Abstract
Ribosome-associated quality control (RQC) represents a rescue pathway in eukaryotic cells that is triggered upon translational stalling. Collided ribosomes are recognized for subsequent dissociation followed by degradation of nascent peptides. However, endogenous RQC-inducing sequences and the mechanism underlying the ubiquitin-dependent ribosome dissociation remain poorly understood. Here, we identified SDD1 messenger RNA from Saccharomyces cerevisiae as an endogenous RQC substrate and reveal the mechanism of its mRNA-dependent and nascent peptide−dependent translational stalling. In vitro translation of SDD1 mRNA enabled the reconstitution of Hel2-dependent polyubiquitination of collided disomes and, preferentially, trisomes. The distinct trisome architecture, visualized using cryo-EM, provides the structural basis for the more-efficient recognition by Hel2 compared with that of disomes. Subsequently, the Slh1 helicase subunit of the RQC trigger (RQT) complex preferentially dissociates the first stalled polyubiquitinated ribosome in an ATP-dependent manner. Together, these findings provide fundamental mechanistic insights into RQC and its physiological role in maintaining cellular protein homeostasis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data Availability
The sequencing data for ribosome profiling experiments have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO series accession number GSE131214. Cryo-EM maps and the respective molecular models have been deposited with accession codes EMD-10262 and PDB 6SNT for the SDD1-stalled 80S ribosome and EMD-10315 and PDB 6SV4 for the SDD1-stalled trisome, respectively. MS results have been deposited in Proteomics Identification Database (PRIDE) with accession number PXD017196.
References
Brandman, O. & Hegde, R. S. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 23, 7–15 (2016).
Inada, T. The ribosome as a platform for mRNA and nascent polypeptide quality control. Trends Biochem. Sci. 42, 5–15 (2017).
Joazeiro, C. A. P. Ribosomal stalling during translation: providing substrates for ribosome-associated protein quality control. Annu. Rev. Cell Dev. Biol. 33, 343–368 (2017).
Ozsolak, F. et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143, 1018–1029 (2010).
Arthur, L. L. & Djuranovic, S. PolyA tracks, polybasic peptides, poly-translational hurdles. Wiley Interdiscip. Rev. RNA 9, e1486 (2018).
Inada, T. & Aiba, H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 24, 1584–1595 (2005).
Ito-Harashima, S., Kuroha, K., Tatematsu, T. & Inada, T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 21, 519–524 (2007).
Pisareva, V. P., Skabkin, M. A., Hellen, C. U., Pestova, T. V. & Pisarev, A. V. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 30, 1804–1817 (2011).
Becker, T. et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482, 501–506 (2012).
Shoemaker, C. J., Eyler, D. E. & Green, R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330, 369–372 (2010).
Brandman, O. et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054 (2012).
Lyumkis, D. et al. Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex. Proc. Natl Acad. Sci. USA 111, 15981–15986 (2014).
Shao, S., Brown, A., Santhanam, B. & Hegde, R. S. Structure and assembly pathway of the ribosome quality control complex. Mol. Cell 57, 433–444 (2015).
Shen, P. S. et al. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347, 75–78 (2015).
Bengtson, M. H. & Joazeiro, C. A. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473 (2010).
Sundaramoorthy, E. et al. ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation. Mol. Cell 65, 751–760.e4 (2017).
Sitron, C. S., Park, J. H. & Brandman, O. Asc1, Hel2, and Slh1 couple translation arrest to nascent chain degradation. RNA 23, 798–810 (2017).
Matsuo, Y. et al. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 8, 159 (2017).
Garzia, A. et al. The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat. Commun. 8, 16056 (2017).
Juszkiewicz, S. & Hegde, R. S. Initiation of quality control during poly(A) translation requires site-specific ribosome ubiquitination. Mol. Cell 65, 743–750.e4 (2016).
Juszkiewicz, S. et al. ZNF598 Is a Quality Control Sensor of Collided Ribosomes. Mol. Cell 72, 469–481.e7 (2018).
Ikeuchi, K. et al. Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 38, e100276 (2019).
Letzring, D. P., Wolf, A. S., Brule, C. E. & Grayhack, E. J. Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1. RNA 19, 1208–1217 (2013).
Dimitrova, L. N., Kuroha, K., Tatematsu, T. & Inada, T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 284, 10343–10352 (2009).
Choe, Y.-J. et al. Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature 531, 191–195 (2016).
