Abstract
Research and practice in critical care medicine have long been defined by syndromes, which, despite being clinically recognizable entities, are, in fact, loose amalgams of heterogeneous states that may respond differently to therapy. Mounting translational evidence—supported by research on respiratory failure due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection—suggests that the current syndrome-based framework of critical illness should be reconsidered. Here we discuss recent findings from basic science and clinical research in critical care and explore how these might inform a new conceptual model of critical illness. De-emphasizing syndromes, we focus on the underlying biological changes that underpin critical illness states and that may be amenable to treatment. We hypothesize that such an approach will accelerate critical care research, leading to a richer understanding of the pathobiology of critical illness and of the key determinants of patient outcomes. This, in turn, will support the design of more effective clinical trials and inform a more precise and more effective practice at the bedside.
Similar content being viewed by others
Main
A 66-year-old woman is admitted to the intensive care unit (ICU) with fever, cough and difficulty breathing. She is diagnosed with pneumonia, intubated and placed on mechanical ventilation. The following day, her chest X-ray reveals bilateral infiltrates, and arterial blood gas analysis shows severe hypoxemia. Her treating clinicians consider what to do next.
Were this patient admitted in 2019, her management might have been beset by more questions than answers. She has both sepsis, a syndrome of life-threatening organ dysfunction in the face of infection, and acute respiratory distress syndrome (ARDS), a syndrome of respiratory failure associated with lung injury and impaired gas exchange. Both of these syndromes have been the subject of many epidemiological and interventional studies, yet little of the resulting evidence is clinically actionable. There are no specific treatments for her sepsis beyond antmicrobials1, and the ventilation strategies used to treat ARDS might reasonably be applied to any patient in the ICU2.
Were she admitted today—and depending on geography and time of year—her condition might well be the result of critical Coronavirus Disease 2019 (COVID-19). She would still meet diagnostic criteria for both sepsis and ARDS and would ostensibly face a similar degree of therapeutic uncertainty. However, in the last few years, many large randomized trials have provided a wellspring of evidence suggesting that a patient in her condition is likely to benefit from corticosteroids3 and interleukin-6 receptor antagonists4,5 but that treatments for milder disease, including remdesivir6 and systemic anticoagulation7, are unlikely to provide substantial benefit. To the great relief of many, the once arid landscape of clinical evidence in critical care has begun to germinate.
In what follows, we examine how advances in translational critical care brought us to this inflection point in our field and how these advances stand to fundamentally alter the way that we conceptualize and classify critical illness.
A new era in translational critical care research
The field of critical care medicine can be described by three stages of development (Fig. 1). In the first stage (‘Foundations’, c. 1955–1980s), mechanical ventilation and continuous monitoring of physiologic parameters were introduced to the care of the critically ill, along with higher nurse-to-patient ratios, standardized practices and an emerging recognition of critical care as a standalone medical specialty. These technological advances provided the basis for a physiology-based understanding of the host response to injury and saved the lives of patients who might otherwise have died. Critical illness was defined as organ-level pathophysiology (for example, shock and respiratory failure), and the delivery of intensive care services was centered on maintaining organ-level homeostasis (for example, assisted breathing and circulatory support).
The first era, Foundations, spans from the founding of the discipline in the 1950s and 1960s to roughly the mid-1980s. In the second era, Acceleration, critical illness was better characterized through formal syndrome definitions and quantitative descriptions of illness severity. Outcomes improved, although few clinical studies yielded actionable results. A third era, Precision, is now emerging, based on a growing body of translational findings that reveal substantial biological heterogeneity within current critical care disease concepts. Parsing this heterogeneity to identify precise mechanisms of disease—along with ways to identify these clinically—will lead to more precise treatments and greater efficiency of care. Delineating these mechanisms and translating them to practice will be central tasks in critical care research in the coming decades. CVP, central venous pressure.
A second stage of development in the critical care field (‘Acceleration’, c. 1980s–2020) arose alongside advances in translational research that proffered an improved understanding of the pathophysiology of the host response. In this era, the field acquired structure, with the advent of quantitative scoring systems and standardized syndrome definitions. These included the APACHE score8 as well as definitions for systemic inflammatory response syndrome (SIRS), sepsis9 and ARDS10. Together, these laid the groundwork for rigorous clinical and translational studies, which, in combination with better organization and inter-disciplinary collaboration, led to tremendous improvements in outcomes for critically ill patients.
In recent years, emerging evidence has begun to suggest that, although initially useful in research and practice, current disease concepts do not sufficiently capture the full complexity of critical illness11,12. Advances in -omics science, data science and machine learning have generated evidence of heterogeneity in common ICU syndromes. Gene expression data from the blood of both pediatric and adult patients with sepsis have been used with hierarchical clustering algorithms to discover and validate distinct subsets of patients with shared transcriptomic responses to severe infection13,14,15,16,17,18,19. Similarly, latent class analysis (another statistical method to identify subgroups in populations) has been used with clinical and biomarker data from patients with ARDS to reveal hypo-inflammatory and hyper-inflammatory subtypes20,21,22. These findings clearly resonate with the day-to-day experience of clinicians caring for critically ill patients who, despite sharing common diagnoses, nonetheless exhibit substantial variability in clinical course and outcome19,20,22,23,24,25. There is an increasingly compelling need to reconsider the prevailing approach to the classification of critical illness26,27,28.
