Abstract
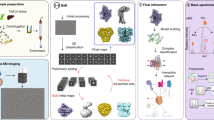
X-ray crystallography often requires non-native constructs involving mutations or truncations, and is challenged by membrane proteins and large multicomponent complexes. We present here a bottom-up endogenous structural proteomics approach whereby near-atomic-resolution cryo electron microscopy (cryoEM) maps are reconstructed ab initio from unidentified protein complexes enriched directly from the endogenous cellular milieu, followed by identification and atomic modeling of the proteins. The proteins in each complex are identified using cryoID, a program we developed to identify proteins in ab initio cryoEM maps. As a proof of principle, we applied this approach to the malaria-causing parasite Plasmodium falciparum, an organism that has resisted conventional structural-biology approaches, to obtain atomic models of multiple protein complexes implicated in intraerythrocytic survival of the parasite. Our approach is broadly applicable for determining structures of undiscovered protein complexes enriched directly from endogenous sources.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The atomic models and the cryoEM density maps are deposited to the Protein Data Bank and the Electron Microscopy Data Bank, with the accession numbers 6PEV, 6PEW, EMD-20333 and EMD-20334. For raw image data, please contact the corresponding author. The proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the MassIVE partner repository with the dataset identifier PDX014263.
Code availability
cryoID is an open source program under the MIT license, available for download at github (https://github.com/EICN-UCLA/cryoID).
References
Cheng, Y. Single-particle cryo-EM at crystallographic resolution. Cell 161, 450–457 (2015).
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013).
Liu, H. et al. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329, 1038–1043 (2010).
McMullan, G., Chen, S., Henderson, R. & Faruqi, A. R. Detective quantum efficiency of electron area detectors in electron microscopy. Ultramicroscopy 109, 1126–1143 (2009).
Clough, G. M. R. N. & Kirkland, A. I. in Journal of Physics: Conference Series Vol. 522 (IOP Publishing, 2013).
Zhang, X., Jin, L., Fang, Q., Hui, W. H. & Zhou, Z. H. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell 141, 472–482 (2010).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290-296 (2017).
Scheres, S. H. W. A bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012).
Scheres, S. H. W. RELION: implementation of a bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Ho, C. M. et al. Malaria parasite translocon structure and mechanism of effector export. Nature 561, 70–75 (2018).
Niedzialkowska, E. et al. Protein purification and crystallization artifacts: the tale usually not told. Protein Sci. 25, 720–733 (2016).
Osipiuk, J., Walsh, M. A. & Joachimiak, A. Crystal structure of MboIIA methyltransferase. Nucleic Acids Res. 31, 5440–5448 (2003).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 66, 213–221 (2010).
Porebski, P. J., Cymborowski, M., Pasenkiewicz-Gierula, M. & Minor, W. Fitmunk: improving protein structures by accurate, automatic modeling of side-chain conformations. Acta Crystallogr. D. Biol. 72, 266–280 (2016).
PDB statistics: overall growth of released structures per year, https://www.rcsb.org/stats/growth/overall (2018).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Bai, X. C. et al. An atomic structure of human gamma-secretase. Nature 525, 212–217 (2015).
Jin, P. et al. Electron cryo-microscopy structure of the mechanotransduction channel NOMPC. Nature 547, 118–122 (2017).
Zhou, R. et al. Recognition of the amyloid precursor protein by human gamma-secretase. Science 363, eaaw0930 (2019).
Gardner, M. J. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 (2002).
Hall, N. et al. A comprehensive survey of the plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 307, 82–86 (2005).
Waters, A. P. Genome-informed contributions to malaria therapies: feeding somewhere down the (pipe)line. Cell Host Microbe 3, 280–283 (2008).
Newby, Z. E. et al. Crystal structure of the aquaglyceroporin PfAQP from the malarial parasite Plasmodium falciparum. Nat. Struct. Mol. Biol. 15, 619–625 (2008).
Li, H. et al. Structure- and function-based design of Plasmodium-selective proteasome inhibitors. Nature 530, 233–236 (2016).
Xie, S. C. et al. The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis. Nat. Microbiol. 4, 1990–2000 (2019).
Sivaraman, K. K. et al. X-ray crystal structure and specificity of the Plasmodium falciparum malaria aminopeptidase PfM18AAP. J. Mol. Biol. 422, 495–507 (2012).
Eisenberg, D., Gill, H. S., Pfluegl, G. M. & Rotstein, S. H. Structure-function relationships of glutamine synthetases. Biochim. Biophys. Acta 1477, 122–145 (2000).
