Abstract
Three highly pathogenic β-coronaviruses have crossed the animal-to-human species barrier in the past two decades: SARS-CoV, MERS-CoV and SARS-CoV-2. To evaluate the possibility of identifying antibodies with broad neutralizing activity, we isolated a monoclonal antibody, termed B6, that cross-reacts with eight β-coronavirus spike glycoproteins, including all five human-infecting β-coronaviruses. B6 broadly neutralizes entry of pseudotyped viruses from lineages A and C, but not from lineage B, and the latter includes SARS-CoV and SARS-CoV-2. Cryo-EM, X-ray crystallography and membrane fusion assays reveal that B6 binds to a conserved cryptic epitope located in the fusion machinery. The data indicate that antibody binding sterically interferes with the spike conformational changes leading to membrane fusion. Our data provide a structural framework explaining B6 cross-reactivity with β-coronaviruses from three lineages, along with a proof of concept for antibody-mediated broad coronavirus neutralization elicited through vaccination. This study unveils an unexpected target for next-generation structure-guided design of a pan-β-coronavirus vaccine.
Similar content being viewed by others
Main
Four coronaviruses mainly associated with common cold-like symptoms are endemic in humans, namely OC43, HKU1, NL63 and 229E, while three highly pathogenic zoonotic coronaviruses have emerged in the past two decades, leading to epidemics and a pandemic. Severe acute respiratory syndrome coronavirus (SARS-CoV) was discovered in Guangdong Province in China in 2002 and spread to five continents through air travel routes, infecting 8,098 people and causing 774 deaths. No cases were reported after 20041,2. In 2012, Middle East respiratory syndrome coronavirus (MERS-CoV) emerged in the Arabian Peninsula, where it still circulates. It was exported to 27 countries, infecting a total of 2,494 individuals and claiming 858 lives as of January 2020 according to the World Health Organization3. A recent study further suggested that undetected zoonotic MERS-CoV transmissions are currently occurring in Africa4. A novel coronavirus, named SARS-CoV-2, was associated with an outbreak of severe pneumonia in Hubei Province, China, at the end of 2019 and has since infected over 121 million people and claimed more than 2.6 million lives worldwide during the ongoing COVID-19 pandemic5,6.
SARS-CoV and SARS-CoV-2 probably originated in bats5,7,8,9,10, with masked palm civets and racoon dogs acting as intermediate amplifying and transmitting hosts for SARS-CoV11,12,13. Although MERS-CoV was also suggested to have originated in bats, repeated zoonotic transmissions occurred from dromedary camels14,15. The identification of numerous coronaviruses in bats, including viruses related to SARS-CoV-2, SARS-CoV and MERS-CoV, along with evidence of spillovers of SARS-CoV-like viruses to humans, strongly indicates that future coronavirus emergence events will continue to occur5,7,8,9,10,16,17,18,19,20.
The coronavirus spike (S) glycoprotein mediates entry into host cells and comprises two functional subunits mediating attachment to host receptors (S1 subunit) and membrane fusion (S2 subunit)21,22,23,24,25,26,27. As the S homotrimer is prominently exposed at the viral surface and is the main target of neutralizing antibodies (Abs), it is a focus of therapeutic and vaccine design efforts28. We previously showed that the SARS-CoV-2 receptor-binding domain (RBD, part of the S1 subunit) is immunodominant, comprises multiple distinct antigenic sites, and is the target of 90% of the neutralizing activity present in COVID-19 convalescent plasma29. Accordingly, monoclonal Abs (mAbs) with potent neutralizing activity have been identified against the SARS-CoV-2, SARS-CoV and MERS-CoV RBDs and shown to protect against viral challenge in vivo29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46. The isolation of S309 from an individual recovered from SARS-CoV, which neutralizes SARS-CoV-2 and SARS-CoV through recognition of a conserved RBD epitope, demonstrated that potent neutralizing mAbs could inhibit β-coronaviruses belonging to different lineage B (sarbecovirus) clades31. An optimized version of S309 has successfully completed phase 3 clinical trials in the United States. Whereas a few other SARS-CoV-2 cross-reactive mAbs have been identified from either SARS-CoV convalescent individuals47,48,49,50 or immunization of transgenic mice42, the vast majority of SARS-CoV-2 S-specific mAbs isolated exhibit narrow binding specificity and neutralization breadth. The SARS-CoV-2 N-terminal domain (NTD) is also the target of potent neutralizing and protective mAbs, driving evolution of this highly variable domain35,51,52,53,54.
Although the COVID-19 pandemic has accelerated the development of SARS-CoV-2 vaccines at an unprecedented pace, worldwide deployment to achieve community protection is expected to take many more months. Based on the available data, it appears unlikely that infection or vaccination will provide durable pan-coronavirus protection due to the immunodominance of the RBD and the waning of Ab responses, leaving the human population vulnerable to the emergence of genetically distinct coronaviruses29,55. The availability of mAbs and other reagents cross-reacting with and broadly neutralizing distantly related coronaviruses is key for pandemic preparedness to enable the detection, prophylaxis and therapy against zoonotic pathogens that might emerge in the future.
We report the isolation of a mAb cross-reacting with the S glycoproteins of at least eight β-coronaviruses from lineages A, B and C, including all five human-infecting β-coronaviruses. This mAb, designated B6, broadly inhibits entry of viral particles pseudotyped with the S glycoprotein of lineage C (MERS-CoV and HKU4) and lineage A (OC43) coronaviruses, providing proof of concept of mAb-mediated broad β-coronavirus neutralization. A cryo-EM structure of MERS-CoV S bound to B6 reveals that the mAb recognizes a linear epitope in the stem helix within a highly dynamic region of the S2 fusion machinery. Crystal structures of B6 in complex with MERS-CoV S, SARS-CoV/SARS-CoV-2 S, OC43 S and HKU4 S stem helix peptides, combined with binding and membrane fusion assays, reveal an unexpected binding mode to a cryptic epitope, delineate the molecular basis of cross-reactivity and rationalize the observed binding affinities for distinct coronaviruses. Collectively, our data indicate that B6 sterically interferes with the S conformational changes leading to membrane fusion, and identify a key target for next-generation structure-guided design of pan-coronavirus vaccines.
Results
Isolation of a broadly neutralizing coronavirus mAb
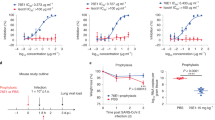
To elicit cross-reactive Abs targeting conserved coronavirus S epitopes, we immunized mice twice with the prefusion-stabilized MERS-CoV S ectodomain trimer and once with the prefusion-stabilized SARS-CoV S ectodomain trimer (Fig. 1a). We subsequently generated hybridomas from immunized animals and implemented a selection strategy to identify those secreting Abs recognizing both MERS-CoV S and SARS-CoV S, but not their respective S1 subunits (which are much less conserved than the S2 subunit21,23), the shared foldon trimerization domain or the His tag. We identified and sequenced a mAb, designated B6, that bound prefusion MERS-CoV S (lineage C) and SARS-CoV S (lineage B) trimers, the two immunogens used, as well as SARS-CoV-2 S (lineage B) and OC43 S (lineage A) trimers with nanomolar to picomolar avidities. Specifically, B6 bound most tightly to MERS-CoV S (Fig. 1b), followed by OC43 S (with one order of magnitude lower apparent affinity, Fig. 1c) and SARS-CoV/SARS-CoV-2 S (with three orders of magnitude reduced apparent affinity, Fig. 1d,e). These results show that B6 is a broadly reactive mAb recognizing at least four distinct S glycoproteins distributed across three lineages of the β-coronavirus genus.
a, Mouse immunization and B6 mAb selection scheme. MERS-CoV and SARS-CoV S1 subunits fused to human Fc and the respiratory syncytial virus fusion glycoprotein (RSV F) ectodomain trimer fused to a foldon and a His tag were used as decoys during selection. b–e, Binding of MERS-CoV S (b), OC43 S (c), SARS-CoV S (d) and SARS-CoV-2 S (e) ectodomain trimers to the B6 mAb immobilized at the surface of biolayer interferometry biosensors. A representative example of two independent experiments is shown. Data were analyzed with the ForteBio software, and global fits are shown as dashed lines. The vertical dotted lines correspond to the transition between the association and dissociation phases. Approximate apparent equilibrium dissociation constants (KD,app) are reported due to the binding avidity resulting from the trimeric nature of S glycoproteins. f–h, B6-mediated neutralization of VSV particles pseudotyped with MERS-CoV S (n = 2 independent experiments) (f), OC43 S (n = 2 independent experiments) (g) and HKU4 S (n = 2 independent experiments) (h). Experiments were performed with two independent mAb and pseudotyped virus preparations. Data are presented as mean values ± s.d. for one representative experiment with five technical replicates for MERS-CoV S (f), one representative experiment with six technical replicates for OC43 (g) and one representative experiment with two technical replicates for HKU4 S (h) and were evaluated using a nonlinear sigmoidal regression model with variable Hill slope. Fits are shown as dashed lines. Source data for f–h are available online.