Critical care medicine is now on the cusp of a sea change—a third phase of development (‘Precision’; Fig. 1) defined by advances in translational science. This phase stands to be more disruptive than those preceding and will require a wholesale reconfiguration of existing classification frameworks.
Critical illness syndromes
Most of the illnesses treated in the ICU are clinical syndromes. Conditions like sepsis, ARDS, acute kidney injury, delirium and even chronic critical illness are characterized not by any particular biopsy feature, genetic mutation, microbial culture or serologic test but, rather, by collections of signs and symptoms that together paint the picture of a clinically recognizable entity. As a result, critical illness syndromes are heterogeneous by nature. For instance, sepsis can arise from a multitude of infections, caused by many different pathogens and resulting in different patterns of organ injury. ARDS may arise from either pulmonary triggers (such as pneumonia or aspiration) or non-pulmonary triggers (such as trauma or pancreatitis), and delirium may manifest as both agitation and somnolence. There is also temporal heterogeneity; a patient meeting diagnostic criteria for one syndrome at a given time may progress through different, often disparate, phases. Added to this is the tremendous heterogeneity in the host response to injury from one individual to the next.
Despite their limitations, syndromes enable the objective and reproducible assembly of patient cohorts and, as such, are useful in research and quality improvement. Syndromes can also be ‘prognostic’, meaning they can be used to estimate the likelihood of an outcome. For example, the current clinical criteria for septic shock are associated with a risk of death in excess of 40%29. These criteria do not, however, identify which patients are likely to respond to any specific treatment. Classifiers that exhibit this latter function are often called ‘predictive’. For example, coagulopathy due to thrombocytopenia is likely to improve with platelet transfusion, whereas that which is due to dysfibrinogenemia is not. This inherent limitation in the syndrome-based classification of critical illness arises because current criteria are based on clinical findings rather than on the underlying biological processes that give rise to them. An important question, therefore, is whether our current syndrome-based classification schema is fit for purpose and whether a new approach is needed.
A translational classification of critical illness
Illness classifications have been proposed and revised since antiquity, but, for the most part, the essential components have changed very little. An early taxonomy developed by Linnaeus in the 18th century bears striking resemblance to modern schemas such as the International Classification of Diseases (ICD) system, whereby individual diseases are specified on the basis of signs and symptoms, and the relationships between them are delineated, often as a nested hierarchy.
Important conceptual advances have, nonetheless, been made (Box 1). The TNM staging system in oncology has been useful in framing cancer not as a single disease but as a collection of related conditions whose optimal treatment depends on the extent of their progression. Adapting this concept to the ICU, the PIRO model (predisposition, insult, response, organ dysfunction) was proposed to underscore the notion that response to treatment is affected by more than whether certain syndromic criteria are met; a patient’s outcome is also strongly influenced by their baseline physiology, the nature of the precipitating insult and the way in which various organ systems respond30.
The PIRO model was an important early step toward acknowledging heterogeneity in critical illness. But translational and clinical evidence accrued in the last decade has deepened understanding of the complexity of critical illness and its biological determinants, compelling us to revisit the nosology of critical care. To best capitalize on these discoveries, a new framework must accommodate complexity and heterogeneity and must also establish a closer correspondence between diagnosis and treatment. In other words, critical illness classification should be not only prognostic, as syndromes are, but predictive as well, allowing researchers and practitioners to focus on measures that stand to improve outcomes.
Conceptually, a new classification system should encompass the inciting illness event, the physiologic disturbances produced and the treatments that could return the affected system(s) to a state of health. We advance a new concept here that begins with insults—events that instigate an acute departure from some baseline level of homeostasis, with the potential to elicit critical illness. Insults are myriad and diverse. Infection, trauma, stroke, hemorrhage, overdose and major surgery—all of these represent an abrupt change in baseline physiology, and all are common reasons for ICU admission. Insults give rise to perturbations in bodily systems that, in turn, lead to disease states, organ dysfunction and clinically overt morbidity.
The basis of this model is a more direct correspondence between insults and the pathophysiologic states they engender. This is achieved by placing the insult, along with its physiologic consequences and potential treatments, in a causal pathway. Causality is a key feature here and an important change from current syndromic classifications. For example, although we know that fluids will generally be helpful in septic shock, and low driving pressures during mechanical ventilation will be helpful in ARDS, the heterogeneity of these conditions limits the causal inferences that can be made, thereby hindering the clinical actionability of these principles in the treatment of any individual patient.
To enhance the precision of diagnosis in critical care, we invoke the concept of a treatable trait—a specific physiologic derangement characterized by biomarkers that portend a predictable response to a particular therapy31. Although biomarkers are often understood to refer to specialized laboratory tests, usually from blood or tissue, our use of the term here is more broadly construed. In the context of a treatable trait, we use the term ‘biomarker’ to mean any observable trait that corresponds with the biological abnormality of interest and that underpins a prediction around how a patient will respond to treatment. As such, biomarkers may include transcriptomic features derived from RNA sequencing, virulence factors identified by pathogen genomics, features seen on advanced imaging studies or even imbalances in the autonomic nervous system identified by millisecond-scale changes in heart rate variability. They may also include simple and routinely measured clinical variables, such as oxygen saturation, hemoglobin levels and glucose concentrations, which currently serve as usable biomarkers by enabling predictions about the effects of oxygen titration, transfusion and insulin therapy, respectively. The particular modality used is of secondary importance; what matters is that the trait can be measured, that it corresponds with the insult or physiological process causing harm and that it can be linked to treatment response.