Kostova, K. K. et al. CAT-tailing as a fail-safe mechanism for efficient degradation of stalled nascent polypeptides. Science 357, 414–417 (2017).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
D’Orazio, K. N. et al. The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during No Go Decay. Elife 8, e49117 (2019).
Wilson, D. N., Arenz, S. & Beckmann, R. Translation regulation via nascent polypeptide-mediated ribosome stalling. Curr. Opin. Struct. Biol. 37, 123–133 (2016).
Schmeing, T. M., Huang, K. S., Strobel, S. A. & Steitz, T. A. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 438, 520–524 (2005).
Lehmann, J. Induced fit of the peptidyl-transferase center of the ribosome and conformational freedom of the esterified amino acids. RNA 23, 229–239 (2017).
Behrmann, E. et al. Structural snapshots of actively translating human ribosomes. Cell 161, 845–857 (2015).
Shao, S. et al. Decoding mammalian ribosome-mRNA states by translational GTPase complexes. Cell 167, 1229–1240.e15 (2016).
Ito, K., Chiba, S. & Pogliano, K. Divergent stalling sequences sense and control cellular physiology. Biochem. Biophys. Res. Commun. 393, 1–5 (2010).
Bhushan, S. et al. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol. Cell 40, 138–146 (2010).
Presta, L. G. & Rose, G. D. Helix signals in proteins. Science 240, 1632–1641 (1988).
Kuroha, K. et al. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep. 11, 956–961 (2010).
Ikeuchi, K. & Inada, T. Ribosome-associated Asc1/RACK1 is required for endonucleolytic cleavage induced by stalled ribosome at the 3′ end of nonstop mRNA. Sci. Reports 6, 28234 (2016).
Shoemaker, C. J. & Green, R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl Acad. Sci. USA 108, E1392–E1398 (2011).
Tsuboi, T. et al. Dom34:Hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol. Cell 46, 518–529 (2012).
Joazeiro, C. A. P. Mechanisms and functions of ribosome-associated protein quality control. Nat. Rev. Mol. Cell Biol. 20, 368–383 (2019).
Tesina, P. et al. Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts. EMBO J. 39, e103365 (2019).
Chandrasekaran, V. et al. Mechanism of ribosome stalling during translation of a poly(A) tail. Nat. Struct. Mol. Biol. 26, 1132–1140 (2019).
Tang, T. T. L., Stowell, J. A. W., Hill, C. H. & Passmore, L. A. The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases. Nat. Struct. Mol. Biol. 26, 433–442 (2019).
Gamble, C. E., Brule, C. E., Dean, K. M., Fields, S. & Grayhack, E. J. Adjacent codons act in concert to modulate translation efficiency in yeast. Cell 166, 679–690 (2016).
Longtine, M. S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998).
Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004).
Ingolia, N. T., Brar, G. A., Rouskin, S., McGeachy, A. M. & Weissman, J. S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 (2012).
Waters, M. G. & Blobel, G. Secretory protein translocation in a yeast cell-free system can occur posttranslationally and requires ATP hydrolysis. J. Cell Biol. 102, 1543–1550 (1986).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Acknowledgements
We thank O. Berninghausen, H. Sieber, C. Ungewickell and S. Rieder for technical support. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants grant no. OD018174. This study was supported by MEXT/JSPS KAKENHI grant nos. JP18H03977 (T.I.), JP19H05281 (T.I.), JP19K06481 (Y.M.), JP18H03993 (Y.S.) and JP18H05498 (Y.S.); AMED under grant nos. JP19gm1110010 (T.I.) and JP18gm1110003 (Y.S); Research Grants in the Natural Sciences from the Takeda Foundation (T.I.) and by Research Grants in the Medical Sciences from Kato Memorial Bioscience Foundation (Y.M.) and by a grant of the Deutsche Forschungsgemeinschaft (DFG; BE1814/15-1) to R. Beckmann and a PhD fellowship from Boehringer Ingelheim Fonds to R. Buschauer.