To evaluate the neutralization potency and breadth of B6, we assessed S-mediated entry into cells of either vesicular stomatitis virus (VSV)56 or murine leukemia virus (MLV)21,57 pseudotyped with MERS-CoV S, OC43 S, SARS-CoV S, SARS-CoV-2 S and HKU4 S in the presence of varying concentrations of mAbs. We determined half-maximal inhibitory concentrations of 1.7 ± 0.9 µg ml−1, 4.0 ± 0.9 µg ml−1 and 2.4 ± 0.9 µg ml−1 for MERS-CoV S, OC43 S and HKU4 S pseudotyped viruses, respectively (Fig. 1f–h) whereas no neutralization was observed for SARS-CoV S and SARS-CoV-2 S (Extended Data Fig. 1). B6 therefore broadly neutralizes S-mediated entry of pseudotyped viruses harboring β-coronavirus S glycoproteins from lineages A and C, but not from lineage B, putatively due to lower-affinity binding (Fig. 1b–e).
B6 targets a linear epitope in the fusion machinery
To identify the epitope recognized by B6, we determined a cryo-EM structure of the MERS-CoV S glycoprotein in complex with the B6 Fab fragment at an overall resolution of 2.5 Å, although the region comprising B6 was highly dynamic and resolved to lower than 12–15 Å (Fig. 2a,b, Extended Data Fig. 2 and Table 1). Three-dimensional (3D) classification of the cryo-EM data revealed incomplete Fab saturation, with one to three B6 Fabs bound to the MERS-CoV S trimer, and a marked conformational dynamic of bound B6 Fabs, yielding a continuum of conformations (Extended Data Fig. 3). Although these two factors compounded the local resolution of the S–B6 interface, we identified that the B6 epitope resides in the stem helix (that is, downstream from the connector domain and before the heptad-repeat 2 region) within the S2 subunit (fusion machinery) (Fig. 2a,b). Our 3D reconstructions further suggest that B6 binding disrupts the stem helix quaternary structure, which is presumed to form a three-helix bundle (observed in the NL63 S58 and SARS-CoV/SARS-CoV-2 S structures21,24,38,59,60,61,62), but not maintained in the B6-bound MERS-CoV S structure (Fig. 2a).
a,b, Molecular surface representation of a composite model of the B6-bound MERS-CoV S cryo-EM structure and of the B6-bound MERS-CoV S stem helix peptide crystal structure shown from the side (a) and viewed from the viral membrane (b). MERS-CoV S protomers are colored pink, cyan and gold, and the B6 Fab heavy and light chains are colored purple and magenta, respectively. The composite model was generated by docking the crystal structure of B6 bound to the MERS-CoV S stem helix into the cryo-EM map. c, Identification of a conserved 15-residue sequence spanning the stem helix. Residue numberings for MERS-CoV S and SARS-CoV-2 S are indicated on the top and bottom of the alignment, respectively. Sequence alignment was performed using Multalin103 and visualized using ESPript3.0 (ref. 104). d,e, Binding of 0.1 µM B6 mAb (d) or 1 µM B6 Fab (e) to biotinylated coronavirus S stem helix peptides immobilized at the surface of biolayer interferometry biosensors. Control: no peptide immobilized. One representative example of two independent experiments with three technical replicates is shown.
Based on our cryo-EM structure, we identified a conserved 15-residue sequence at the C terminus of the last residue resolved in previously reported MERS-CoV S structures38,60,63,64 and confirmed by biolayer interferometry that it encompasses the B6 epitope using synthetic MERS-CoV S biotinylated peptides (Fig. 2c–e and Extended Data Fig. 4). We further found that B6 bound to the corresponding stem helix peptides from all known human-infecting β-coronaviruses: SARS-CoV-2 and SARS-CoV, where the sequence is strictly conserved among the two viruses, OC43 and HKU1, as well as mouse hepatitis virus (MHV) and two MERS-CoV-related bat viruses (HKU4 and HKU5) in mAb and Fab formats (Fig. 2d,e). B6 interacted most efficiently with the MERS-CoV S peptide, likely due to its major role in elicitation of this mAb, followed by all other coronavirus peptides tested, which bound with comparable affinities, except for HKU1 which interacted more weakly than other stem helix peptides.
B6 recognizes a conserved epitope in the stem helix
To obtain an atomic-level understanding of the broad B6 cross-reactivity, we determined four crystal structures of the B6 Fab in complex with peptide epitopes derived from MERS-CoV S (residues 1230–1244), SARS-CoV S (residues 1129–1143)/SARS-CoV-2 S (residues 1147–1161), OC43 S (residues 1232–1246) and HKU4 S (residues 1231–1245), at resolutions ranging from 1.4 to 1.8 Å (Fig. 3a–f, Extended Data Fig. 5 and Table 2). In all four structures, the stem helix epitope folds as an amphipathic ɑ-helix resolved for residues 1230–1240 (MERS-CoV S numbering), irrespective of the peptide length used for co-crystallization. B6 interacts with the helical epitope through shape complementarity, hydrogen bonding and salt bridges using complementarity determining regions CDRH1-H3, framework region 3, CDRL1 and CDRL3 to bury ~600 Å2 at the paratope–epitope interface. The stem helix docks its hydrophobic face, lined by residues F1231MERS-CoV, L1235MERS-CoV, F1238MERS-CoV and F1239MERS-CoV, into a hydrophobic groove formed by B6 heavy chain residues Y35, W49, V52 and L61, as well as light chain Y103 (Figs. 2c and 3a,b,d). Moreover, B6 binding leads to the formation of a salt bridge triad, involving residue D1236MERS-CoV, CDRH3 residue R104 and CDRL1 residue H33.
a, Crystal structure of the B6 Fab (surface rendering) in complex with the MERS-CoV S stem helix peptide. b,c, Crystal structures of the B6 Fab bound to the MERS-CoV S (b) or HKU4 S (c) stem helix show a conserved network of interactions, except for the substitution of D1236MERS-CoV with E1237HKU4, which preserves the salt bridge triad formed with CDRH3 (H3) residue R104 and CDRL1 (L1) residue H33. d–f, Crystal structures of the B6 Fab bound to the MERS-CoV S (d), OC43 S (e) or SARS-CoV-2 S (f) stem helix, showcasing the conservation of the paratope–epitope interface except for the conservative substitution of F1238MERS-CoV with W1240OC43 or Y1137SARS-CoV/Y1155SARS-CoV-2. The B6 heavy and light chains are colored purple and magenta, respectively, and only selected regions are shown in b–f for clarity. The coronavirus S stem helix peptides are rendered in ribbon representation and colored gold, with interacting side chains shown in stick representation.
Comparison of the B6-bound structures of MERS-CoV, HKU4, SARS-CoV/SARS-CoV-2 and OC43 S stem helix peptides explains the broad mAb cross-reactivity with β-coronavirus S glycoproteins, as shape complementarity is maintained through strict conservation of three out of four hydrophobic residues, whereas F1238MERS-CoV is conservatively substituted with Y1137SARS-CoV/Y1155SARS-CoV-2 or W1240 OC43/W1237HKU1 (our structures demonstrate that all three aromatic side chains are accommodated by B6). Furthermore, the D1236MERS-CoV-mediated salt bridge triad is preserved, including with a non-optimal E1237HKU4 side chain, with the exception of S1235HKU1, which abrogates these interactions and explains the dampened B6 binding to the HKU1 peptide (Figs. 2c–e and 3b–f). B6 heavy chain residue L61 and CDRL1 residue H33 are mutated from the germline and make major contributions to epitope recognition, highlighting the key contribution of affinity maturation to the cross-reactivity of this mAb.