Evidence suggests that disparate insults may give rise to shared molecular patterns of injury. Influential work by the Inflammation and the Host Response to Injury (NCT00257231) program replicated clinical observations of pathophysiologic similarities across critical illness syndromes, by showing that molecular signatures in trauma and burn injuries include activation of similar infection-related and inflammation-related pathways32. This work has recently been extended, revealing molecular similarities between bacterial sepsis and COVID-19 viral sepsis33 as well as between ARDS and pancreatitis34. These observations suggest that some signals might be generalizable across different forms of critical illness, precipitated by very different insults.
Such findings hint at a previously uncharacterized richness in the biological determinants of critical illness. Rather than a one-to-one correspondence between insult and disease state, a one-to-many relationship or even a many-to-many relationship is likely more appropriate. As traditional hierarchical models of classification cannot easily represent such a system, we offer the circular model shown in Fig. 2 to depict the precise biological processes that characterize a disease mechanism shared among different illness states, irrespective of the insult from which they arise. This configuration better accommodates the complexity of critical illness by acknowledging that certain states may be reached through different causal paths, and that, although the insult itself is important, it is the resultant physiologic state that may better characterize a patient’s current status.
Individual insults and biological abnormalities are combined in a circular model that accommodates connections between entities. In this example, four insults are portrayed (infection, trauma, surgery and pancreatitis). The same biological abnormality (represented by interconnecting bands) can arise from multiple different insults; for example, certain shared inflammation-mediated pathways may underpin each of the four insults shown.
To illustrate the potential utility of a model thus construed, consider the role of Toll-like receptor (TLR) signaling in critical illness. TLR pathways contribute to the inflammatory response and are known to be activated by various triggers, both exogenous (for example, bacterial endotoxin) and endogenous (for example, heme and hyaluronic acid)35. Indeed, upregulation of TLR pathways has been identified through gene expression profiling in the settings of both trauma32 and sepsis36. However, given the heterogeneity of these clinical syndromes—as well as differences in the genetic determinants of the immune response to TLR activation37—the extent of TLR-mediated inflammation likely varies among patients. This biological heterogeneity may, in part, explain why inhibiting TLR-mediated inflammation does not appear to be an effective treatment for cohorts defined by diagnostic criteria for severe sepsis38. We might, however, hypothesize that this approach will be helpful in a subset of patients with sepsis with more pronounced dysregulation of TLR signaling. Moreover, we might also hypothesize that a subset of trauma patients who manifest maladaptive TLR pathway upregulation will also benefit from this approach, even though their illness state arose from a different insult. Answering this question would require a clinical trial in which patients are prospectively enrolled based on a treatable trait—in this case TLR upregulation—rather than on a clinical syndrome, such as sepsis or trauma.
TLR signaling may also play an important role in the host response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Rapid whole-exome sequencing of probands with COVID-19 have identified deletions in the TLR7 gene that were associated with an extreme critical illness phenotype39. Although TLR signaling is implicated here as well, the nature of the derangement is different; loss-of-function variants lead to an impaired interferon-mediated response to the virus, and subsequent critical COVID-19. Rather than a TLR antagonist, we might reasonably hypothesize that a TLR agonist (such as imiquimod) would be effective in these cases. This would be a different treatable trait, one that might be shared by other conditions, including certain skin cancers40.
The conceptual critical care model we describe here has yet to be validated in prospective clinical trials; doing so will require studies that recruit patients based on treatable traits rather than syndrome criteria. However, early evidence for the feasibility and efficacy of this approach is mounting. For example, in oncology, the I-SPY platform uses molecular profiling of breast tumors to identify specific subtypes most likely to respond to certain treatments, such as the tyrosine kinase inhibitor neratinib41. This approach, often called predictive enrichment, is used to evaluate many breast cancer subtypes derived from tumor gene expression data, often coupled with adaptive randomization, a type of treatment allocation strategy that adjusts the randomization ratios according to interim results. The I-SPY consortium has recently expanded to launch I-SPY COVID, a phase 2 clinical trial platform designed to use adaptive randomization to rapidly evaluate the viability of new COVID-19 therapies, with those deemed potentially viable graduated to larger definitive trials42.
Within critical care, randomized trials are beginning to explore the use of predictive enrichment to reduce the heterogeneity of treatment effect seen when recruitment is based strictly on syndromic criteria. One example is the EUPHRATES study, which examined the use of polymyxin B hemoperfusion in patients with septic shock43. This therapy is designed to remove bacterial endotoxin from the circulation, and so, rather than enrolling all patients meeting syndrome criteria for septic shock, the investigators randomized only those patients with high baseline levels of circulating endotoxin. The EUPHRATES experience demonstrates the feasibility of using a biomarker to rapidly identify a specific subgroup of patients expected to be most responsive to treatment. It also illustrates the challenges in identifying treatable traits. With no difference in mortality seen between the treatment and placebo arms, this study highlights the importance of defining appropriate subgroups, developing predictive biomarkers and devising realistic measures of treatment response.
In many ways, recent COVID-19 clinical trials have also demonstrated the potential viability of using a treatable trait concept to disambiguate critical illness syndromes and increase the yield of actionable evidence. The role of corticosteroids in treating ARDS remains uncertain, but many patients with ARDS arising from COVID-19 appear to respond favorably to this treatment3. Here, a positive polymerase chain reaction (PCR) test for the SARS-CoV-2 virus might be seen as a biomarker for a subtype of ARDS with a greater than average likelihood of responding favorably to corticosteroid therapy. Adding further nuance is the predictive importance of dynamic patient factors, such as timing with respect to the initial insult and the severity of the resulting illness. Corticosteroids for COVID-19 appear to be most effective in those who are sickest and when given at a later phase of illness. With the success of the RECOVERY3 and REMAP-CAP4 studies, COVID-19 research also increased our familiarity with adaptive randomization.
