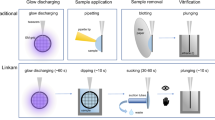
Abstract
Most protein particles prepared in vitreous ice for single-particle cryo-electron microscopy (cryo-EM) are adsorbed to air–water or substrate–water interfaces, which can cause the particles to adopt preferred orientations. By using a rapid plunge-freezing robot and nanowire grids, we were able to reduce some of the deleterious effects of the air–water interface by decreasing the dwell time of particles in thin liquid films. We demonstrated this by using single-particle cryo-EM and cryo-electron tomography (cryo-ET) to examine hemagglutinin, insulin receptor complex, and apoferritin.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
Single-particle half-maps, full sharpened maps, and masks for insulin-bound insulin receptor (200-ms or 600-ms spot-to-plunge time) and hemagglutinin (100 ms) have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-7788, EMD-7791, and EMD-7792, respectively. The full single-particle collection of hemagglutinin with 100-ms and 500-ms spot-to-plunge times has been deposited in the Electron Microscopy Pilot Image Archive (EMPIAR) with accession codes EMPIAR-10175 and EMPIAR-10197, respectively. Single-particle cryo-ET tomograms have been deposited in the EMDB with accession codes EMD-7623, EMD-7624, EMD-7625, EMD-7150, EMD-7627, EMD-7628, EMD-7629, and EMD-7630. Single-particle cryo-ET tilt series, cryo-ET tilt-series alignment runs with Appion-Protomo, cryo-ET tomograms, and apoferritin particle picks have been deposited in the EMPIAR with accession codes EMPIAR-10169, EMPIAR-10170, EMPIAR-10171, EMPIAR-10141, EMPIAR-10172, EMPIAR-10129, EMPIAR-10173, and EMPIAR-10174.
References
Nogales, E. Nat. Methods 13, 24–27 (2016).
Earl, L. A., Falconieri, V., Milne, J. L. & Subramaniam, S. Curr. Opin. Struct. Biol. 46, 71–78 (2017).
Taylor, K. A. & Glaeser, R. M. Science 186, 1036–1037 (1974).
Adrian, M., Dubochet, J., Lepault, J. & McDowall, A. W. Nature 308, 32–36 (1984).
Kühlbrandt, W. Science 343, 1443–1444 (2014).
Merk, A. et al. Cell 165, 1698–1707 (2016).
Noble, A. J. et al. eLife 7, e34257 (2018).
Glaeser, R. M. & Han, B.-G. Biophys. Rep. 3, 1–7 (2017).
Naydenova, K. & Russo, C. J. Nat. Commun. 8, 629 (2017).
Taylor, K. A. & Glaeser, R. M. J. Struct. Biol. 163, 214–223 (2008).
Feng, X. et al. Structure 25, 663–670 (2017).
Ashtiani, D. et al. J. Struct. Biol. 203, 94–101 (2018).
Jain, T., Sheehan, P., Crum, J., Carragher, B. & Potter, C. S. J. Struct. Biol. 179, 68–75 (2012).
Razinkov, I. et al. J. Struct. Biol. 195, 190–198 (2016).
Dandey, V. P. et al. J. Struct. Biol. 202, 161–169 (2018).
Wei, H. et al. J. Struct. Biol. 202, 170–174 (2018).
Scapin, G. et al. Nature 556, 122–125 (2018).
Tan, Y. Z. et al. Nat. Methods 14, 793–796 (2017).
Rice, W. J. et al. bioRxiv Preprint at https://www.biorxiv.org/content/early/2018/04/16/302018 (2018).
Zhang, K. J. Struct. Biol. 193, 1–12 (2016).
Grigorieff, N., Grant, T. & Rohou, A. bioRxiv Preprint at https://www.biorxiv.org/content/early/2018/01/31/257618 (2018).
Hu, M. et al. bioRxiv Preprint at https://www.biorxiv.org/content/early/2018/05/23/329169 (2018).