Mechanism of B6-mediated neutralization
We set out to elucidate the molecular basis of the B6-mediated broad neutralization of multiple coronaviruses from lineages A and C and lack of inhibition of lineage B coronaviruses. Our biolayer interferometry data indicate that, although the B6 mAb interacted efficiently with the stem helix peptide of all but one of the coronaviruses evaluated (HKU1, Fig. 2d,e), the SARS-CoV-2 S and SARS-CoV S ectodomain trimers bound to B6 with three orders of magnitude reduced avidities compared to MERS-CoV S (Fig. 1b–e). Although the B6 epitope is not resolved in any prefusion coronavirus S structures determined so far, the stem helix region directly upstream is resolved to a much greater extent for SARS-CoV-2 S and SARS-CoV S, indicating a rigid structure21,38,59,60,61,62 compared to MERS-CoV S38,60,63,64, OC43 S65, HKU1 S25 or MHV S23 (Fig. 4a–c). Furthermore, we determined B6 Fab binding affinities of 0.3 µM and 1.5 µM for MERS-CoV S and OC43 S, respectively, whereas SARS-CoV S recognition was too weak to quantitate accurately (Extended Data Fig. 6). These findings, along with the largely hydrophobic nature of the B6 epitope, which is expected to be occluded in the center of a three-helix bundle (Fig. 4c; as is the case for the region directly N-terminal to it), suggest that B6 recognizes a cryptic epitope and that binding to S trimers is modulated (at least in part) by the quaternary structure of the stem. The reduced conformational dynamics of the SARS-CoV-2 S and SARS-CoV S stem helix quaternary structure is expected to limit B6 accessibility to its cryptic epitope relative to other coronavirus S glycoproteins. This hypothesis is supported by the correlation between neutralization potency and binding affinity observed here for B6, which probably explains the lack of neutralization of lineage B β-coronaviruses.
a, Cryo-EM map of prefusion SARS-CoV-2 S (EMD-21452) filtered at a resolution of 6 Å to emphasize the intact trimeric stem helix bundle. b, Cryo-EM map of the MERS-CoV S–B6 complex showing a disrupted stem helix bundle. c, Model of B6-induced S stem movement obtained through comparison of the apo SARS-CoV-2 S (EMD-21452)21 and B6-bound MERS-CoV S structures. d, MERS-CoV S binding to DPP4 in the presence of mAb B6 analyzed by ELISA. Data are presented as mean values ± s.d. (n = 3 technical replicates). e, Inhibition of MERS-CoV S-mediated cell–cell fusion in the presence of mAb B6. ΔS corresponds to cells not transfected with MERS-CoV S. AMMO1 is an Epstein–Barr virus gH/gL mAb used as a negative control105. ****Significant difference in the level of fusion (P < 0.001, unpaired t-test). NS, not significant. The data are from two independent experiments with eight technical replicates each for ΔS and the no mAb control and four and eight technical replicates for mAb B6 and AMMO1, respectively. Each dot represents data from one well. f–h, Proposed mechanism of inhibition mediated by the B6 mAb. B6 binds to the hydrophobic core (red) of the stem helix bundle (f) and disrupts its quaternary structure (g). B6 binding probably prevents S2 subunit refolding from the pre- to the postfusion state and blocks viral entry (h). Source data for d and e are available online.
B6 did not interfere with the binding of MERS-CoV S to immobilized DPP4 by ELISA, indicating that this antibody does not inhibit receptor recognition (Fig. 4d). However, B6 inhibited fusion of MERS-CoV S-expressing HEK293E cells with DPP4-expressing HEK293T cells, which suggests that the main mechanism of neutralization is to block membrane fusion (Fig. 4e). Analysis of the postfusion mouse hepatitis S22, SARS-CoV-2 S66 and SARS-CoV S67 structures show that the B6 epitope is buried at the interface with the other two protomers of the rod-shaped trimer. As a result, B6 binding appears to be incompatible with adoption of the postfusion S conformation (Fig. 4f–h), in agreement with the observed inhibition of membrane fusion. Collectively, the data presented here suggest that B6 binding sterically interferes with S fusogenic conformational changes and blocks viral entry through inhibition of membrane fusion, as proposed for fusion machinery-directed mAbs against influenza virus68, ebolavirus69 or HIV70.
Discussion
The high sequence variability of viral glycoproteins was long considered as an unsurmountable obstacle to the development of mAb therapies or vaccines conferring broad protection71. The identification of broadly neutralizing mAbs targeting conserved HIV-1 envelope epitopes from infected individuals brought about a paradigm shift for this virus undergoing extreme antigenic drift70,72,73,74,75,76,77. Heterotypic influenza virus neutralization was also described for human cross-reactive mAbs recognizing the hemagglutinin receptor-binding site or the fusion machinery68,78,79,80,81,82. These findings were paralleled by efforts to identify broadly neutralizing Abs against respiroviruses83, henipaviruses84,85,86, Dengue and Zika viruses87,88,89 or ebolaviruses69,90,91,92.
The genetic diversity of coronaviruses circulating in chiropteran and avian reservoirs, along with the recent emergence of multiple highly pathogenic coronaviruses, showcase the need for vaccines and therapeutics that protect humans against a broad range of viruses. As the S2 fusion machinery contains several important antigenic sites and is more conserved than the S1 subunit, it is an attractive target for broad coronavirus neutralization23,28. Previous studies have described conserved epitopes targeted by neutralizing Abs, such as the fusion peptide or heptad-repeats, as well as a variable loop in the MERS-CoV S connector domain23,50,64,93,94,95,96,97. The discovery of the B6 mAb provides proof of concept of mAb-mediated broad β-coronavirus neutralization and uncovers a previously unknown conserved cryptic epitope that is predicted to be located in the hydrophobic core of the stem helix. B6 cross-reacts with at least eight distinct S glycoproteins from β-coronaviruses belonging to lineages A, B and C, and broadly neutralizes one bat and two human pseudotyped viruses from lineages A and C. B6 could be used for detection or diagnosis of coronavirus infection, and humanized versions of this mAb would represent promising candidate therapeutics against emerging and re-emerging β-coronaviruses from lineages A and C. Our data further suggest that affinity maturation of B6 using SARS-CoV-2 S and SARS-CoV S might enhance recognition of and extend neutralization breadth towards β-coronaviruses from lineage B. Finally, the identification of the conserved B6 epitope paves the way for epitope-focused vaccine design98,99,100 that could elicit pan-β-coronavirus immunity, as supported by the elicitation of the B6 mAb through vaccination and the recent findings that humans and camels infected with MERS-CoV, humans infected with SARS-CoV-2 and humanized mice immunized with a cocktail of coronavirus S glycoproteins produce antibodies targeting an epitope similar to the one targeted by B6101,102.
Methods
Identification of the B6 broadly neutralizing mAb
Four 10-week-old female CD-1 mice (Charles River Labs) were injected twice with 50 µg of MERS-CoV S formulated with Adjuplex at weeks 0 and 2 and once with 50 µg of SARS-CoV S formulated with Adjuplex at week 8, at the Fred Hutchinson Cancer Research Center Antibody Technology Resource (AAALAC accredited). Work performed for this study was in accordance with the Guide for the Care and Use of Laboratory Animals, eighth edition, published by the National Research Council of the National Academies. The mouse husbandry and experiments were approved and supervised by Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee. All aspects of mouse handling and euthanasia were carried out in accordance with the American Veterinary Medical Association (AVMA) guidelines. Three days after the final injection, splenocytes were isolated from high-titer mice and electrofused with the P3X63-Ag8 myeloma cell line (BTX, Harvard Apparatus). Hybridoma supernatants were tested for binding to prefusion SARS-CoV S, MERS-CoV S, SARS-CoV S1 subunit, MERS-CoV S1 subunit and respiratory syncytial virus F (which harbors a foldon motif and a His tag similar to the SARS-CoV S and MERS-CoV S ectodomain trimer constructs) using a high-throughput bead-based binding array. Hybridomas from wells containing supernatants that were positive for binding to prefusion SARS-CoV S and MERS-CoV S, but negative for SARS-CoV S1, MERS-CoV S1 and respiratory syncytial virus F, were subcloned by limiting dilution and rescreened for binding as above. The B6 VH and VL sequences were recovered using the mouse immunoglobulin (Ig) primer set (Millipore) using the protocol outlined in ref. 106, then Sanger sequenced (Genewiz). The VH/VL sequences were codon-optimized and cloned into full-length pTT3-derived immunoglobulin (Ig)-G1 and Ig-L kappa expression vectors containing human constant regions using Gibson assembly105.