In proposing this modernized conceptual model of critical illness, we hasten to add some potential limitations and nuances. First, although the model has direct implications for treatment, it leaves prognosis largely unchanged. Age, for example, may not be a treatable trait, but it is prognostic in most conditions. That said, critical care has no shortage of prognostic models, both general and disease-specific, that fulfil this function well.
Second, although we emphasize some of the key molecular findings that have shown promise in critical care, the critical illness concept proposed here by no means requires that a treatable trait be a molecular or genomic trait. Despite an increasing emphasis on molecular techniques in translational critical care research, there are no guarantees that increasing granularity will lead to tangible gains. Any feature that distinguishes a specific pathophysiologic process with causal links to treatment effects can serve this function.
Third, the discussion of a new conceptual model of critical illness raises some questions as to the fate of the critical illness syndromes that have, for decades, steered the field through a period of remarkable advancement. These are bedrock concepts in the modern ICU, and they are deeply ingrained in our systems of prognostication, record-keeping, disease surveillance, epidemiology, administration, quality improvement and research. It remains to be seen whether the field is ready for a wholesale shift away from syndromes or whether they will be retained in some capacity.
Lastly, the model proposed here is but one among many possible ways forward. Although we think that the principles outlined above address many of the challenges facing critical care, our overarching objective is to bring these challenges to light and suggest how progress might be made in addressing them.
The next phase of critical care
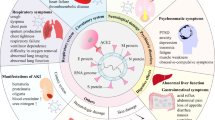
Upon arrival in the ICU, our patient is found to have a PCR-positive nasopharyngeal swab for the SARS-CoV-2 virus, worsening hypoxemia, decreased urine output and confusion. An echocardiogram reveals mild left ventricular dysfunction, and her D-dimer levels (a marker of blood clotting) are markedly elevated. By current standards, we might diagnose several syndromes—ARDS, sepsis, acute kidney injury, delirium and disseminated intravascular coagulation—each of which may be treated with different types of supportive care. These treatments may conflict with one another, and the lack of precision in our diagnoses makes it difficult to predict how she will respond to any of them.
A new conceptual model developed on the principles described above would support a more efficient approach in which syndrome labels are de-emphasized in favor of more precise biological descriptors. Genome sequencing may reveal that our patient has an allelic variant that puts her at much higher risk of severe lung inflammation than age-matched and sex-matched counterparts with the same presentation44,45. Transcriptome profiling could reveal her organ dysfunction to be largely the result of TNF/IL-1-mediated inflammation46, with little contribution from microvascular thrombosis. Heart rate variability analysis may reveal changes in autonomic function that portend delirium47. Moreover, these pathophysiologic features might not be confined to COVID-19 alone and may be seen in critical illness states arising from entirely different insults. These features will be understood as treatable traits, evoking a specific therapeutic course. The genetic polymorphism may be targeted with a known pharmacologic agent, she may be more likely to benefit from the inhibition of certain inflammatory pathways, and a sympatholytic medication may prove better than an anti-psychotic at preventing and treating agitation.
How do we get there?
The gulf between aspiration and achievement is wide. Many share the conviction that we need to move beyond syndromic characterization of the diseases of critical illness and to develop disease models based on shared biology48,49,50,51. Position papers and consensus conferences will be useful in cultivating and refining key concepts. But meaningful progress will also require concerted effort directed toward technical considerations as well. An overall approach to addressing the challenges is shown in Fig. 3 and must focus on theoretical and practical considerations across a range of key domains.
At the top of the figure, the circular model shows how different insults can give rise to shared biological abnormalities, with each gray triangle representing a patient with a specific insult. To characterize the patient response to injury, samples are collected at various times (blue dots) and used to generate biological characteristics. Tests may include blood tests, physiologic waveforms and imaging studies, as well as genomic, transcriptomic and proteomic profiling, and may be added to existing data such as age, comorbidities, environmental factors and functional status. The heat map depicts the clustering of these data to identify physiologic states of interest, which may be used to place patients into cohorts or to describe any single patient along a temporal trajectory of injury response. Note that each patient, when assessed at multiple points, may remain in an unchanged physiologic state or move to another. Unsupervised machine learning and other statistical techniques are used for subtype discovery, with supervised machine learning deployed to identify potential biomarkers. These are developed into tests that can be used at the point of care, including as an enrichment strategy for recruitment into prospective trials. Endpoints that directly reflect the response to treatment are defined and may include proximal outcomes that can be located in a causal pathway with the treatment. A physiologic state of interest and its corresponding predictive biomarker constitute a ‘treatable trait’, which, upon demonstration of efficacy in clinical trials, can be integrated into clinical care pathways.
Basic science
The concept of a ‘treatable trait’ generally implies that the underlying mechanism is understood and that the treatment relates to the mechanism. Thus, detailed preclinical work aimed at mechanistic understanding of putative treatable traits must be undertaken in earnest .