Gill, H. S. & Eisenberg, D. The crystal structure of phosphinothricin in the active site of glutamine synthetase illuminates the mechanism of enzymatic inhibition. Biochem.-Us 40, 1903–1912 (2001).
Asano, S., Engel, B. D. & Baumeister, W. In situ cryo-electron tomography: a post-reductionist approach to structural biology. J. Mol. Biol. 428, 332–343 (2016).
Beck, M. & Baumeister, W. Cryo-Electron tomography: can it reveal the molecular sociology of cells in atomic detail? Trends Cell Biol. 26, 825–837 (2016).
Albert, S. et al. Proteasomes tether to two distinct sites at the nuclear pore complex. Proc. Natl. Acad. Sci. USA 114, 13726–13731 (2017).
Mahamid, J. et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972 (2016).
Mosalaganti, S. et al. In situ architecture of the algal nuclear pore complex. Nat. Commun. 9, 2361 (2018).
Bai, R., Wan, R., Yan, C., Lei, J. & Shi, Y. Structures of the fully assembled saccharomyces cerevisiae spliceosome before activation. Science 360, 1423–1429 (2018).
Zhang, X. et al. Structures of the human spliceosomes before and after release of the ligated exon. Cell Res. 29, 274–285 (2019).
Liu, S. et al. Structure of the yeast spliceosomal postcatalytic P complex. Science 358, 1278–1283 (2017).
Agafonov, D. E. et al. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 351, 1416–1420 (2016).
Galej, W. P. et al. Cryo-EM structure of the spliceosome immediately after branching. Nature 537, 197–201 (2016).
Haselbach, D. et al. Structure and conformational dynamics of the human spliceosomal B(act) complex. Cell 172, 454–464 e411 (2018).
Nguyen, T. H. D. et al. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 A resolution. Nature 530, 298–302 (2016).
Kaiser, P. & Wohlschlegel, J. Identification of ubiquitination sites and determination of ubiquitin-chain architectures by mass spectrometry. Methods Enzymol. 399, 266–277 (2005).
Kelstrup, C. D., Young, C., Lavallee, R., Nielsen, M. L. & Olsen, J. V. Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. J. Proteome Res. 11, 3487–3497 (2012).
Tabb, D. L., McDonald, W. H. & Yates, J. R. 3rd DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 (2002).
Xu, T. et al. ProLuCID: an improved sequest-like algorithm with enhanced sensitivity and specificity. J. Proteom. 129, 16–24 (2015).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zhang, K. Gautomatch plugin (GitHub, 2016); https://github.com/scipion-em/scipion-em-gautomatch
Pettersen, E. F. et al. UCSF chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of coot. Acta Crystallogr. D. 66, 486–501 (2010).
Coordinators, N. R. Database resources of the national center for biotechnology information. Nucleic Acids Res. 44, D7–D19 (2016).
Aurrecoechea, C. et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37, D539–D543 (2009).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. 66, 12–21 (2010).
Schrodinger, L. L. C. The PyMOL Molecular Graphics System, Version 1.8 (2015).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EMEM density maps. Nat. Methods 11, 63-65 (2014).
Rotkiewicz, P. & Skolnick, J. Fast procedure for reconstruction of full-atom protein models from reduced representations. J. Comput. Chem. 29, 1460–1465 (2008).
Swint-Kruse, L. & Brown, C. S. Resmap: automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics 21, 3327–3328 (2005).
Acknowledgements
This research was supported in part by grants from National Institutes of Health (R01GM071940/AI094386/DE025567 to Z.H.Z. and K99/R00 HL133453 to J.R.B.). C.M.H. acknowledges funding from the Ruth L. Kirschstein National Research Service Award (AI007323). X.L. acknowledges funding from the China Scholarship Council (CSC). We thank the UCLA Proteome Research Center for assistance in mass spectrometry and acknowledge the use of resources in the Electron Imaging Center for Nanomachines supported by UCLA and grants from NIH (S10RR23057, S10OD018111 and U24GM116792) and NSF (DBI-1338135 and DMR-1548924).