Cell lines
HEK293F cells were obtained from Thermo Fisher Scientific (R79007), HEK293T cells (CRL-11268) and HRT-18 cells (CRL-11663) from ATCC, Huh7 cells from the National Institutes of Biomedical Innovation, Health and Nutrition (JCRB0403) and HEK293-6E cells from the National Research Council of Canada (RRID:CVCL_HF20). HEK293T-ACE2 cells were provided by J. Bloom107 and HEK293T-T7 cells were provided by R. Longnecker108. Cell lines were not tested for mycoplasma contamination.
Protein expression and purification
MERS-CoV 2 proline (2P) S, OC43 S, SARS-CoV 2P S and SARS-CoV-2 2P S were produced as previously described21,38,65. Briefly, all ectodomains were produced in HEK293F cells (Thermo Fisher Scientific, R79007) grown in suspension using FreeStyle 293 expression medium (Life Technologies) at 37 °C in a humidified 8% (vol/vol) CO2 incubator rotating at 130 r.p.m. The cultures were transfected using 293fectin (Thermo Fisher Scientific) with cells grown to a density of 106 cells per ml and cultivated for three days. The supernatants were collected and cells resuspended for another three days, yielding two harvests. For MERS-CoV 2P S, SARS-CoV 2P S and SARS-CoV-2 2P S, clarified supernatants were purified using a 5-ml Cobalt affinity column (Takara). HCoV-OC43 S was purified using a StrepTrap HP column (GE Healthcare). Purified proteins were concentrated, flash-frozen in Tris-saline (50 mM Tris, pH 8.0 (25 °C), 150 mM NaCl) and stored at −80 °C. The MERS-CoV S1-Fc and SARS-CoV S1-Fc have been described previously109 and were produced as before for the prefusion S trimers and purified using protein A affinity chromatography.
For mAb B6 production, 250 µg of B6 heavy chain and 250 µg of B6 light chain encoding plasmids were co-transfected per liter of suspended HEK293F culture using 293-Free transfection reagent (Millipore Sigma) according to the manufacturer’s instructions. Cells were transfected at a density of 106 cells per ml. Expression was carried out for six days, after which cells and cellular debris were removed by centrifugation at 4,000g followed by filtration through a 0.22-µm filter. Clarified cell supernatant containing recombinant mAbs was passed over Protein A agarose resin (Thermo Fisher Scientific). The Protein A resin was extensively washed with 25 mM phosphate pH 7.4, 150 mM NaCl (PBS) and eluted with IgG elution buffer (Thermo Fisher Scientific). Purified B6 was extensively dialyzed against PBS, concentrated, flash-frozen and stored at −80 °C.
DS-Cav1-foldon-SpyTag110 was produced by lentiviral transduction of HEK293F cells using the Daedalus system111. Lentivirus was produced by transient transfection of HEK293T cells (ATCC CRL-11268) using linear 25-kDa polyethyleneimine (PEI; Polysciences). Briefly, 4 × 106 cells were plated onto 10-cm tissue culture plates. After 24 h, 3 mg of psPAX2, 1.5 mg of pMD2G (Addgene plasmids 12260 and 12259, respectively) and 6 mg of lentiviral vector plasmid were mixed in 500 ml of diluent (5 mM HEPES, 150 mM NaCl, pH 7.5) and 42 ml of PEI (1 mg ml−1) and incubated for 15 min. The DNA–PEI complex was then added to the plate, dropwise. Lentivirus was collected 48 h post-transfection and concentrated 100× by centrifugation at 8,000g for 18 h. Transduction of the target cell line was carried out in 125-ml shake flasks containing 10 × 106 cells in 10 ml of growth medium, then 100 μl of 100× lentivirus was added to the flask and the cells were incubated with 225 r.p.m. oscillation at 37 °C in 8% CO2 for 4–6 h, after which 20 ml of growth medium was added to the shake flask. Transduced cells were expanded every other day to a density of 1 × 106 cells per ml until a final culture size of 4 l was reached. The medium was collected after 17 days of total incubation after measuring the final cell concentration (~5 × 106 cells per ml) and viability (~90% viable). Culture supernatant was collected by low-speed centrifugation to remove cells from the supernatant. NaCl and NaN3 were added to final concentrations of 250 mM and 0.02%, respectively. The supernatant was loaded over one 5-ml HisTrap FF Crude column (GE Healthcare) at 5 ml min−1 by an ÄKTA Pure system (GE Healthcare). The 5-ml HisTrap column was washed with 10 column volumes of wash buffer (2× Gibco 14200-075 PBS, 5 mM imidazole, pH 7.5) followed by six column volumes of elution buffer (2× Gibco 14200-075 PBS, 150 mM imidazole, pH 7.5). The nickel elution was applied to a HiLoad 16/600 Superdex 200 prep-grade column (GE Healthcare) and run in dPBS (Gibco 14190-144) with 5% glycerol (Thermo BP229-1) to further purify the target protein by size-exclusion chromatography. The purified protein was snap-frozen in liquid nitrogen and stored at −80 °C.
As previously described109, for human DPP4, HEK293F cells were grown in suspension using FreeStyle 293 expression medium (Life Technologies) at 37 °C in a humidified 5% CO2 incubator on a Celltron shaker (Infors HT) rotating at 130 r.p.m. (for 1-l culture flasks). The cell density was adjusted to 1.5 × 106 cells per ml, 24 h before transfection, and grown overnight to reach ~2.5 × 106 cells per ml before transfection. The cells were centrifuged at 1,250 r.p.m. for 5 min and resuspended in fresh medium to the same density (2.5 × 106 cells per ml). Suspension cells (200 ml) were transfected with 400 μg of pCD5-hDPP4-Fc vector109. The cultures were swirled for 5 min on a shaker in the culture incubator before adding 9 μg ml−1 of linear 25-kDa PEI solution (Polysciences). At 24 h after transfection, cells were diluted 1:1 in medium and cultivated for six days. Clarified cell supernatant was concentrated 10-fold using a Vivaflow system (Sartorius, 10-kDa cutoff). DDP4 was purified by affinity purification using a Protein A column (GE Life Sciences) followed by size-exclusion chromatography using a Superdex 200 10/300 GL column (GE Life Sciences) equilibrated in 20 mM Tris-HCl, pH 7.5 and 100 mM NaCl. Trypsin was used to remove the Fc tag in a reaction mixture of 7 mg of recombinant DPP4-Fc and 5 μg trypsin in 100 mM Tris-HCl, pH 8.0 and 20 mM CaCl2, incubated for 16 h at 25 °C. The mixture was re-loaded on a Protein A column to remove uncleaved protein and the Fc tag and DPP4 were further purified by size-exclusion chromatography using a Superdex 75 10/300 GL column (GE Life Sciences) equilibrated in 20 mM Tris-HCl, pH 7.5, and 100 mM NaCl. The purified protein was concentrated to ~10 mg ml−1 and stored at −80 °C.
Kinetics of B6 mAb binding to coronavirus S proteins
The avidities of complex formation between B6 mAbs and selected coronavirus S proteins were determined in PBS supplemented with 0.005% Tween20 and 0.1% BSA (PBSTB) at 30 °C and 1,000 r.p.m. shaking on an Octet RED instrument (ForteBio). Curve fitting was performed using a 1:1 binding model and the ForteBio data analysis software. KD ranges were determined with a global fit. AHC biosensors (ForteBio) were hydrated in water and subsequently equilibrated in PBSTB buffer. B6 mAbs (10 μg ml−1) were loaded to the biosensors to a shift of ~1 nm. The system was then equilibrated in PBSTB buffer for 300 s before immersing the sensors in the respective coronavirus S protein (0–218 nM) for up to 600 s, before dissociation in buffer for an additional 600 s.
Binding of B6 to different synthetic coronavirus S stem peptides
Analysis of B6 binding to selected biotinylated coronavirus S stem helix peptides was performed in PBS supplemented with 0.005% Tween20 (PBST) at 30 °C and 1,000 r.p.m. shaking on an Octet RED instrument (ForteBio).