Biomarker development
On a practical level, operationalizing the treatable trait concept will, in some cases, necessitate the development of novel biomarkers that can be used in the ICU environment. This will require close collaboration with clinical chemists and laboratory experts to create validated assays that can be run in a clinical lab, respecting both the multifocal nature of critical care and the rapid turnaround times needed to inform decision-making. Assays run on readily available samples such as blood, urine, exhaled gases or even physiologic signals are more likely to be adopted than more invasive assays, such as tissue biopsies. Similarly, tests based on faster modalities, such as PCR or molecular barcoding platforms, will see greater uptake than more cumbersome sequencing technologies. Developing viable biomarker assays will involve addressing many hurdles, including identifying physiologically important disease states, describing the appropriate clinical interpretation of test results and satisfying regulatory requirements. Entirely new technologies will undoubtedly be explored to meet the exigencies of finding biomarkers of treatable traits in the ICU.
Outcome measures
Outcomes must be devised that can readily determine whether biomarker-informed treatment has been effective. Current outcomes such as mortality, organ support-free days and coarse measures of neurologic function may lack the necessary specificity to adjudicate the success of a given treatment. For instance, a patient with COVID-19 may respond favorably to corticosteroids, only to succumb later to a pulmonary embolism or bacterial coinfection. We must consider the relative importance of intermediate outcomes as well as outcomes that may not be considered patient-centered by current standards52.
Data integration
The noise resulting from large numbers of variables, the confounding effects of differing approaches to treatment and healthcare delivery and the diminishing realistic size of individual effects all argue for the integration of data on a grand scale and over a sustained period of time. Data from electronic health records, next-generation sequencing and multi-omics biology provide the substrate, and data science and enhanced statistical and machine learning approaches provide the methods. The precedents set by the Framingham Heart Study53, the Human Genome Project54 or the insights in particle physics generated by the large Hadron collider all speak to the power of the creation, curation and sharing of large amounts of data.
Novel trial designs
Causal inference is challenged by confounding. Randomization provides the most reliable means of reducing confounding, thereby establishing causality. Large randomized clinical trials, therefore, provide powerful but under-used opportunities for causal inference, whereas emerging methods, such as Mendelian randomization55, enable more robust inferences of causality from random biologic variability. The use of platform trials, which incorporate adaptive designs that evaluate multiple treatments, has shown promise in efficiently weighing the effectiveness of multiple different treatments and can accommodate heterogeneity in the study population56, as evidenced by the success of the RECOVERY and REMAP-CAP studies3,7.
National and international collaboration of investigator-led research consortia
Large-scale, multi-national and multi-institutional collaborations such as CERN, LIGO and the Human Genome Project are becoming more common. The move toward open science and the creation of shared data repositories emphasize the will and provide the platforms for collaboration. Collaboration among national clinical research groups is increasing in areas such as emerging infectious diseases, cancer, stroke and thrombosis. In critical care, the International Forum for Acute Care Trialists (InFACT) has provided a forum for early discussions on the staging and stratification of critical illness. Collaboration at the scale needed to address the challenge is becoming possible.
Conclusion
The management of patients with cancer was transformed in 1933 by the creation of the Union for International Cancer Control and by the development of the TNM staging system, first proposed by Pierre Denoix in the 1940s57,58. The treatment of cardiovascular disease has been shaped by the Framingham Heart Study, with its comprehensive characterization of the natural history of a disease over time53. A similar approach will be needed to reframe critical illness. Owing to the rapid changes and multi-organ manifestations seen in critical illness, it is likely to be more complicated and to take a correspondingly greater effort than the precedents of oncology and cardiology. It is achievable, but it will require collaboration at a global scale—in reaching agreement on terminology and approaches to taxonomy, in creating shared data repositories to test and validate models and in incorporating models into randomized trials to evaluate causal inference. For all the upheaval it has created, COVID-19 has shown that such an aspiration in global research collaboration is not only desirable but also possible.
References
Marshall, J. C. Why have clinical trials in sepsis failed? Trends Mol. Med. 20, 195–203 (2014).
Writing Group for the PReVENT Investigators. Effect of a low vs intermediate tidal volume strategy on ventilator-free days in intensive care unit patients without ARDS: a randomized clinical trial. JAMA 320, 1872–1880 (2018).
The Recovery Collaborative Group. Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 384, 693–704 (2021).
REMAP-CAP Investigators et al. Interleukin-6 receptor antagonists in critically ill patients with Covid-19. N. Engl. J. Med. 384, 1491–1502 (2021).
WHO Rapid Evidence Appraisal for COVID-19 Therapies (REACT) Working Group et al. Association between administration of IL-6 antagonists and mortality among patients hospitalized for COVID-19: a meta-analysis. JAMA 326, 499–518 (2021).
Beigel, J. H. et al. Remdesivir for the treatment of Covid-19—final report. N. Engl. J. Med. 383, 1813–1826 (2020).
REMAP-CAP Investigators et al. Therapeutic anticoagulation with heparin in critically ill patients with Covid-19. N. Engl. J. Med. 385, 777–789 (2021).
Knaus, W. A., Draper, E. A., Wagner, D. P. & Zimmerman, J. E. APACHE II: a severity of disease classification system. Crit. Care Med. 13, 818–829 (1985).
Bone, R. C. et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest 101, 1644–1655 (1992).
Bernard, G. R. et al. Report of the American-European consensus conference on ARDS: definitions, mechanisms, relevant outcomes and clinical trial coordination. The Consensus Committee. Intensive Care Med. 20, 225–232 (1994).
Leligdowicz, A. & Matthay, M. A. Heterogeneity in sepsis: new biological evidence with clinical applications. Crit. Care 23, 80 (2019).
