Abstract
Dynein-2 assembles with polymeric intraflagellar transport (IFT) trains to form a transport machinery that is crucial for cilia biogenesis and signaling. Here we recombinantly expressed the ~1.4-MDa human dynein-2 complex and solved its cryo-EM structure to near-atomic resolution. The two identical copies of the dynein-2 heavy chain are contorted into different conformations by a WDR60−WDR34 heterodimer and a block of two RB and six LC8 light chains. One heavy chain is steered into a zig-zag conformation, which matches the periodicity of the anterograde IFT-B train. Contacts between adjacent dyneins along the train indicate a cooperative mode of assembly. Removal of the WDR60−WDR34−light chain subcomplex renders dynein-2 monomeric and relieves autoinhibition of its motility. Our results converge on a model in which an unusual stoichiometry of non-motor subunits controls dynein-2 assembly, asymmetry, and activity, giving mechanistic insight into the interaction of dynein-2 with IFT trains and the origin of diverse functions in the dynein family.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Cryo-EM maps are available from the EMDB under accession codes EMD-4918 (dynein-2 tail domain) and EMD-4917 (dynein-2 motor domains). Coordinates are available from the RCSB Protein Data Bank under accession codes PDB 6RLB (dynein-2 tail domain), PDB 6RLA (dynein-2 motor domains), and PDB 6SC2 (dynein-2, docked into subtomogram average of the anterograde IFT-B train29 (EMDB-4303)). All other data supporting the conclusions of this manuscript are available from the corresponding author upon reasonable request.
References
Rosenbaum, J. L. & Witman, G. B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3, 813–825 (2002).
Pazour, G. J., Wilkerson, C. G. & Witman, G. B. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J. Cell Biol. 141, 979–992 (1998).
Porter, M. E., Bower, R., Knott, J. A., Byrd, P. & Dentler, W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell 10, 693–712 (1999).
Pazour, G. J., Dickert, B. L. & Witman, G. B. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 144, 473–481 (1999).
Signor, D. et al. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J. Cell Biol. 147, 519–530 (1999).
Nachury, M. V. & Mick, D. U. Establishing and regulating the composition of cilia for signal transduction. Nat. Rev. Mol. Cell Biol. 20, 389–405 (2019).
King, S. M. Axonemal dynein arms. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a028100 (2016).
Ye, F., Nager, A. R. & Nachury, M. V. BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J. Cell Biol. 217, 1847–1868 (2018).
Lechtreck, K.-F. et al. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol. 187, 1117–1132 (2009).
Cao, M. et al. Uni-directional ciliary membrane protein trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding. eLife 4, e05242 (2015).
Vuolo, L., Stevenson, N. L., Heesom, K. J. & Stephens, D. J. Dynein-2 intermediate chains play crucial but distinct roles in primary cilia formation and function. eLife 7, e39655 (2018).
Jensen, V. L. et al. Role for intraflagellar transport in building a functional transition zone. EMBO Rep. 19, e45862 (2018).
Huangfu, D. & Anderson, K. V. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl Acad. Sci. USA 102, 11325–11330 (2005).
Schmidts, M. et al. Mutations in the gene encoding IFT dynein complex component WDR34 cause Jeune asphyxiating thoracic dystrophy. Am. J. Hum. Genet. 93, 932–944 (2013).
Dagoneau, N. et al. DYNC2H1 mutations cause asphyxiating thoracic dystrophy and short rib-polydactyly syndrome, type III. Am. J. Hum. Genet. 84, 706–711 (2009).
Merrill, A. E. et al. Ciliary abnormalities due to defects in the retrograde transport protein DYNC2H1 in short-rib polydactyly syndrome. Am. J. Hum. Genet. 84, 542–549 (2009).
Huber, C. et al. WDR34 mutations that cause short-rib polydactyly syndrome type III/severe asphyxiating thoracic dysplasia reveal a role for the NF-κB pathway in cilia. Am. J. Hum. Genet. 93, 926–931 (2013).