Author information
Authors and Affiliations
Contributions
Experiments were designed by Y.M., P.T., T.B., R. Beckmann and T.I.; Y.M. performed in vitro experiments and analyzed ribosome profiling data sets; P.T. prepared cryo-EM samples and, together with T.B., processed the cryo-EM data. R. Buschauer, P.T. and J.C. built molecular models, and P.T., T.B., and R. Buschauer analyzed the structures. S.N. performed cis-element analysis of Sdd1; M.M. performed initial screening of RQC targets and uL4 mutant analysis; O.S. performed 25S rRNA mutant analysis; K.I. prepared the library of ribosome profiling; A.E. performed MS analysis together with Y.S.; the manuscript was written by Y.M., P.T., T.B., R. Beckmann and T.I.; all authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Anke Sparmann was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Screening of RQC targets.
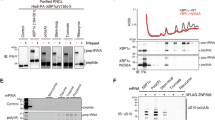
a, List of genes comprising the ‘CGACGA’ di-codons. Extended sequence fragments involving 14 codons both up- and down-stream of the respective ‘CGACGA’ di-codon were used as candidates in the screen. Tandem arginine coded by ‘CGACGA’ di-codons is indicated by red characters. Other basic amino acid residues are indicated by blue characters. b, Schematic drawing of the reporter system used in the screen. c, Scheme of the RQC targets validation procedure. The amounts of full-length products depend on the degree of translational arrest and frequency of read through. The level of RQC induction is reflected in the amount of arrest products in ltn1∆ cells. d, Immunoblotting of protein products of the reporters (V5-Rluc-X-Fluc-HA) including the respective candidate sequences (X) listed in (a) using anti-V5 antibody. e, Immunoblotting of the products of HA-SDD1 or HA-SDD1-V5 in the indicated strains using anti-HA antibody or anti-eEF2 antibodies. f, Immunoblotting of protein products of the GFP-X-FLAG-HIS3 reporter was detected by immunoblotting in ltn1∆rqc2∆ strain using anti-GFP or anti-eEF2 antibodies.
Extended Data Fig. 2 SDD1 reporter mRNA, purification of the stalled RNCs and processing scheme of the monosome fraction.
a, Schematic representation of the SDD1 mRNA reporter used for the structural studies. b, In vitro translation reaction (IVT) using a yeast translation extract from a ski2Δ strain and subsequent affinity purification of His-tagged ribosome-nascent chain complexes. Fractions representing time points of the IVT together with input (IN), flow through (FT), wash 1 (W1), elution (E) and negative control without added mRNA (c-) were visualized by immunoblotting using anti-HA antibody. c, The eluate was loaded on a 10–50% sucrose gradient and fractionated. Peaks representing 80S and trisomes were collected and concentrated using a sucrose cushion. Resuspended pellets were used for cryo-EM sample preparation. d, 3D classification and processing scheme of the monosome fraction stalled on the SDD1 mRNA reporter (SDD-RNCs). The first refined map was sorted into 6 classes. Classes 3 and 5 represented a vast majority of programmed ribosomal particles exhibiting the non-rotated POST state with P/P (class 5) and E/E (class 3) site tRNAs. These classes were joined and further sub-classified using a mask around the P site tRNA and the entry path of mRNA into the decoding center. This yielded a subclass with strong density for both the mRNA and P site tRNA which was further refined and processed as indicated.
Extended Data Fig. 3 Tunnel interactions and PTC conformation in the SDD1 RNC.
a, Schematic representation of the Sdd1 nascent chain and its interactions with the peptide exit tunnel. b, Molecular model of the peptidyl-transferase center and the positions observed for U2954 and U2875. The clash of the Sdd1 nascent chain with the induced position of U2875 is indicated with a red cross.
Extended Data Fig. 4 Mutational analysis of Y201 and stalling efficiency of SDD1 constructs.
a, Mutational analysis of Y201 in Sdd1 (related to Fig. 3). Sdd1-Y201 mutants in WT and ltn1∆ strains were detected by immunoblotting. b, Mutation analysis of the basic residues near the PTC (related to Fig. 3). c, Time course of peptidyl-tRNA production as an indicator of ribosomal stalling (related to Fig. 4); The upper peptidyl-tRNA band were shifted to lower band (peptide) after RNase treatment.