Prescott, H. C., Calfee, C. S., Thompson, B. T., Angus, D. C. & Liu, V. X. Toward smarter lumping and smarter splitting: rethinking strategies for sepsis and acute respiratory distress syndrome clinical trial design. Am. J. Respir. Crit. Care Med. 194, 147–155 (2016).
Wong, H. R. et al. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 7, 34 (2009).
Wong, H. R. et al. Genomic expression profiling across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum. Crit. Care Med. 37, 1558–1566 (2009).
Wong, H. R., Freishtat, R. J., Monaco, M., Odoms, K. & Shanley, T. P. Leukocyte subset-derived genomewide expression profiles in pediatric septic shock. Pediatr. Crit. Care Med. 11, 349–355 (2010).
Wong, H. R. et al. Toward a clinically feasible gene expression-based subclassification strategy for septic shock: proof of concept. Crit. Care Med. 38, 1955–1961 (2010).
Wong, H. R. et al. Validation of a gene expression-based subclassification strategy for pediatric septic shock. Crit. Care Med. 39, 2511–2517 (2011).
Maslove, D. M., Tang, B. M. & McLean, A. S. Identification of sepsis subtypes in critically ill adults using gene expression profiling. Crit. Care 16, R183 (2012).
Sweeney, T. E. et al. Unsupervised analysis of transcriptomics in bacterial sepsis across multiple datasets reveals three robust clusters. Crit. Care Med. 46, 915–925 (2018).
Calfee, C. S. et al. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir. Med. 2, 611–620 (2014).
Calfee, C. S. et al. Distinct molecular phenotypes of direct vs indirect ARDS in single-center and multicenter studies. Chest 147, 1539–1548 (2015).
Famous, K. R. et al. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am. J. Respir. Crit. Care Med. 195, 331–338 (2017).
Cohen, M. J. et al. Identification of complex metabolic states in critically injured patients using bioinformatic cluster analysis. Crit. Care 14, R10 (2010).
Davenport, E. E. et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir. Med. 4, 259–271 (2016).
Scicluna, B. P. et al. Classification of patients with sepsis according to blood genomic endotype: a prospective cohort study. Lancet Respir. Med. 5, 816–826 (2017).
Reilly, J. P., Christie, J. D. & Meyer, N. J. Fifty years of research in ARDS. Genomic contributions and opportunities. Am. J. Respir. Crit. Care Med. 196, 1113–1121 (2017).
Seymour, C. W. et al. Precision medicine for all? Challenges and opportunities for a precision medicine approach to critical illness. Crit. Care 21, 257 (2017).
Calfee, C. S. Opening the debate on the new sepsis definition. Precision medicine: an opportunity to improve outcomes of patients with sepsis. Am. J. Respir. Crit. Care Med. 194, 137–139 (2016).
Singer, M. et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 315, 801–810 (2016).
Marshall, J. C. The PIRO (predisposition, insult, response, organ dysfunction) model: toward a staging system for acute illness. Virulence 5, 27–35 (2014).
Russell, C. D. & Baillie, J. K. Treatable traits and therapeutic targets: goals for systems biology in infectious disease. Curr. Opin. Syst. Biol. 2, 140–146 (2017).
Xiao, W. et al. A genomic storm in critically injured humans. J. Exp. Med. 208, 2581–2590 (2011).
Sweeney, T. E. et al. Validation of inflammopathic, adaptive, and coagulopathic sepsis endotypes in Coronavirus Disease 2019. Crit. Care Med. 49, e170–e178 (2021).
Neyton, L. P. A. et al. Molecular patterns in acute pancreatitis reflect generalizable endotypes of the host response to systemic injury in humans. Ann. Surg. 275, e453–e462 (2022).
Lorne, E., Dupont, H. & Abraham, E. Toll-like receptors 2 and 4: initiators of non-septic inflammation in critical care medicine? Intensive Care Med. 36, 1826–1835 (2010).
Maslove, D. M. & Wong, H. R. Gene expression profiling in sepsis: timing, tissue, and translational considerations. Trends Mol. Med. 20, 204–213 (2014).
Kim, S. et al. Characterizing the genetic basis of innate immune response in TLR4-activated human monocytes. Nat. Commun. 5, 5236 (2014).
Opal, S. M. et al. Effect of eritoran, an antagonist of MD2-TLR4, on mortality in patients with severe sepsis: the ACCESS randomized trial. JAMA 309, 1154–1162 (2013).
van der Made, C. I. et al. Presence of genetic variants among young men with severe COVID-19. JAMA 324, 663–673 (2020).
Migden, M. R., Chang, A. L. S., Dirix, L., Stratigos, A. J. & Lear, J. T. Emerging trends in the treatment of advanced basal cell carcinoma. Cancer Treat. Rev. 64, 1–10 (2018).
Park, J. W. et al. Adaptive randomization of neratinib in early breast cancer. N. Engl. J. Med. 375, 11–22 (2016).
I-SPY Covid Consortium. Clinical trial design during and beyond the pandemic: the I-SPY COVID trial. Nat. Med. 28, 9–11 (2022).
Dellinger, R. P. et al. Effect of targeted polymyxin B hemoperfusion on 28-day mortality in patients with septic shock and elevated endotoxin level: the EUPHRATES randomized clinical trial. JAMA 320, 1455–1463 (2018).
Pairo-Castineira, E. et al. Genetic mechanisms of critical illness in COVID-19. Nature 591, 92–98 (2021).