Biotinylated stem peptide (1 µg ml−1; 15- or 16-residue-long stem peptide-PEG6-Lys-biotin synthesized by GenScript) was loaded on SA biosensors to a threshold of 0.5 nm. The system was then equilibrated in PBST for 300 s before immersing the sensors in 0.1 µM B6 mAbs or 1 µM B6 Fabs, respectively, for 300 s prior to dissociation in buffer for 300 s.
Kinetics of B6 Fab binding to different coronavirus S proteins
The rate constants of binding (kon) and dissociation (koff) for the complex between the B6 Fabs and selected coronavirus S proteins were measured in PBST at 30 °C and 1,000 r.p.m. shaking on an Octet RED instrument (ForteBio). Global curve fitting was performed using a 1:1 binding model and the ForteBio data analysis software. For MERS-CoV S and SARS-CoV S, HIS1K or Ni-NTA biosensors (ForteBio) were hydrated in water and subsequently equilibrated in PBST buffer, then 20 μg ml−1 SARS-CoV S or 10 μg ml−1 MERS-CoV S, respectively, were loaded to the biosensors for up to 1,800 s (shift of 1–4 nm). The system was equilibrated in PBST for 300 s before immersing the sensors in B6 Fabs (0–16 µM) for up to 1,800 s prior to dissociation in buffer for 1,800 s. For OC43 S, ARG2 biosensors were hydrated in water then activated for 300 s with an NHS-EDC solution (ForteBio) before amine coupling, then 20 μg ml−1 OC43 was amine-coupled to AR2G (ForteBio) sensors in 10 mM acetate pH 6.0 (ForteBio) for 300 s and then quenched with 1 M ethanolamine (ForteBio) for 300 s. The system was equilibrated in PBST for 300 s before immersing the sensors in B6 Fabs (0–4 µM) for 75 s prior to dissociation in buffer for 75 s.
Pseudovirus entry assays
Production of OC43 S pseudotyped VSV virus and the neutralization assay were performed as described previously65,112. Briefly, HEK293T cells at ~70–80% confluency were transfected with the pCAGGS expression vectors encoding full-length OC43 S with a truncation of the 17 C-terminal residues (to increase cell surface expression levels) along with fusion to a flag tag and the Fc-tagged bovine coronavirus hemagglutinin esterase protein at molar ratios of 8:1. At 48 h after transfection, cells were transduced with VSV∆G/Fluc (bearing the Photinus pyralis firefly luciferase)56 at a multiplicity of infection of 1. After 24 h, the supernatant was collected and filtered through a 0.45-μm membrane. Pseudotyped VSV virus was titrated on a monolayer of HRT-18 (ATCC, CRL-11663) cells. In the virus neutralization assay, serially diluted mAbs were pre-incubated with an equal volume of virus at room temperature for 1 h, then the samples were transfered to the HRT-18 cells, and further incubated at 37 °C. After 20 h, cells were washed once with PBS, lysed with cell lysis buffer (Promega), and firefly luciferase expression was measured on a Berthold Centro LB 960 plate luminometer using d-luciferin as a substrate (Promega). The percentage of infectivity was calculated as the ratio of luciferase readout in the presence of mAbs normalized to luciferase readout in the absence of mAbs, and the half-maximal inhibitory concentrations (IC50) were determined using four-parameter logistic regression (GraphPad Prism v8.0).
MERS-CoV S, HKU4 S, SARS-CoV S and SARS-CoV-2 S pseudotyped VSV viruses were prepared using HEK293T cells seeded in 10-cm dishes in DMEM supplemented with 10% FBS, 1% PenStrep and transfected with plasmids encoding for the corresponding S glycoprotein (24 μg per dish) using Lipofectamine 2000 (Life Technologies), according to the manufacturer’s instructions. One day post-transfection, cells were infected with VSV(G*ΔG-luciferase). After 2 h, infected cells were washed four times with DMEM before medium supplemented with anti-VSV-G antibody was added (I1-mouse hybridoma supernatant diluted 1:50; ATCC CRL-2700). Particles were collected 18 h post-inoculation, clarified from cellular debris by centrifugation at 2,000g for 5 min, and concentrated 10 times using a 30-kDa cutoff membrane. HKU4 S pseudotyped VSV was additionally primed with 20 µg ml−1 trypsin for 15 min at 37 °C, and the reaction was stopped using 100 µg ml−1 soybean trypsin inhibitor.
MERS-CoV S, SARS-CoV S and SARS-CoV-2 S MLV pseudotypes were prepared as previously described21.
For viral neutralization, Huh7 cells (National Institutes of Biomedical Innovation, Health and Nutrition JCRB0403; for MERS-CoV S pseudotyped virus), stable 293T cells expressing ACE2107 (for SARS-CoV and SARS-CoV-2 pseudotyped virus) or 293T cells transiently transfected with Tylonycteris bat DPP4 (GenBank AZO92860.1; pcDNA3.1_tDPP4), in DMEM supplemented with 10% FBS, 1% PenStrep, were seeded at 40,000 cells per well into clear-bottom white-walled 96-well plates and cultured overnight at 37 °C. Two- or threefold serial dilutions of B6 mAbs were prepared in DMEM and pseudotyped viruses were added 1:1 to each B6 dilution (for VSV viruses in the presence of anti-VSV-G antibody from I1-mouse hybridoma supernatant diluted 50 times). After a 45-min incubation at 37 °C, 40 µl of the mixture was added to the cells and, 2 h post-infection, 40 μl of DMEM was added to the cells. After 18 h (for VSV viruses) or 72 h (for MLV viruses) 50 μl per well of One-Glo-EX substrate (Promega) was added to the cells and incubated in the dark for 5–10 min before reading on a Varioskan Lux plate reader (Thermo Fisher).
Cryo-electron microscopy sample preparation and data collection
Lacey carbon copper grids (400 mesh) were coated with a thin layer of continuous carbon using a carbon evaporator, then 1 mg ml−1 MERS-CoV S was incubated with 100 mM neuraminic acid (to promote the closed trimer conformation), 150 mM Tris pH 8 (25 °C) and 150 mM NaCl for 16 h at 4 °C. A twofold molar excess of B6 Fabs over MERS-CoV S protomer was then added to the solution and incubated for 1 h at 37 °C. The sample was diluted to 0.2 mg ml−1 S protein with 100 mM neuraminic acid, 150 mM Tris pH 8 (25 °C) and 150 mM NaCl, before 3 µl of sample was applied onto a freshly glow-discharged grid. Plunge freezing was performed using a TFS Vitrobot Mark IV system (blot force, −1; blot time, 2.5 s; humidity, 100%; temperature, 25 °C). Data were acquired using an FEI Titan Krios transmission electron microscope operated at 300 kV and equipped with a Gatan K2 Summit direct detector and Gatan Quantum GIF energy filter, operated in zero-loss mode with a slit width of 20 eV. Automated data collection was carried out using Leginon113 at a nominal magnification of ×130,000 with a pixel size of 0.525 Å. The dose rate was adjusted to 8 counts pixel−1 s−1, and each video was acquired in super-resolution mode fractionated in 50 frames of 200 ms. A total of 2,180 micrographs were collected in a single session with a defocus range between −0.5 and −3.0 μm.
Cryo-electron microscopy data processing
Video frame alignment, estimation of the microscope contrast-transfer function parameters, particle picking and extraction were carried out using Warp114. Particle images were extracted with a box size of 800 pixels2 binned to 400 pixels2, yielding a pixel size of 1.05 Å. Two rounds of reference-free 2D classification were performed using Relion3.0115 to select well-defined particle images. Subsequently, two rounds of 3D classification with 50 iterations each (angular sampling of 7.5° for 25 iterations and 1.8° with local search for 25 iterations), using the previously reported closed MERS-CoV S structure without the G4 Fab (PDB 5W9J) as initial model, were carried out using Relion, without imposing symmetry.
For the high-resolution map, particle images were subjected to Bayesian polishing116 before performing non-uniform refinement, defocus refinement and non-uniform refinement again in cryoSPARC117. Finally, two rounds of global contrast transfer function (CTF) refinement of beam-tilt, trefoil and tetrafoil parameters were performed before a final round of non-uniform refinement to produce the final map with a resolution of 2.5 Å.
For the lower-resolution map, one additional round of focused classification in Relion, with 50 iterations, using a broad mask covering the region of interest (B6/stem), was carried out to further separate distinct B6 Fab conformations. 3D refinements of the best subclasses were carried out using homogeneous refinement in cryoSPARC117. Reported resolutions are based on the gold-standard Fourier shell correlation (FSC) of 0.143 criterion and Fourier shell correlation curves were corrected for the effects of soft masking by high-resolution noise substitution118.