McInerney-Leo, A. M. et al. Short-rib polydactyly and Jeune syndromes are caused by mutations in WDR60. Am. J. Hum. Genet. 93, 515–523 (2013).
Schmidts, M. et al. TCTEX1D2 mutations underlie Jeune asphyxiating thoracic dystrophy with impaired retrograde intraflagellar transport. Nat. Commun. 6, 7074 (2015).
Taylor, S. P. et al. Mutations in DYNC2LI1 disrupt cilia function and cause short rib polydactyly syndrome. Nat. Commun. 6, 7092 (2015).
Kessler, K. et al. DYNC2LI1 mutations broaden the clinical spectrum of dynein-2 defects. Sci. Rep. 5, 11649 (2015).
Gholkar, A. A. et al. Tctex1d2 associates with short-rib polydactyly syndrome proteins and is required for ciliogenesis. Cell Cycle 14, 1116–1125 (2015).
Taschner, M. & Lorentzen, E. The intraflagellar transport machinery. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a028092 (2016).
Chien, A. et al. Dynamics of the IFT machinery at the ciliary tip. eLife 6, e28606 (2017).
Toropova, K., Mladenov, M. & Roberts, A. J. Intraflagellar transport dynein is autoinhibited by trapping of its mechanical and track-binding elements. Nat. Struct. Mol. Biol. 24, 461–468 (2017).
Yi, P., Li, W.-J., Dong, M.-Q. & Ou, G. Dynein-driven retrograde intraflagellar transport is triphasic in C. elegans sensory cilia. Curr. Biol. 27, 1448–1461.e7 (2017).
Mijalkovic, J., Prevo, B., Oswald, F., Mangeol, P. & Peterman, E. J. G. Ensemble and single-molecule dynamics of IFT dynein in Caenorhabditis elegans cilia. Nat. Commun. 8, 14591 (2017).
Wingfield, J. L. et al. IFT trains in different stages of assembly queue at the ciliary base for consecutive release into the cilium. eLife 6, e26609 (2017).
Jordan, M. A., Diener, D. R., Stepanek, L. & Pigino, G. The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat. Cell Biol. 20, 1250–1255 (2018).
Mijalkovic, J., van Krugten, J., Oswald, F., Acar, S. & Peterman, E. J. G. Single-molecule turnarounds of intraflagellar transport at the C. elegans ciliary tip. Cell Reports 25, 1701–1707.e2 (2018).
Pedersen, L. B., Geimer, S. & Rosenbaum, J. L. Dissecting the molecular mechanisms of intraflagellar transport in chlamydomonas. Curr. Biol. 16, 450–459 (2006).
Reck-Peterson, S. L., Redwine, W. B., Vale, R. D. & Carter, A. P. The cytoplasmic dynein transport machinery and its many cargoes. Nat. Rev. Mol. Cell Biol. 19, 382–398 (2018).
Rompolas, P., Pedersen, L. B., Patel-King, R. S. & King, S. M. Chlamydomonas FAP133 is a dynein intermediate chain associated with the retrograde intraflagellar transport motor. J. Cell Sci. 120, 3653–3665 (2007).
Asante, D., Stevenson, N. L. & Stephens, D. J. Subunit composition of the human cytoplasmic dynein-2 complex. J. Cell Sci. 127, 4774–4787 (2014).
Gibbons, B. H., Asai, D. J., Tang, W. J., Hays, T. S. & Gibbons, I. R. Phylogeny and expression of axonemal and cytoplasmic dynein genes in sea urchins. Mol. Biol. Cell 5, 57–70 (1994).
Mikami, A. et al. Molecular structure of cytoplasmic dynein 2 and its distribution in neuronal and ciliated cells. J. Cell Sci. 115, 4801–4808 (2002).