Kousathanas, A. et al. Whole genome sequencing reveals host factors underlying critical Covid-19. Nature https://doi.org/10.1038/s41586-022-04576-6 (2022).
Lee, J. S. et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 5, eabd1554 (2020).
Oh, J. et al. Prediction and early detection of delirium in the intensive care unit by using heart rate variability and machine learning. Physiol. Meas. 39, 035004 (2018).
Marshall, J. C. et al. Measures, markers, and mediators: towards a staging system for clinical sepsis. Crit. Care Med. 31, 1560–1567 (2003).
Wu, A. C. et al. Current status and future opportunities in lung precision medicine research with a focus on biomarkers. An American Thoracic Society/National Heart, Lung, and Blood Institute research statement. Am. J. Respir. Crit. Care Med. 198, e116–e136 (2018).
Maslove, D. M., Lamontagne, F., Marshall, J. C. & Heyland, D. K. A path to precision in the ICU. Crit. Care 21, 79 (2017).
Matthay, M. A. et al. Acute respiratory distress syndrome. Nat. Rev. Dis. Prim. 5, 18 (2019).
Gajic, O., Ahmad, S. R., Wilson, M. E. & Kaufman, D. A. Outcomes of critical illness: what is meaningful? Curr. Opin. Crit. Care 24, 394–400 (2018).
Mahmood, S. S., Levy, D., Vasan, R. S. & Wang, T. J. The Framingham Heart Study and the epidemiology of cardiovascular disease: a historical perspective. Lancet 383, 999–1008 (2014).
Green, E. D., Watson, J. D. & Collins, F. S. Human Genome Project: twenty-five years of big biology. Nature 526, 29–31 (2015).
Emdin, C. A., Khera, A. V. & Kathiresan, S. Mendelian randomization. JAMA 318, 1925–1926 (2017).
Berry, S. M., Connor, J. T. & Lewis, R. J. The platform trial: an efficient strategy for evaluating multiple treatments. JAMA 313, 1619–1620 (2015).
Gospodarowicz, M. et al. History and international developments in cancer staging. Cancer Prev. Cont. 2, 262–268 (1998).
Mackillop, W. J., O’Sullivan, B. & Gospodarowicz, M. The role of cancer staging in evidence-based medicine. Cancer Prev. Cont. 2, 269–277 (1998).
Bakhirev, A. G., Vasef, M. A., Zhang, Q. Y., Reichard, K. K. & Czuchlewski, D. R. Fluorescence immunophenotyping and interphase cytogenetics (FICTION) detects BCL6 abnormalities, including gene amplification, in most cases of nodular lymphocyte-predominant Hodgkin lymphoma. Arch. Pathol. Lab. Med. 138, 538–542 (2014).
Seymour, C. W. et al. Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. JAMA 321, 2003–2017 (2019).
Shankar-Hari, M. & Rubenfeld, G. D. Population enrichment for critical care trials: phenotypes and differential outcomes. Curr. Opin. Crit. Care 25, 489–497 (2019).
Wong, H. R. & Marshall, J. C. Leveraging transcriptomics to disentangle sepsis heterogeneity. Am. J. Respir. Crit. Care Med. 196, 258–260 (2017).
Sweeney, T. E. & Khatri, P. Generalizable biomarkers in critical care: toward precision medicine. Crit. Care Med. 45, 934–939 (2017).
Kitsios, G. D. et al. Host–response subphenotypes offer prognostic enrichment in patients with or at risk for acute respiratory distress syndrome. Crit. Care Med. 47, 1724–1734 (2019).
Wong, H. R. et al. Combining prognostic and predictive enrichment strategies to identify children with septic shock responsive to corticosteroids. Crit. Care Med. 44, e1000–e1003 (2016).
Wong, H. R. et al. Developing a clinically feasible personalized medicine approach to pediatric septic shock. Am. J. Respir. Crit. Care Med. 191, 309–315 (2015).
Wong, H. R., Hart, K. W., Lindsell, C. J. & Sweeney, T. E. External corroboration that corticosteroids may be harmful to septic shock endotype a patients. Crit. Care Med. 49, e98–e101 (2021).
Antcliffe, D. B. et al. Transcriptomic signatures in sepsis and a differential response to steroids. From the VANISH randomized trial. Am. J. Respir. Crit. Care Med. 199, 980–986 (2019).
Acknowledgements
The authors would like to acknowledge the pioneering work of Hector Wong, whose leadership was instrumental in establishing the use of genome-wide analysis to more richly describe the heterogeneity of host responses to infection. His mentorship, generosity, collegiality, vision and tireless dedication will be deeply missed. Development of the concepts shaping this Perspective was facilitated through meetings of the Staging and Stratification Working Group of the International Forum for Acute Care Trialists.
Author information
Authors and Affiliations
Contributions
B.T. and J.C.M. conceived of the idea, with all authors making important conceptual contributions and refining the writing of the manuscript. D.M.M., B.T., M.S.-H., P.R.L. and J.C.M. formed the primary writing group.