Stagg, S. et al. Microsc. Microanal. 11, 1072–1073 (2005).
Zheng, S. Q. et al. Nat. Methods 14, 331–332 (2017).
Voss, N. R., Yoshioka, C. K., Radermacher, M., Potter, C. S. & Carragher, B. J. Struct. Biol. 166, 205–213 (2009).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. Nat. Methods 14, 290–296 (2017).
Saxton, W. O., Baumeister, W. & Hahn, M. Ultramicroscopy 13, 57–70 (1984).
Suloway, C. et al. J. Struct. Biol. 167, 11–18 (2009).
Mastronarde, D. N. Microsc. Microanal. 9, 1182CD (2003).
Noble, A. J. & Stagg, S. M. J. Struct. Biol. 192, 270–278 (2015).
Winkler, H. & Taylor, K. A. Ultramicroscopy 106, 240–254 (2006).
Grant, T. & Grigorieff, N. eLife 4, e06980 (2015).
Agulleiro, J.-I. & Fernandez, J.-J. J. Struct. Biol. 189, 147–152 (2015).
Agulleiro, J. I. & Fernandez, J. J. Bioinformatics 27, 582–583 (2011).
Castaño-Díez, D., Kudryashev, M., Arheit, M. & Stahlberg, H. J. Struct. Biol. 178, 139–151 (2012).
Acknowledgements
The authors thank R. Glaeser at Lawrence Berkeley National Laboratory for helpful discussions, K. Kelley at the New York Structural Biology Center (NYSBC) for apoferritin purification, and the Electron Microscopy Group at NYSBC for microscope calibration and assistance. This work was supported by the NIH National Institute of General Medical Sciences (grant F32GM128303 to A.J.N.) and the Agency for Science, Technology and Research Singapore (Y.Z.T.). This work was performed at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the NYSBC, supported by grants from the Simons Foundation (SF349247), NYSTAR, and the NIH National Institute of General Medical Sciences (GM103310), with additional support from the Agouron Institute (F00316) and NIH (OD019994).
Author information
Authors and Affiliations
Contributions
A.J.N. collected and aligned tilt series and analyzed cryo-ET data. H.W. and V.P.D. performed the Spotiton experiments and the cryo-EM experiments. H.W. made nanowire grids and collected tilt series. H.W., V.P.D., and Y.Z.T. analyzed single-particle cryo-EM data. A.J.N., H.W., V.P.D., Z.Z., C.S.P., and B.C. conceived and designed the experiments. A.J.N. and B.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
B.C. and C.S.P. have a commercial relationship with TTP Labtech, a company that will produce a commercially available Spotiton instrument.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Particle distribution and concentrations of apoferritin (15 mg/mL = 20,530 particles/µm3 in solution) with 0.5 mM TCEP prepared using Spotiton and nanowire grids with spot-to-plunge time of 500 ms.
Particle picking and depictions were performed in Dynamo35. Left depiction is a side-view aligned roughly perpendicular to the air-water interfaces. Right depiction is a top-down view with a tomographic slice included. Particles were manually picked according to their estimated locations relative to the air-water interface and carbon. Calculated particle densities (particles/µm3) are: 1,668 non-adsorbed in the hole and 48,281 adsorbed to the air-water interfaces. Particle surface density at the air-water interface is 531 particles/µm2. Particle concentration calculations are estimated to be accurate to within 5%. Scale: 100 pixels is ~94 nm. n = 1 independent particle counting of this tomogram.
Supplementary Figure 2 3D reconstructions of hemagglutinin with varying plunge times.