Grissom, P. M., Vaisberg, E. A. & McIntosh, J. R. Identification of a novel light intermediate chain (D2LIC) for mammalian cytoplasmic dynein 2. Mol. Biol. Cell 13, 817–829 (2002).
Hamada, Y., Tsurumi, Y., Nozaki, S., Katoh, Y. & Nakayama, K. Interaction of WDR60 intermediate chain with TCTEX1D2 light chain of the dynein-2 complex is crucial for ciliary protein trafficking. Mol. Biol. Cell 29, 1628–1639 (2018).
Patel-King, R. S., Gilberti, R. M., Hom, E. F. Y. & King, S. M. WD60/FAP163 is a dynein intermediate chain required for retrograde intraflagellar transport in cilia. Mol. Biol. Cell 24, 2668–2677 (2013).
Roberts, A. J. Emerging mechanisms of dynein transport in the cytoplasm versus the cilium. Biochem. Soc. Trans. 46, 967–982 (2018).
Urnavicius, L. et al. Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature 554, 202–206 (2018).
Williams, J. C. et al. Structural and thermodynamic characterization of a cytoplasmic dynein light chain-intermediate chain complex. Proc. Natl Acad. Sci. USA 104, 10028–10033 (2007).
Rapali, P. et al. Directed evolution reveals the binding motif preference of the LC8/DYNLL hub protein and predicts large numbers of novel binders in the human proteome. PLoS One 6, e18818 (2011).
DiMaio, F. et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat. Methods 12, 361–365 (2015).
Schmidt, H., Zalyte, R., Urnavicius, L. & Carter, A. P. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature 518, 435–438 (2015).
Erdős, G. et al. Novel linear motif filtering protocol reveals the role of the LC8 dynein light chain in the Hippo pathway. PLoS Comput. Biol. 13, e1005885 (2017).
Clark, S. A., Jespersen, N., Woodward, C. & Barbar, E. Multivalent IDP assemblies: Unique properties of LC8-associated, IDP duplex scaffolds. FEBS Lett. 589, 2543–2551 (2015).
Hou, Y. & Witman, G. B. Dynein and intraflagellar transport. Exp. Cell Res. 334, 26–34 (2015).
Ichikawa, M., Watanabe, Y., Murayama, T. & Toyoshima, Y. Y. Recombinant human cytoplasmic dynein heavy chain 1 and 2: observation of dynein-2 motor activity in vitro. FEBS Lett. 585, 2419–2423 (2011).
Schroeder, C. M., Ostrem, J. M. L., Hertz, N. T. & Vale, R. D. A Ras-like domain in the light intermediate chain bridges the dynein motor to a cargo-binding region. eLife 3, e03351 (2014).
Hou, Y., Pazour, G. J. & Witman, G. B. A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol. Biol. Cell 15, 4382–4394 (2004).
Reck, J. et al. The role of the dynein light intermediate chain in retrograde IFT and flagellar function in Chlamydomonas. Mol. Biol. Cell 27, 2404–2422 (2016).
Zhang, K. et al. Cryo-EM reveals how human cytoplasmic dynein is auto-inhibited and activated. Cell 169, 1303–1314.e18 (2017).
Perrone, C. A. et al. A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in chlamydomonas and Mammalian cells. Mol. Biol. Cell 14, 2041–2056 (2003).
Tsurumi, Y., Hamada, Y., Katoh, Y. & Nakayama, K. Interactions of the dynein-2 intermediate chain WDR34 with the light chains are required for ciliary retrograde protein trafficking. Mol. Biol. Cell 30, 658–670 (2019).
Li, W., Yi, P. & Ou, G. Somatic CRISPR-Cas9-induced mutations reveal roles of embryonically essential dynein chains in Caenorhabditis elegans cilia. J. Cell Biol. 208, 683–692 (2015).