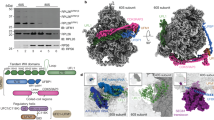
Extended Data Fig. 5 Trisome dataset processing scheme and detailed view on the inter-ribosomal interactions.
a, 3D classification and processing scheme of the trisome fraction stalled on the SDD1 mRNA reporter (SSD1 trisome) as described in Extended Data Fig. 2b. The first refined map was sorted into 8 classes. Class 5 represented the first stalled ribosome with a density of one neighboring ribosome near the mRNA exit site as observed in the monosome dataset. Class 7 represented the last ribosome with a neighbor density only at mRNA entry site and ES27 in L1 position (this position would directly clash and excludes neighboring ribosome present at the mRNA exit site). These classes were further processed individually as indicated in the scheme using stepwise box extension and re-centering. This yielded two consensus refinements showing overlapping positions of individual ribosomes. Individual ribosomes were resolved via multi-body refinement and fitted into the consensus refinement position. b, General overview of the trisome assembly using molecular models. An interaction area of the three respective 40S heads is depicted in detail showing individual RACK1 (Asc1), uS10 and uS3 proteins colored according to the color scheme of their respective ribosomes. Direct interaction between RACK1 molecules was only observed between second and third ribosome. Direct interaction between RACK1-1 and uS10-2 as well as between RACK1-2 and uS10-2 was observed, analogous to the situation reported in the disome22. Interaction between RACK1 and uS3 of the following ribosome is possible and would be mediated by the C-terminus of RACK1. This interface brings six copies or Hel2 ubiquitination target proteins (uS3 1-3 and uS10 1-3) in close vicinity.
Extended Data Fig. 6 In vitro ribosome splitting assay by RQT complex.
a, Scheme of the in vitro splitting assay. b, SDS-PAGE of the purified Rqc2 used in this assay. c, Time course of the in vitro splitting assay before (left) and after addition of purified wild-type RQT complex and ATP (middle and right). Purified RNCs were incubated with the purified RQT complex and ATP for 10, 30 min (as described in panel (a)), then separated in sucrose density gradient and detected by UV absorbance at 254 nm wavelength. Tagged uS10 in each fraction was detected by immunoblotting using anti-HA antibody. d, Comparison of ribosome UV profiles obtained after individual splitting assay reactions (related to Fig. 6).
Extended Data Fig. 7 Analysis of canonical splitting factors and their activity in the RQT-mediated ribosome splitting assays.
a, SDS-PAGE of the reaction mixtures used for in vitro splitting assay. b–d, The peak intensity-based quantification by mass spectrometry analysis: RNCs (b), RNCs with RQT complex (c), and RNCs with Rli1 (d); Detection below the limit of quantification and not detected are indicated by <l.o.q and n.d., respectively. Mass spectrometry data is available in Source Data. e-g, In vitro splitting assay results. Purified RNCs after splitting reaction were separated by sucrose density gradient and detected by UV absorbance at 254 nm wavelength. Tagged uS10 in each fraction was detected by immunoblotting using anti-HA antibody: RNCs (e), RNCs with ATP (f), and RNCs with Rli1 and ATP (g).
Extended Data Fig. 8 Local resolution and Fourier shell correlation plots of the SDD1 stalled 80S and trisome structures.
Cryo-EM density maps filtered and colored according to local resolution as estimated by Relion with Fourier Shell Correlation (FSC) plots for the refined 2.8 Å 80S map (a) as well as the individually refined ribosomes of the trisome map (b).
Extended Data Fig. 9 Map to model Fourier shell correlation (FSC) curves.
Map to model FSC plots of the SDD1 stalled 80S ribosome (a) and trisome (b) as calculated by Phenix.
Supplementary information
Supplementary Table
List of materials used for this study
Source data
Source Data Fig. 1
Unprocessed western blots
Source Data Fig. 3
Unprocessed western blots
Source Data Fig. 4
Unprocessed western blots
Source Data Fig. 6
Unprocessed western blots
Source Data Fig. 7
Unprocessed western blots
Source Data Extended Data Fig. 7
Statistical data
Rights and permissions
About this article
Cite this article
Matsuo, Y., Tesina, P., Nakajima, S. et al. RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1. Nat Struct Mol Biol 27, 323–332 (2020). https://doi.org/10.1038/s41594-020-0393-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-0393-9
This article is cited by
-
Transient disome complex formation in native polysomes during ongoing protein synthesis captured by cryo-EM
Nature Communications (2024)
-
Transcriptional profile of ribosome-associated quality control components and their associated phenotypes in mammalian cells
Scientific Reports (2024)
-
Recognition of an Ala-rich C-degron by the E3 ligase Pirh2
Nature Communications (2023)
-
Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2
Nature Communications (2023)
-
B. subtilis MutS2 splits stalled ribosomes into subunits without mRNA cleavage
The EMBO Journal (2023)