Corresponding author
Ethics declarations
Competing interests
R.M.B. reports having served on advisory boards for Merck and Genentech. M.B. is a stockholder of SmartDyeLivery, a company developing nanodrugs for sepsis. T.G.B. has no direct conflicts of interest. His employer, Emory University, collects a stipend for his service as Editor-in-Chief of Critical Care Medicine from the Society of Critical Care Medicine. Emory University also collects a stipend from the US Government for his service as senior advisor to the Biomedical Advanced Research and Development Authority. C.S.C. receives grant funding from the NIH, US Food and Drug Administration, US Department of Defense, Quantum Leap Healthcare Collaborative and Roche-Genentech and has provided consulting services for Quark, Vasomune, Gen1e Life Sciences, Cellenkos, and Janssen. A.C.G. is supported by an NIHR Research Professorship (RP-2015-06-018) and the NIHR Imperial Biomedical Research Centre. Outside of this work he has received personal fees from 30 Respiratory paid to his institution. Outside the submitted work, D.F.M. reports personal fees from consultancy for GlaxoSmithKline, Boehringer Ingelheim, Bayer, Novartis, Sobi and Eli Lilly and from sitting on a Data Monitoring and Ethics Committee for trials undertaken by Vir Biotechnology and Faron Pharmaceuticals. In addition, his institution has received funds from grants from the National Institute for Health and Care Research (NIHR), the Wellcome Trust, Innovate-UK, the Medical Research Council (MRC) and the Northern Ireland HSC R&D Division. D.F.M. also holds a patent for an anti-inflammatory treatment issued to Queen’s University Belfast. D.F.M. was Director of Research for the UK Intensive Care Society (term ended in June 2021) and is NIHR/MRC Efficacy and Mechanism Evaluation Programme Director. D.K.M. reports grants from the European Union, the NIHR and the Canadian Institute for Advanced Research supporting the submitted work; grants from GlaxoSmithKline and Lantmannen; and consulting fees from Calico, GlaxoSmithKline, Lantmannen, NeuroTrauma Sciences and Integra Neurosciences outside the submitted work. L.L.M. reports grants and contracts from the NIGMS, NICHD, and NIAID (NIH), as well as subcontracts from Inflammatix and Beckman-Coulter through awards from BARDA (HHS). In addition, L.L.M. participates in clinical studies supported by RevImmune, Immunex, Faraday and Beckman-Coulter. J.A.R. reports patents owned by the University of British Columbia that are related to (1) the use of PCSK9 inhibitor(s) in sepsis and (2) the use of vasopressin in septic shock, as well as (3) a patent owned by Ferring Pharmaceuticals for the use of selepressin in septic shock. J.A.R. is an inventor on these patents. J.A.R. was a founder, director and shareholder in Cyon Therapeutics and is a shareholder in Molecular You Corp. J.A.R. is no longer actively consulting for any industry. J.A.R. reports receiving consulting fees in the last 3 years from SIB Therapeutics (developing a sepsis drug), Ferring Pharmaceuticals (manufactures vasopressin and is developing selepressin) and PAR Pharma (sells prepared bags of vasopressin). J.A.R. was a funded member of the Data and Safety Monitoring Board of a National Institutes of Health-sponsored trial of plasma in COVID-19 (PASS-IT-ON) (2020–2021). J.A.R. reports having received an investigator-initiated grant from Grifols (titled ‘Is HBP a mechanism of albuminʼs efficacy in human septic shock?’) that was provided to and administered by the University of British Columbia. J.A.R. has received four grants for COVID-19 research from the Canadian Institutes of Health Research and two grants from the St. Paul’s Foundation. J.A.R. was also a non-funded science advisor and member of the Government of Canada COVID-19 Therapeutics Task Force (2020–2021). N.I.S. reports research funding from the National Institutes of Health, Luminos, Inflammatix and Google and is a consultant for Diagnostic Robotics. M.S. has served on advisory boards for infection and sepsis-related projects from Abbott, AM Pharma, Aptarion, Biotest, Biomerieux, deePull, Pfizer, Roche, Safeguard Biosystems and Spiden, and from Deltex Medical, Fresenius and Nestle outside the submitted work. M.S. holds a patent owned by University College London related to a sulphide-releasing molecule for ischaemia-reperfusion injury and a pending patent for phytosterol use in sepsis. UCL also holds shares in Deltex Medical and receives research funding from the Medical Research Council, Wellcome Trust, NIHR, Innovate UK, the European Commission and the UCL Technology Fund. T.E.S. is stockholder in, and employee of, Inflammatix, which is developing a rapid test for sepsis endotypes. Outside the submitted work, B.T.T. reports personal fees from consultancy for Bayer and Genentech. B.V. reports being an NHMRC Investigator Fellow.
Peer review
Peer review information
Nature Medicine thanks Robert Stevens and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary handling editor: Karen O’Leary, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Maslove, D.M., Tang, B., Shankar-Hari, M. et al. Redefining critical illness. Nat Med 28, 1141–1148 (2022). https://doi.org/10.1038/s41591-022-01843-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-022-01843-x
This article is cited by
-
Monocyte state 1 (MS1) cells in critically ill patients with sepsis or non-infectious conditions: association with disease course and host response
Critical Care (2024)
-
Identifying hub genes of sepsis-associated and hepatic encephalopathies based on bioinformatic analysis—focus on the two common encephalopathies of septic cirrhotic patients in ICU
BMC Medical Genomics (2024)
-
Activation of senescence in critically ill patients: mechanisms, consequences and therapeutic opportunities
Annals of Intensive Care (2024)
-
Association between tranexamic acid administration and mortality based on the trauma phenotype: a retrospective analysis of a nationwide trauma registry in Japan
Critical Care (2024)
-
Circulating N-lactoyl-amino acids and N-formyl-methionine reflect mitochondrial dysfunction and predict mortality in septic shock
Metabolomics (2024)