Blisnick, T. et al. The intraflagellar transport dynein complex of trypanosomes is made of a heterodimer of dynein heavy chains and of light and intermediate chains of distinct functions. Mol. Biol. Cell 25, 2620–2633 (2014).
Vijayachandran, L. S. et al. Gene gymnastics: synthetic biology for baculovirus expression vector system engineering. Bioengineered 4, 279–287 (2013).
Schlager, M. A., Hoang, H. T., Urnavicius, L., Bullock, S. L. & Carter, A. P. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 33, 1855–1868 (2014).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
van Heel, M., Harauz, G., Orlova, E. V., Schmidt, R. & Schatz, M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Lopéz-Blanco, J. R. & Chacón, P. iMODFIT: efficient and robust flexible fitting based on vibrational analysis in internal coordinates. J. Struct. Biol. 184, 261–270 (2013).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Wang, S., Sun, S., Li, Z., Zhang, R. & Xu, J. Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput. Biol. 13, e1005324 (2017).
Jones, D. T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 (1999).
Zimmermann, L. et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Kon, T. et al. The 2.8 Å crystal structure of the dynein motor domain. Nature 484, 345–350 (2012).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
We thank H. Mitchison, C. Moores, S. Webb, and G. Zanetti for comments on the manuscript; Diamond Light Source for cryo-EM facilities at the UK national electron bio-imaging centre (eBIC) supported by the Wellcome Trust, MRC and BBSRC; N. Lukoyanova, J. van Rooyen, A. Sielbert and D. Clare for help with cryo-EM data collection; and D. Houldershaw for computational support. This work was funded by Wellcome Trust and Royal Society (104196/Z/14/Z), BBSRC (BB/P008348/1), and Royal Society (RG170260) grants to A.J.R; Wellcome Trust (WT100387) and MRC grants (MC_UP_A025_1011) to A.P.C; and Wellcome Trust (079605/Z/06/Z) and BBSRC (BB/L014211/1) grants supporting cryo-EM equipment at Birkbeck.
Author information
Authors and Affiliations
Contributions
K.T: investigation, methodology, visualization, writing of original draft. R.Z: investigation, methodology. M.M: investigation, methodology. A.G.M: investigation, writing - review and editing. A.P.C: investigation, methodology, funding acquisition, supervision, writing - review and editing. A.J.R: conceptualization, investigation, methodology, funding acquisition, supervision, visualization, writing of original draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Katarzyna Marcinkiewicz was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
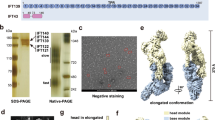
Supplementary Figure 1 Recombinant expression and flexibility of the human dynein-2 complex.
(a) Assembly strategy for the dynein-2 expression plasmid. See Supplementary Table 1 for subunit nomenclature in different organisms. In mammals there are two paralogs for the light chains LC8 (1 and 2), RB (1 and 2), and TCTEX (1 and 3), in addition to the TCTEX-related protein TCTEX1D2. (b) Organisation of expression cassettes in the final plasmid. Each gene is flanked by a polH promoter and SV40 terminator. DHC2 contains an N-terminal ZZ tag and TEV cleavage site used for purification, followed by a SNAPf tag. (c) Size exclusion chromatogram of a dynein-2 complex preparation after affinity purification and TEV cleavage. V0; void volume. (d) SDS-PAGE of the peak fraction in (c). Mass spectrometry confirmed the presence of the indicated subunits. (e) Negative stain class averages reveal flexibility between the tail and motor domains of the dynein-2 complex. Particles are aligned on motor domains and classified to reveal varying tail domain positions. The angle between the C2 symmetry axis in the motor domains (dashed line) and the dimerization domain in the tail (blue dot) is shown. At high tail-motor angles, the TCTEX-TCTEX1D2 region of the tail is in close proximity to the motor domain. (f) Polar histogram plot of the angles between the tail and motor domains. A total of 1,120 particles, classified into 27 averages were analysed. Bins = 10 degrees. Arrows mark the median tail angle in isolated dynein-2 (gray) and the angle when dynein-2 is bound to the anterograde IFT-B train (maroon). (g) Overlay of the median tail angle in isolated dynein-2 and when IFT-bound. All subunits except DHC2 are omitted for clarity.