Cryo-electron microscopy model building and analysis
UCSF Chimera119 and Coot120 were used to fit atomic models into the cryo-EM maps. The MERS-CoV S EM structure in complex with 5-N-acetyl neuraminic acid (PDB 6Q04, residues 18–1224) and the B6–MERS-CoV15 (residues 1230–1240) crystal structure were fit into the cryo-EM map. Subsequently, the linker connecting the stem helix to the rest of the MERS-CoV S ectodomain (residues 1225–1229) was manually built using Coot. N-linked glycans were hand-built into the density where visible, and the models were refined and relaxed using Rosetta using both sharpened and unsharpened maps121,122. Models were analyzed using MolProbity123, EMringer124, Phenix125 and privateer126 to validate the stereochemistry of both the protein and glycan components. Figures were generated using UCSF Chimera.
Crystallization and structure determination
The stem peptides were dissolved in DMSO to a concentration of 20 mM. All crystallization experiments were performed at 23 °C in hanging drop vapor diffusion experiments with initial concentrations of 20 mg ml−1 B6 Fabs and 1.5-fold molar excess of peptide. Crystal trays were set up with a mosquito device using 100 nl of mother liquor solution and 100 or 150 nl of B6–peptide complex solution, respectively. Crystals of B6–OC4315 appeared after several weeks in 0.2 M potassium thiocyanate and 20% (wt/vol) PEG3350, B6–MERS-CoV15 in 0.2 M MgCl2 and 20% (wt/vol) PEG3350, B6–HKU415 in 0.6 M NaCl, 0.1 M MES-NaOH pH 6.5 and 20% (wt/vol) PEG 4000, and B6–SARS-CoV/SARS-CoV-216 in 0.2 M KCl and 20% (wt/vol) PEG3350. Crystals were cryoprotected by addition of glycerol to a final concentration of 25% (vol/vol) and flash-cooled in liquid nitrogen. Diffraction data were collected at Beamline 5.0.1 of the Advanced Light Source. All data were integrated, indexed and scaled using XDS127 and Aimless128. The structures were solved by molecular replacement using Phaser129 and the S230 Fab (PDB 6NB8) or B6 Fab without ligand as a search model. Model building was performed with Coot120 and structure refinement with Buster130 and Phenix125. The ligand was built in density only after the Fab was fully refined to avoid any bias. Validation was performed with MolProbity123 and Phenix125.
Enzyme-linked immunosorbent assay
A 20 µl volume of human DDP4 ectodomain was coated on 384-well ELISA plates at 3 ng µl−1 for 16 h at 4 °C, then 10 µg ml−1 MERS-CoV S ectodomain was pre-incubated with 0.01–200 µg ml−1 mAb B6 at room temperature for 16 h. The plates were washed with a 405 TS microplate washer (BioTek Instruments) then blocked with 80 µl of SuperBlock (PBS) blocking buffer (Thermo Scientific) for 1 h at 37 °C. The plates were washed and 25 µl of premix MERS-CoV S–mAb complex was added to the plates and incubated for 1 h at 37 °C. The plates were washed and then incubated with 25 µl of 1:1,000 diluted mouse anti-His tag IgG-HRP (R&D Systems #MAB050H). The plates were washed and then 25 µl of Substrate TMB Microwell peroxidase (Seracare 5120-0083) was added for 5 min at room temperature. The colorimetric reaction was stopped by addition of 25 µl of 1 M HCl. Absorption at 450 nm was read on a Varioskan Lux plate reader (Thermo Scientific). The data were normalized using the signal of 10 µg ml−1 MERS-CoV S ectodomain without mAb B6 and plotted using Prism v8.0.
Virus-free syncytia assay
pTT3-MERS-CoV S plasmid codon-optimized cDNA corresponding to the full-length MERS-CoV S protein (GenBank AHI48572.1) was synthesized by Twist Biosciences and cloned into the pTT3 expression vector.
HEK293T-T7 cells were seeded onto six-well plates at a density of 3 × 105 cells per well and 293E cells were split into 12.5 ml at 0.5 × 106 cells per ml in 125-ml flasks. After 24 h, the 293T-T7 cells were transfected with 2 µg dpp4 DNA using Genjuice transfection reagent (Millipore Sigma, cat. no. 70967). The 293E cells were co-transfected with 3.125 µg of pT7EMCLuc, which encodes the luciferase gene under the control of the T7 promoter, and 3.125 µg of the pTT3-MERS-CoV S, or co-transfected with 3.125 µg of pT7EMCLuc and 3.125 µg of empty pTT3 using 293-Free transfection reagent (Millipore Sigma, cat. no. 72181).
After 24 h, 293E cells co-transfected with pT7EMCLuc/MERS-CoV S were diluted to 6 × 105 cells per ml in FreeStyle 293 medium (Thermo Fisher, cat. no. 12338018) and 25 µl per well was added to a 96-well plate (Greiner, cat. no. 655090). Immediately afterward, 25 µl per well of medium containing 40 µg ml−1 of B6 or AMMO1 were added to quadruplicate wells. Medium containing no mAbs was added to eight wells as a control.
As a control for non-specific fusion, 293E cells co-transfected with pT7EMCLuc/pTT3 (ΔS) cells were diluted to 6 × 105 cells per ml in FreeStyle 293 medium and 25 µl per well was added to a 96-well plate. The transfected 293T-T7 cells were dissociated from culture plates using Accutase (Biolegend, cat. no. 423201), washed twice with cDMEM, resuspended at a density of 3 × 105 cells per ml in cDMEM, and 50 µl was added to every well. After 24 h, the medium was aspirated, and the cells were lysed in 100 µl of Steady-Glo luciferase reagent. The luciferase activity was read on a SpectraMax i3x fluorimeter. Fusion was determined by the relative luciferase activity in the presence of AMMO1 or B6 relative to the average luciferase activity measured in 293E cells expressing pT7EMCLuc+ MERS-CoV S and 293 T7 cells, which was set to 100%.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The atomic coordinates and cryo-EM maps have been deposited to the Protein Data Bank and Electron Microscopy Data Bank with the following accession nos.: B6–MERS-CoV S, PDB 7M5E, EMD-23674 and EMD-23672; B6–OC43, PDB 7M51; B6–HKU4, PDB 7M52; B6–SARS-CoV-2, PDB 7M53; B6–MERS-CoV, PDB 7M55. Source data are provided with this paper.
References
Drosten, C. et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967–1976 (2003).
Ksiazek, T. G. et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1953–1966 (2003).
Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D. & Fouchier, R. A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012).
Mok, C. K. P. et al. T-cell responses to MERS coronavirus infection in people with occupational exposure to dromedary camels in Nigeria: an observational cohort study. Lancet Infect. Dis. 21, 385–395 (2020).
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Zhu, N. et al. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 382, 727–733 (2020).
Ge, X. Y. et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503, 535–538 (2013).
Yang, X. L. et al. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J. Virol. 90, 3253–3256 (2015).
Li, W. et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 310, 676–679 (2005).
Hu, B. et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 13, e1006698 (2017).
Wang, M. et al. SARS-CoV infection in a restaurant from palm civet. Emerg. Infect. Dis. 11, 1860–1865 (2005).
Guan, Y. et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302, 276–278 (2003).
Kan, B. et al. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 79, 11892–11900 (2005).
Haagmans, B. L. et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 14, 140–145 (2014).
Memish, Z. A. et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 19, 1819–1823 (2013).
Menachery, V. D. et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 21, 1508–1513 (2015).
Menachery, V. D. et al. SARS-like WIV1-CoV poised for human emergence. Proc. Natl Acad. Sci. USA 113, 3048–3053 (2016).
Anthony, S. J. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. Mbio 8, 2 (2017).
Li, H. et al. Human–animal interactions and bat coronavirus spillover potential among rural residents in Southern China. Biosaf. Health 1, 84–90 (2019).
Wang, N. et al. Serological evidence of bat SARS-related coronavirus infection in humans, China. Virol. Sin. 33, 104–107 (2018).
Walls, A. C. et al. Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292 (2020).
Walls, A. C. et al. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl Acad. Sci. USA 114, 11157–11162 (2017).
Walls, A. C. et al. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature 531, 114–117 (2016).
Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020).