Author information
Authors and Affiliations
Contributions
C.M.H., A.W.P.F. and Z.H.Z. initiated the project; J.R.B. cultured and harvested parasite material; C.M.H. purified the sample from parasite pellets, screened purified samples by negative stain, optimized sample freezing conditions for cryoEM, acquired and processed the cryoEM data, interpreted the structures, designed the endogenous structural proteomics workflow, helped design cryoID and wrote the paper; M.L. built and refined the atomic models and helped interpret the structures; J.A.W. performed the mass spectrometry; C.M.H., X.L. and M.L. designed the cryoID workflow. X.L. developed and benchmarked cryoID and helped write the paper. X.L. and C.M.H. worked with T.C.T. to write and optimize the Phenix tool sequence_from_map. Z.H.Z. supervised the cryoEM aspects of the project, interpreted the structures and wrote the paper; D.E.G. supervised parasitology aspects of the project. A.W.P.F., M.L., T.C.T., J.R.B., J.A.W. and D.E.G. helped edit the paper; all authors approved the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Allison Doerr was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Fig. 1 Evaluation of sucrose gradient fractions by SDS–PAGE and negative-stain EM.
a, Silver-stained SDS–PAGE of fractions from P. falciparum lysate sucrose gradient fractionation. b–e, Comparison of non-promising (b) and promising (d) negative-stain TEM images and their corresponding 2D class averages (c,e).
Supplementary Fig. 2
Manual inspection in Coot via the cryoID GUI.
Supplementary Fig. 3 Representative results from benchmarking of cryoID using simulated data.
a–d, The test results for query conditions (m,n) of 4,8 (a), 4,10 (b) 4,15 (c) and 4,30 (d) are shown. In each case, the top 10 candidates identified by cryoID are shown. The bar shown for each candidate represents 1,000 independent runs using unique sets of queries randomly generated from full length P. falciparum protein sequences (one-sided statistics test: P(f) < 0.005, n = 1,000, P value < 0.0067, α < 0.01). In each case, the bar shown for the correct protein is indicated with a star. The gap between the correct protein and the next closest match is indicated. The query condition shown in a is suboptimal, as cryoID identified multiple protein candidates that exhibit 100% identity with the queries. In optimal query conditions, shown in b–d, a clear gap is visible in percentage identity between the correct protein candidate (100% identity with queries) and the next closest matching candidate. This gap increases as m and n increase.
Supplementary Fig. 4 Endogenous cryoEM structures of the P. falciparum 20S proteasome in two conformations were obtained from the same proof-of-principle fraction analyzed and presented in our manuscript.
a–h, The first structure is of a full 20S proteasome containing all 14 α and all 14 β subunits (pink, a–d), while in the second structure (indigo, e–h), four β subunits appear to be disordered (the density corresponding to the β2 and β5 subunits is broken). Full surface front (a,e) and side (c,g) views and central slice front (b,f) and side (d,h) views of the two structures are shown. Models for the two structures are shown superposed with the maps in the central slice views. In g and h, a copy of the map has been low-pass filtered and is displayed as a translucent blue envelope, superposed over the unfiltered map, in order to show the disordered density.
Supplementary Fig. 5 Comparison of glutamine synthetase from P. falciparum (by endogenous cryoEM) and S. enterica (by X-ray crystallography).
The active sites, shown in light pink in both models, is well conserved. One region in which the two structures diverge is highlighted in red in the P. falciparum structure and green in the S. enterica structure.
Supplementary Information
Supplementary Information
Supplementary Figures 1–5, Supplementary Tables 1–4 and 6–7 and Supplementary Notes 1 and 2
Supplementary Table 5
Results of the mass spectrometry analysis of the sucrose gradient fractionated P. falciparum lysate.
Supplementary Video 1
Details of the Plasmodium falciparum M18 aspartyl aminopeptidase (PfM18AA) cryoEM Density Map. A 360° view of the cryoEM density map and atomic model of the PfM18AA dodecamer, including detailed cutaway views of the density map overlaid with the model.
Supplementary Video 2
Details of the Plasmodium falciparum glutamine synthetase (PfGS); cryoEM Density Map. A 360° view of the cryoEM density map and atomic model of the PfGS dodecamer, including detailed views of side-chain densities (mesh) superposed with the PfGS atomic model.
Rights and permissions
About this article
Cite this article
Ho, CM., Li, X., Lai, M. et al. Bottom-up structural proteomics: cryoEM of protein complexes enriched from the cellular milieu. Nat Methods 17, 79–85 (2020). https://doi.org/10.1038/s41592-019-0637-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0637-y
This article is cited by
-
Distinct evolution of type I glutamine synthetase in Plasmodium and its species-specific requirement
Nature Communications (2023)
-
Architecture of the baculovirus nucleocapsid revealed by cryo-EM
Nature Communications (2023)
-
Native structure of mosquito salivary protein uncovers domains relevant to pathogen transmission
Nature Communications (2023)
-
Structures and comparison of endogenous 2-oxoglutarate and pyruvate dehydrogenase complexes from bovine kidney
Cell Discovery (2022)
-
Model building of protein complexes from intermediate-resolution cryo-EM maps with deep learning-guided automatic assembly
Nature Communications (2022)