Supplementary Figure 2 Cryo-EM structure determination of dynein-2.
(a) Representative cryo-EM micrograph of dynein-2 particles (arrowheads; black – tail, white – motor domain). (b) Class averages of dynein-2 tail (top row) and motor domains (bottom row) in different orientations. (c) Fourier shell correlation (FSC) plots for the tail and motor domain reconstructions. Resolution at FSC=0.143 is marked. (d) Euler angle distribution of particles in the tail and (e) motor domain reconstructions. Cylinder height and color represent number of particles (blue to red = low to high). For the motor domain reconstruction, a hemisphere is shown as C2 symmetry was applied. (f) Tail and (g) motor domain reconstructions colored by local resolution as calculated by Relion. (h) – (j) Focused refinement of different regions in the tail domain. For each region, the mask applied to the reference during 3D refinement is shown on the left, with the refined map on the right.
Supplementary Figure 3 Map and model quality for the dynein-2 tail and motor domains.
(a) Cryo-EM density and models for the dynein-2 tail subunits. The overall tail map (Supplementary Fig. 2f) is shown, with the exception of the lower portion of DHC2-B and its associated LIC3, for which a map from focused refinement is shown (Supplementary Fig. 2j). See Fig. 1 for enlarged density examples. (b) Real-space correlation coefficient (CC) between the map and model for each subunit, calculated in Phenix (phenix.validation_cryoem). (c) Dynein-2 motor domain reconstruction (top) and refined atomic model in ribbon representation (bottom). AAA+ domains, linker and C-terminal domain (CTD) are labeled. (d) Example density in the motor domain map shown in mesh representation. Model is shown in stick representation. (e) Density at the AAA1–4 nucleotide binding sites suggests ADP at sites AAA1, 3 and 4 and ATP at AAA2.
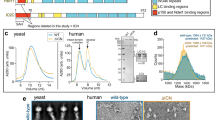
Supplementary Figure 4 Dynein-2 intermediate chains, WDR60 and WDR34.
(a) Secondary structure elements and primary sequence of WDR34 (left) and WDR60 (right). Beta sheets are numbered according to the blade of the β-propeller domain to which they belong. LC8 binding β-sheets are shown as yellow arrows and RB-binding α-helices are in pink. *denotes regions of WDR34 involved in LC8 and RB binding as mapped by Tsurumi, Y. et al. (Mol. Biol. Cell. 30, 658-670, 2019). #denotes the region of WDR60 involved in TCTEX1D2 binding as mapped by Hamada, Y. et al. (Mol. Biol. Cell. 29, 1628–1639, 2018). Putatively disordered N-terminal region of WDR60 is not shown. (b) Cryo-EM density for the WDR34 and WDR60 ß-propellers with models in ribbon representation. Three large inserts in WDR60 (teal, arrowheads) allow unambiguous distinction from WDR34. (c) 3D classes showing varying position of density assigned to TCTEX1/TCTEX1D2 (orange).
Supplementary Figure 5 Contrasting architectures of dynein-2 and -1.