Kirchdoerfer, R. N. et al. Pre-fusion structure of a human coronavirus spike protein. Nature 531, 118–121 (2016).
Ke, Z. et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 588, 498–502 (2020).
Turoňová, B. et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science 370, 203–208 (2020).
Tortorici, M. A. & Veesler, D. Structural insights into coronavirus entry. Adv. Virus Res. 105, 93–116 (2019).
Piccoli, L. et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell 183, 1024–1042 (2020).
Tortorici, M. A. et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 370, 950–957 (2020).
Pinto, D. et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020).
Brouwer, P. J. M. et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369, 643–650 (2020).
Rogers, T. F. et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963 (2020).
Seydoux, E. et al. Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity 53, 98–105.e5 (2020).
Liu, L. et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020).
Barnes, C. O. et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020).
Barnes, C. O. et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 182, 828–842 (2020).
Walls, A. C. et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell 176, 1026–1039 (2019).
Rockx, B. et al. Escape from human monoclonal antibody neutralization affects in vitro and in vivo fitness of severe acute respiratory syndrome coronavirus. J. Infect. Dis. 201, 946–955 (2010).
Rockx, B. et al. Structural basis for potent cross-neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge. J. Virol. 82, 3220–3235 (2008).
Corti, D. et al. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc. Natl Acad. Sci. USA 112, 10473–10478 (2015).
Wang, C. et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 11, 2251 (2020).
Hansen, J. et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 369, 1010–1014 (2020).
Zost, S. J. et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449 (2020).
Alsoussi, W. B. et al. A potently neutralizing antibody protects mice against SARS-CoV-2 infection. J. Immunol. 205, 915–922 (2020).
Hassan, A. O. et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell 182, 744–753 (2020).
ter Meulen, J. et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 3, e237 (2006).
Yuan, M. et al. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633 (2020).
Huo, J. et al. Neutralisation of SARS-CoV-2 by destruction of the prefusion spike. Cell Host Microbe 28, 445–454 (2020).
Wec, A. Z. et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 369, 731–736 (2020).
McCallum, M. et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell https://doi.org/10.1016/j.cell.2021.03.028 (2021).
Chi, X. et al. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science 369, 650–655 (2020).
Suryadevara, N. et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell https://doi.org/10.1016/j.cell.2021.03.029 (2021).
Cerutti, G. et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe https://doi.org/10.1016/j.chom.2021.03.005 (2021).
Edridge, A. W. D. et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 26, 1691–1693 (2020).
Kaname, Y. et al. Acquisition of complement resistance through incorporation of CD55/decay-accelerating factor into viral particles bearing baculovirus GP64. J. Virol. 84, 3210-9 (2010).
Millet, J. K. & Whittaker, G. R. Murine leukemia virus (MLV)-based coronavirus spike-pseudotyped particle production and infection. Bio Protoc. 6, e2035 (2016).
Walls, A. C. et al. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 23, 899–905 (2016).
Kirchdoerfer, R. N. et al. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 8, 15701 (2018).
Yuan, Y. et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 8, 15092 (2017).
Gui, M. et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 27, 119–129 (2017).
Cao, L. et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 370, 426–431 (2020).
Park, Y. J. et al. Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nat. Struct. Mol. Biol. 26, 1151–1157 (2019).
Pallesen, J. et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl Acad. Sci. USA 114, E7348–E7357 (2017).
Tortorici, M. A. et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 26, 481–489 (2019).
Cai, Y. et al. Distinct conformational states of SARS-CoV-2 spike protein. Science 369, 1586–1592 (2020).
Fan, X., Cao, D., Kong, L. & Zhang, X. Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein. Nat. Commun. 11, 3618 (2020).
Corti, D. et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333, 850–856 (2011).
King, L. B. et al. Cross-reactive neutralizing human survivor monoclonal antibody BDBV223 targets the ebolavirus stalk. Nat. Commun. 10, 1788 (2019).
Kong, R. et al. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352, 828–833 (2016).
Corti, D. & Lanzavecchia, A. Broadly neutralizing antiviral antibodies. Annu. Rev. Immunol. 31, 705–742 (2013).
Walker, L. M. et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289 (2009).
Huang, J. et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412 (2012).
Walker, L. M. et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011).
Wu, X. et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 (2010).
Zhou, T. et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817 (2010).
Scheid, J. F. et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636–640 (2009).
Whittle, J. R. et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl Acad. Sci. USA 108, 14216–14221 (2011).
Ekiert, D. C. et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489, 526–532 (2012).
Ekiert, D. C. et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333, 843–850 (2011).
Dreyfus, C. et al. Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348 (2012).
Kallewaard, N. L. et al. Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell 166, 596–608 (2016).
Corti, D. et al. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature 501, 439–443 (2013).
Dang, H. V. et al. An antibody against the F glycoprotein inhibits Nipah and Hendra virus infections. Nat. Struct. Mol. Biol. 26, 980–987 (2019).
Mire, C. E. et al. A cross-reactive humanized monoclonal antibody targeting fusion glycoprotein function protects ferrets against lethal Nipah virus and Hendra virus infection. J. Infect. Dis. 221, S471–S479 (2020).
Zhu, Z. et al. Potent neutralization of Hendra and Nipah viruses by human monoclonal antibodies. J. Virol. 80, 891–899 (2006).
Barba-Spaeth, G. et al. Structural basis of potent Zika–Dengue virus antibody cross-neutralization. Nature 536, 48–53 (2016).
Dejnirattisai, W. et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with Dengue virus. Nat. Immunol. 16, 170–177 (2015).
Rouvinski, A. et al. Recognition determinants of broadly neutralizing human antibodies against Dengue viruses. Nature 520, 109–113 (2015).
Bornholdt, Z. A. et al. Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science 351, 1078–1083 (2016).
Flyak, A. I. et al. Broadly neutralizing antibodies from human survivors target a conserved site in the Ebola virus glycoprotein HR2-MPER region. Nat. Microbiol. 3, 670–677 (2018).
West, B. R. et al. Structural basis of pan-Ebolavirus neutralization by a human antibody against a conserved, yet cryptic epitope. Mbio 9, e01674-18 (2018).
Daniel, C. et al. Identification of an immunodominant linear neutralization domain on the S2 portion of the murine coronavirus spike glycoprotein and evidence that it forms part of complex tridimensional structure. J. Virol. 67, 1185–1194 (1993).
Zhang, H. et al. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J. Virol. 78, 6938–6945 (2004).
Poh, C. M. et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun. 11, 2806 (2020).
Elshabrawy, H. A., Coughlin, M. M., Baker, S. C. & Prabhakar, B. S. Human monoclonal antibodies against highly conserved HR1 and HR2 domains of the SARS-CoV spike protein are more broadly neutralizing. PLoS ONE 7, e50366 (2012).
Zheng, Z. et al. Monoclonal antibodies for the S2 subunit of spike of SARS-CoV-1 cross-react with the newly-emerged SARS-CoV-2. Eur. Surveill. 25, 2000291 (2020).
Azoitei, M. L. et al. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science 334, 373–376 (2011).
Correia, B. E. et al. Proof of principle for epitope-focused vaccine design. Nature 507, 201–206 (2014).
Sesterhenn, F. et al. De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science 368, eaay5051 (2020).
Wang, C. et al. A conserved immunogenic and vulnerable site on the coronavirus spike protein delineated by cross-reactive monoclonal antibodies. Nat. Commun. 12, 1715 (2021).
Song, G. et al. Cross-reactive serum and memory B cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Preprint at bioRxiv https://doi.org/2020.09.22.308965 (2020).
Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 (1988).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Snijder, J. et al. An antibody targeting the fusion machinery neutralizes dual-tropic infection and defines a site of vulnerability on Epstein–Barr virus. Immunity 48, 799–811 (2018).
Siegel, R. W. Antibody affinity optimization using yeast cell surface display. Methods Mol. Biol. 504, 351–383 (2009).
Crawford, K. H. D. et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12, 513 (2020).
Omerović, J., Lev, L. & Longnecker, R. The amino terminus of Epstein–Barr virus glycoprotein gH is important for fusion with epithelial and B cells. J. Virol. 79, 12408–12415 (2005).
Raj, V. S. et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495, 251–254 (2013).
McLellan, J. S. et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342, 592–598 (2013).
Bandaranayake, A. D. et al. Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors. Nucleic Acids Res. 39, e143 (2011).
