nat. Biotechnol 37, 430–435 (2019).
Kim, D. et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 34, 863–868 (2016).
Kim, D. & Kim, J. S. DIG-seq: a genome-wide CRISPR off-target profiling method using chromatin DNA. Genome Res. 28, 1894–1900 (2018).
Crosetto, N. et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods 10, 361–365 (2013).
Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186 (2012).
Bae, S., Park, J. & Kim, J. S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014).
Zhang, Y., Li, J. & Gao, C. Generation of stable transgenic rice (Oryza sativa L.) by Agrobacterium-mediated transformation. Curr. Protoc. Plant Biol. 1, 235–246 (2016).
Wei, F. J. et al. Somaclonal variation does not preclude the use of rice transformants for genetic screening. Plant J. 85, 648–659 (2016).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Genomes Project Consortium . et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Wilm, A. et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 40, 11189–11201 (2012).
Kim, S. et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594 (2018).
Shan, Q., Wang, Y., Li, J. & Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 9, 2395–2410 (2014).
Shan, Q. et al. Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol. Plant 6, 1365–1368 (2013).
Hiei, Y. & Komari, T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 3, 824–834 (2008).
Acknowledgements
We thank Y. Zhang (Department of Plant and Environmental Sciences, University of Copenhagen) for critical revision of the manuscript. This work was supported by grants from the National Natural Science Foundation of China (31788103), the Strategic Priority Research Program of the Chinese Academy of Sciences (Precision Seed Design and Breeding, XDA24020102), the National Key Research and Development Program of China (2016YFD0101804) and the R&D Program in Key Areas of Guangdong Province (2018B020202005).
Author information
Authors and Affiliations
Contributions
S.J. performed the experiments; S.J. designed figures; C.G. supervised the project; S.J., Q.G. and C.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Keiji Nishida, Pengcheng Wei and Huanbin Zhou for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Jin, S. et al. Science 364, 292–295 (2019): https://doi.org/10.1126/science.aaw7166
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2, Supplementary Notes 1–3 and Supplementary Tables 1–4.
Rights and permissions
About this article
Cite this article
Jin, S., Gao, Q. & Gao, C. An unbiased method for evaluating the genome-wide specificity of base editors in rice. Nat Protoc 16, 431–457 (2021). https://doi.org/10.1038/s41596-020-00423-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-00423-y
This article is cited by
-
Conditional knockdown of OsMLH1 to improve plant prime editing systems without disturbing fertility in rice
Genome Biology (2024)
-
Optimized prime editing in monocot plants using PlantPegDesigner and engineered plant prime editors (ePPEs)
Nature Protocols (2023)
-
Split complementation of base editors to minimize off-target edits
Nature Plants (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.