From left to right: 3D reconstruction of hemagglutinin (C3 symmetry applied) from the given number of particles, orientation direction distribution plot (C1 symmetry applied), directional FSC plot (C3 symmetry applied). The directional FSC plots are created by calculating the FSC from 100 solid angles as a way to account for orientation when reporting resolution18; the histogram shows how many solid angles report a resolution in a particular range, which is reported as ‘Percentage of Per Angle FSC (%)’. (a) The full 100 ms spot-to-plunge time dataset of ~130,000 particles shows a relatively isotropic distribution, a sphericity of 0.895, and a resolution of 3.77 Å. (b) ~15,000 particles from the 100 ms spot-to-plunge time dataset (normalized to the number of particles available in the 500 ms dataset in (c)) shows a somewhat isotropic distribution, a sphericity of 0.795, and a resolution of 4.03 Å. (c) ~15,000 particles from the 500 ms spot-to-plunge time dataset shows a somewhat isotropic distribution, a sphericity of 0.771, and a resolution of 4.75 Å. (d) ~15,000 particles from the CP3-plunged sample shows a severely anisotropic distribution, an erroneous sphericity of 0.794, and an erroneous resolution of 3.41 Å. For a and b, n = 2 independent experiments. For c, n = 3 independent experiments. For d, n > 10 independent experiments.
Supplementary Figure 3 Single-particle cryo-EM of insulin-bound insulin receptor with spot-to-plunge times of 200 ms and 600 ms.
As shown in Figure 1, some side-views of insulin receptor are missing in the class averages of the 600 ms spot-to-plunge preparation. This resulted in a reduced sphericity of the directional FSC (0.811 vs. 0.849 for the 200 ms spot-to-plunge time) and worse resolution in the calculated map (6.09 Å vs. 4.93 Å for 200 ms). The greater anisotropy associated with for the longer spot-to-plunge time is reflected in the stretching artifacts in the 3D reconstruction. For a, n = 2 independent experiments. For b, n > 10 independent experiments.
Supplementary Figure 4 Particle distribution and concentrations of apoferritin (15 mg/mL = 20,530 particles/µm3 in solution) with 0.5 mM TCEP prepared using Spotiton and nanowire grids with spot-to-plunge time of 170 ms.
Left depiction is a side-view aligned roughly perpendicular to the air-water interfaces. Right depiction is a top-down view with a tomographic slice included. Particles were manually picked according to their estimated locations relative to the air-water interface and carbon. Calculated particle densities (particles/µm3) are: 31,725 in the hole and 57,878 adsorbed to the air-water interfaces. Particle surface density at the air-water interface is 637 particles/µm2. Particle concentration calculations are estimated to be accurate to within 5%. Scale: 100 pixels is ~94 nm. n = 1 independent particle counting of this tomogram.
Supplementary Figure 5 Particle distribution and concentrations of apoferritin (6 mg/mL = 8,212 particles/µm3 in solution) prepared using Spotiton and nanowire grids with spot-to-plunge time of 400 ms.
Left depiction is a side-view aligned roughly perpendicular to the air-water interfaces. Right depiction is a top-down view with a tomographic slice included. Particles were manually picked according to their estimated locations relative to the air-water interface and carbon. Calculated particle densities (particles/µm3) are: 3,043 in the hole, 195,199 adsorbed to the air-water interfaces, and 7,833 over or adsorbed to carbon. Particle surface density at the air-water interface is 5,092 particles/µm2. Particle concentration calculations are estimated to be accurate to within 5%. Scale: 100 pixels is ~86 nm. n = 1 independent particle counting of this tomogram.
Supplementary Figure 6 Particle distribution and concentrations of apoferritin (6 mg/mL = 8,212 particles/µm3 in solution) prepared using Spotiton and nanowire grids with spot-to-plunge time of 100 ms.
Left depiction is a side-view aligned roughly perpendicular to the air-water interfaces. Right depiction is a top-down view with a tomographic slice included. Particles were manually picked according to their estimated locations relative to the air-water interface and carbon. Calculated particle densities (particles/µm3) are: 17,927 in the hole, 268,423 adsorbed to the air-water interfaces, and 19,072 over or adsorbed to carbon. Particle surface density at the air-water interface is 1,735 particles/µm2. Particle concentration calculations are estimated to be accurate to within 5%. Scale: 100 pixels is ~86 nm. n = 1 independent particle counting of this tomogram.