(a) Cryo-EM maps of dynein-2 tail and motor domains (solid) and an unsharpened map showing the flexible connection between them (transparent). (b) Cryo-EM maps of the dynein-1 tail [EMD-3703] and motor domains [EMD-3698] (solid) and entire molecule [EMD-3705] (transparent) (Zhang, K. et al. Cell. 169, 1303–1314, 2017). Dynein-2 has a strikingly asymmetric architecture compared to dynein-1, which allows dynein-2 to associate with the anterograde IFT-B train by matching its periodicity. DHC2TAIL is also shorter by one N-terminal bundle compared to the dynein-1 heavy chain, which uses this subdomain to engage the Arp filament of dynactin (Zhang, K. et al. Cell. 169, 1303–1314, 2017). (c, d) Comparison of the β-propeller domains of dynein-2’s heterodimeric intermediate chains, WDR60 and WDR34 (c) with dynein-1’s homodimeric intermediate chain (IC2) (d), following alignment based on RB. The WDR60 and WDR34 β-propellers are vertically offset and related by a ~90° rotation, which we propose drives the asymmetry of dynein-2, together with the block of three LC8 dimers which is also unique to dynein-2. (e, f) Comparison of dynein-2’s light-intermediate chain, LIC3 (e), with the dynein-1 light-intermediate chain, LIC2 (f) [EMDB-4171] (Urnavicius, L. et al. Nature. 554, 202–206, 2018). The dynein-1 light-intermediate chain has a long disordered C-terminal extension (dashed line) containing a helix that binds to dynein-1 cargo adaptors (Schroeder, C. M. et al. Elife. 3, e03351, 2014; Lee, I. G. et al. Nat Commun. 9, 986, 2018; Celestino, R. et al. PLoS Biol. 17, e3000100, 2019). In contrast, LIC3’s C-terminal region takes an opposite path and interacts with WDR60 within dynein-2.
Supplementary information
Supplementary Information
Supplementary Figures 1–5, Supplementary Tables 1 and 2, Supplementary Notes 1–3
Supplementary Video 1
Large-scale flexibility of the dynein-2 complex. Series of negative stain EM class averages of the dynein-2 complex showing the variation in tail position with respect to the aligned motor domains. Movie shows 27 class averages, ordered by tail angle, looped 6 times.
Supplementary Video 2
Cryo-EM structure of the dynein-2 complex reveals its marked asymmetry and unusual stoichiometry. Cryo-EM maps of the dynein-2 tail and motor domains are shown in surface representation, rotating about the y-axis, colored by subunit. Connecting density from an unsharpened map is shown as a transparent surface.
Supplementary Video 3
Dynein-2’s intermediate and light chains are required for motor auto-regulation. Microtubule gliding activity of the dynein- 2 holoenzyme and a construct lacking the intermediate and light chain sub-complex (∆IC-LC). Whereas the holoenzyme binds microtubules weakly and exhibits slow microtubule gliding, consistent with the majority of complexes being in an auto-inhibited state, ΔIC-LC drives rapid and continuous microtubule movement. See also Fig. 3e. Movies are shown at 45x real time.
Supplementary Video 4
Dynein-2’s asymmetric structure matches the periodicity of the anterograde IFT-B train. The cryo-EM structure of the dynein-2 complex docked into a ~40 Å sub-tomogram average of the IFT-B train from C. reinhardtii cilia (Jordan, M. et al. Nat. Cell Biol. 20, 1250–1255, 2018) (transparent surface representation). Dynein-2 complexes along the train are shown in alternating surface and cylinder representation for distinction. See also Fig. 4.
Rights and permissions
About this article
Cite this article
Toropova, K., Zalyte, R., Mukhopadhyay, A.G. et al. Structure of the dynein-2 complex and its assembly with intraflagellar transport trains. Nat Struct Mol Biol 26, 823–829 (2019). https://doi.org/10.1038/s41594-019-0286-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0286-y
This article is cited by
-
Transport and barrier mechanisms that regulate ciliary compartmentalization and ciliopathies
Nature Reviews Nephrology (2024)
-
Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport
The EMBO Journal (2024)
-
The molecular structure of IFT-A and IFT-B in anterograde intraflagellar transport trains
Nature Structural & Molecular Biology (2023)
-
A structural model of the core of cilia
Nature (2023)
-
Combinations of deletion and missense variations of the dynein-2 DYNC2LI1 subunit found in skeletal ciliopathies cause ciliary defects
Scientific Reports (2022)