Hulswit, R. J. G. et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl Acad. Sci. USA 116, 2681–2690 (2019).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Tegunov, D. & Cramer, P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Zivanov, J., Nakane, T. & Scheres, S. H. W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Frenz, B. et al. Automatically fixing errors in glycoprotein structures with Rosetta. Structure 27, 134–139 (2019).
Wang, R. Y. et al. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife 5, e17219 (2016).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019).
Agirre, J. et al. Privateer: software for the conformational validation of carbohydrate structures. Nat. Struct. Mol. Biol. 22, 833–834 (2015).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 (2004).
Acknowledgements
We thank H. Tani (University of Toyama) for providing the reagents necessary for preparing VSV pseudotyped viruses and B. Fiala for assisting with protein production. This study was supported by the National Institute of General Medical Sciences (R01GM120553, D.V.); the National Institute of Allergy and Infectious Diseases (DP1AI158186 and HHSN272201700059C, D.V.); a Pew Biomedical Scholars Award (D.V.); an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund (D.V.); a Fast Grant (D.V.); the University of Washington Arnold and Mabel Beckman cryoEM center; the Swiss National Science Foundation (P400PB_183942, M.M.S.); the Pasteur Institute (M.A.T.); the M.J. Murdock Charitable Trust (A.T.M. and B.H.); and beamlines 8.2.1 and 5.0.1 at the Advanced Light Source at Lawrence Berkeley National Laboratory, a US DOE Office of Science User Facility under contract no. DE-AC02-05CH11231.
Author information
Authors and Affiliations
Contributions
A.T.M. and D.V. supervised the research. M.M.S., M.A.T., Y.-J.P., A.C.W., B.G.H., A.T.M. and D.V. designed the experiments. M.M.S., M.A.T., A.C.W., J.B., X.X. and W.d.v.S. expressed and purified proteins. B.G.H. performed the mouse immunization study. M.M.S., A.C.W., L.H., B.G.H., A.T.M. and D.V. discovered the B6 antibody. M.M.S. and L.H. performed the binding and cell fusion experiments. M.M.S., M.A.T., L.H., C.W. and B.-J.B. performed neutralization assays. M.M.S., Y.-J.P., O.J.A., J.Q. and D.V. performed cryo-EM sample preparation, data collection, processing and model building. M.M.S., Y.-J.P. and D.V. performed crystallization, X-ray crystallography data collection, processing and model building. M.M.S. and D.V. prepared the manuscript, with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
M.M.S., M.A.T., Y.J.P., A.C.W., A.T.M. and D.V. are named as inventors on patent applications filed by the University of Washington based on the studies presented in this paper. D.V. is a consultant for Vir Biotechnology Inc. The Veesler laboratory has received an unrelated sponsored research agreement from Vir Biotechnology Inc. The other authors declare no competing interests.
Additional information
Peer review information Nature Structural & Molecular Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available. Anke Sparmann was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 MERS-CoV S, SARS-CoV S and SARS-CoV-2 S pseudotyped virus neutralization.
Neutralization assays of MLV (a-c) or VSV (d–f) particles pseudotyped with (a,d) MERS-CoV S (a, Data shown for 1 representative experiment with 3 replicates (n = 2 independent experiments); d, Data shown for 1 representative experiment with 5 replicates (n = 2 independent experiments)) (b,e) SARS-CoV S (b, Data shown for 1 representative experiment with 3 replicates (n = 2 independent experiments); e, Data shown for 1 representative experiment with 2 replicates (n = 2 independent experiments)) and (c,f) SARS-CoV-2 S (c, Data shown for 1 representative experiment with 2 replicates (n = 2 independent experiments); f, Data shown for 1 representative experiment with 2 replicates (n = 2 independent experiments)) were performed in the presence of the indicated concentration of B6 mAb. Data are presented as mean values ± SD and were evaluated using a non-linear sigmoidal regression model with variable Hill slope. Experiments were performed with two independent mAb and pseudotyped virus preparations.
Extended Data Fig. 2 CryoEM characterization of the B6-bound MERS-CoV S complex.
(a) Representative image of 1,146 cryoEM micrograph of the MERS-CoV S prefusion trimer bound to B6 Fab embedded in vitreous ice. Scale bar: 20 nm. (b) Selected reference-free 2D class averages. Scale bar: 20Å. (c) Fourier shell correlation curves for the reconstructions shown in panels (d) and (e). (d) Reconstruction obtained with all selected particles and applying C3 symmetry colored by local resolution. (e) Reconstruction obtained with a subset of particles obtained through focused classification to improve B6 resolvability colored by local resolution. (f) Zoomed-in view of the MERS-CoV stem bound to the B6 Fab. The position of MERS-CoV S residues 1226-1229 connecting the cryoEM model to the B6/MERS-CoV stem peptide crystal structure is indicated as dashed line.
Extended Data Fig. 3 CryoEM data processing flow chart.
CTFR: per-particle defocus refinement, NUR: non-uniform refinement, HR: homogenous refinement.
Extended Data Fig. 4 Protein sequence alignment of the stem region for selected β-coronavirus S glycoproteins.
The sequence alignment was performed based on MERS-CoV S using the following S protein sequences: MERS-CoV EMC/2012 (GenBank: AFS88936.1), HKU4 (UniProtKB: A3EX94.1), HKU5 (UniProtKB: A3EXD0.1), HKU1 isolate N5 (UniProtKB: Q0ZME7.1), MHV A59 (UniProtKB: P11224.2), OC43 (UniProtKB: Q696P8), SARS-CoV Urbani (GenBank: AAP13441.1), SARS-CoV-2 (NCBI Reference Sequence: YP_009724390.1). Sequence alignment was performed using Multalin and visualized using ESPript3.0. The conserved stem helix recognized by B6 is indicated.
Extended Data Fig. 5 Crystal structures of B6 bound to coronavirus S stem helix peptides.
Stem peptides of (a) MERS-CoV S (b) OC43 S (c) SARS-CoV/SARS-CoV-2 S and (d) HKU4 S are shown in stick representation with carbon atoms colored yellow. B6 is shown in ribbon representation with interacting residues rendered as stick representation in gray. Oxygen and nitrogen atoms are colored red and blue, respectively. The 2Fo-Fc maps for the different peptides are shown as a blue mesh at a contour level of 1 σ.
Extended Data Fig. 6 B6 binding kinetics to different coronavirus S ectodomain trimers.
a–c Binding of B6 to immobilized (a) MERS-CoV S, (b) OC43 S and (c) SARS-CoV S measured by biolayer interferometry. The vertical dotted lines correspond to the transition between the association and dissociation phases. Data are shown for one representative measurement and were analyzed with the OctetBio software. Global fits are shown as dashed lines. We determined dissociation constant (KD) values of 0.28 (0.2) ± 0.001 and 1.50 (1.47) ± 0.01 µM for two independent batches of S protein for MERS-CoV S and OC43 S, respectively (n = 2 independent experiments). The dissociation constant for SARS-CoV S could not be evaluated reliably, however, the predicted affinity is significantly lower compared to the other two S proteins.
Supplementary information
Source data
Source Data Fig. 1
Pseudovirus assay (unprocessed and normalized data).
Source Data Fig. 4
ELISA and membrane fusion assay (unprocessed and normalized data).
Source Data Extended Data Fig. 1
Pseudovirus assay (unprocessed and normalized data).
Rights and permissions
About this article
Cite this article
Sauer, M.M., Tortorici, M.A., Park, YJ. et al. Structural basis for broad coronavirus neutralization. Nat Struct Mol Biol 28, 478–486 (2021). https://doi.org/10.1038/s41594-021-00596-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-021-00596-4
This article is cited by
-
Prefusion-stabilized SARS-CoV-2 S2-only antigen provides protection against SARS-CoV-2 challenge
Nature Communications (2024)
-
Structural insights into the modulation of coronavirus spike tilting and infectivity by hinge glycans
Nature Communications (2023)
-
Targetable elements in SARS-CoV-2 S2 subunit for the design of pan-coronavirus fusion inhibitors and vaccines
Signal Transduction and Targeted Therapy (2023)
-
Enhanced protective efficacy of a thermostable RBD-S2 vaccine formulation against SARS-CoV-2 and its variants
npj Vaccines (2023)
-
Automatic and accurate ligand structure determination guided by cryo-electron microscopy maps
Nature Communications (2023)