Electronic supplementary material
Supplementary Text and Figures
Supplementary Figures 1–6
Supplementary Video 1
Tomogram slice-through of hemagglutinin prepared using Spotiton and nanowire grids with spot-to-plunge time of 800 ms, alongside a cross-sectional depiction. The vast majority of particles are observed to be adsorbed to the air–water interfaces with severe preferred orientation. Slice-through videos were rendered using ‘Slicer’ 3dmod from the IMOD package36 by orienting one plane of particles at the air–water interface to be roughly parallel to the field of view. n = 4 independent experiments.
Supplementary Video 2
Tomogram slice-through of insulin-bound insulin receptor prepared using Spotiton and nanowire grids with spot-to-plunge time of 600 ms, alongside a cross-sectional depiction. The vast majority of particles are observed to be adsorbed to the air–water interfaces. n = 2 independent experiments.
Supplementary Video 3
Tomogram slice-through of apoferritin with 0.5 mM TCEP (15 mg/mL) prepared using Spotiton and nanowire grids with spot-to-plunge time of 500 ms, alongside a cross-sectional depiction. Most particles are observed to be adsorbed to the air–water interfaces, with a few nonadsorbed particles. n = 3 independent experiments.
Supplementary Video 4
Tomogram slice-through of hemagglutinin prepared using Spotiton and nanowire grids with spot-to-plunge time of 100 ms, alongside a cross-sectional depiction. A majority of the particles are observed to be adsorbed to the air–water interfaces. n = 2 independent experiments.
Supplementary Video 5
Tomogram slice-through of insulin-bound insulin receptor prepared using Spotiton and nanowire grids with spot-to-plunge time of 200 ms, alongside a cross-sectional depiction. n = 3 independent experiments.
Supplementary Video 6
Tomogram slice-through of apoferritin with 0.5 mM TCEP (15 mg/mL) prepared using Spotiton and nanowire grids with spot-to-plunge time of 170 ms, alongside a cross-sectional depiction. Approximately 19 times as many nonadsorbed particles are in the field of view as compared to the spot-to-plunge time of 500 ms (Supplementary Video 3). n = 3 independent experiments
Supplementary Video 7
Tomogram slice-through of apoferritin (6 mg/mL) prepared using Spotiton and nanowire grids with spot-to-plunge time of 400 ms, alongside a cross-sectional depiction. Most of the particles are observed to be adsorbed to the air–water interfaces, with a few nonadsorbed particles. n = 5 independent experiments.
Supplementary Video 8
Tomogram slice-through of apoferritin (6 mg/mL) prepared using Spotiton and nanowire grids with spot-to-plunge time of 100 ms, alongside a cross-sectional depiction. Approximately six times as many nonadsorbed particles are in the field of view as compared to the spot-to-plunge time of 400 ms (Supplementary Video 7). n = 6 independent experiments.
Rights and permissions
About this article
Cite this article
Noble, A.J., Wei, H., Dandey, V.P. et al. Reducing effects of particle adsorption to the air–water interface in cryo-EM. Nat Methods 15, 793–795 (2018). https://doi.org/10.1038/s41592-018-0139-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-018-0139-3
This article is cited by
-
Anaerobic cryoEM protocols for air-sensitive nitrogenase proteins
Nature Protocols (2024)
-
Metallo-supramolecular branched polymer protects particles from air-water interface in single-particle cryo-electron microscopy
Communications Biology (2024)
-
Directly visualizing individual polyorganophosphazenes and their single-chain complexes with proteins
Communications Materials (2024)
-
Structural insights into intron catalysis and dynamics during splicing
Nature (2023)
-
Structures of human primosome elongation complexes
Nature Structural & Molecular Biology (2023)