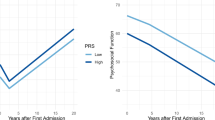
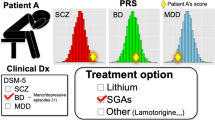
Abstract
Polygenic risk scores (PRS) summarize genetic liability to a disease at the individual level, and the aim is to use them as biomarkers of disease and poor outcomes in real-world clinical practice. To date, few studies have assessed the prognostic value of PRS relative to standards of care. Schizophrenia (SCZ), the archetypal psychotic illness, is an ideal test case for this because the predictive power of the SCZ PRS exceeds that of most other common diseases. Here, we analyzed clinical and genetic data from two multi-ethnic cohorts totaling 8,541 adults with SCZ and related psychotic disorders, to assess whether the SCZ PRS improves the prediction of poor outcomes relative to clinical features captured in a standard psychiatric interview. For all outcomes investigated, the SCZ PRS did not improve the performance of predictive models, an observation that was generally robust to divergent case ascertainment strategies and the ancestral background of the study participants.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Data availability
BioMe data, including both clinical (extracted via NLP) and genetic (PRS and ancestry PCs) features, are available at https://github.com/landiisotta/prs_psychosis/tree/master/data. GPC clinical phenotypic data have been deposited in and will be accessible via the NIMH Repository & Genomics Resource at nimhgenetics.org under Study 76. GPC genotyping and sequencing data have been deposited to dbGaP with accession codes phs001020.v2.p1 and phs002041.v1.p1.
Code availability
Code for data preprocessing and modeling of both BioMe and GPC datasets is available at https://github.com/landiisotta/prs_psychosis.
References
Wray, N. R., Goddard, M. E. & Visscher, P. M. Prediction of individual genetic risk of complex disease. Curr. Opin. Genet. Dev. 18, 257–263 (2008).
International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009).
Collins, F. S. & Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 372, 793–795 (2015).
Green, E. D. et al. Strategic vision for improving human health at The Forefront of Genomics. Nature 586, 683–692 (2020).
Warren, M. The approach to predictive medicine that is taking genomics research by storm. Nature 562, 181–183 (2018).
Lewis, C. M. & Vassos, E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 12, 44 (2020).
Demontis, D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75 (2019).
Levey, D. F. et al. Reproducible genetic risk loci for anxiety: results from ~200,000 participants in the Million Veteran Program. Am. J. Psychiatry 177, 223–232 (2020).
Jansen, I. E. et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2019).
Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 53, 817–829 (2021).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019).
Misganaw, B. et al. Polygenic risk associated with post-traumatic stress disorder onset and severity. Transl. Psychiatry 9, 165 (2019).
The Schizophrenia Working Group of the Psychiatric Genomics Consortium. Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. Preprint at medRxiv https://doi.org/10.1101/2020.09.12.20192922 (2020).
Oetjens, M. T., Kelly, M. A., Sturm, A. C., Martin, C. L. & Ledbetter, D. H. Quantifying the polygenic contribution to variable expressivity in eleven rare genetic disorders. Nat. Commun. 10, 4897 (2019).
Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50, 1219–1224 (2018).
Haas, M. E. et al. Genetic association of albuminuria with cardiometabolic disease and blood pressure. Am. J. Hum. Genet. 103, 461–473 (2018).
Suvisaari, J. et al. Is it possible to predict the future in first-episode psychosis? Front. Psychiatry 9, 580 (2018).
Belbin, G. M. et al. Toward a fine-scale population health monitoring system. Cell 184, 2068–2083 (2021).
Tayo, B. O. et al. Genetic background of patients from a university medical center in Manhattan: implications for personalized medicine. PLoS ONE 6, e19166 (2011).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Pato, M. T. et al. The genomic psychiatry cohort: partners in discovery. Am. J. Med. Genet. B Neuropsychiatr. Genet. 162B, 306–312 (2013).
McGuffin, P., Farmer, A. & Harvey, I. A polydiagnostic application of operational criteria in studies of psychotic illness: development and reliability of the OPCRIT system. Arch. Gen. Psychiatry 48, 764–770 (1991).
Bigdeli, T. B. et al. Contributions of common genetic variants to risk of schizophrenia among individuals of African and Latino ancestry. Mol. Psychiatry 25, 2455–2467 (2020).
Goddard, M. Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 136, 245–257 (2009).
Meuwissen, T. H., Hayes, B. J. & Goddard, M. E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829 (2001).
Zhang, J.-P. et al. Schizophrenia polygenic risk score as a predictor of antipsychotic efficacy in first episode psychosis. Am. J. Psychiatry 176, 21–28 (2019).
Zheutlin, A. B. et al. Penetrance and pleiotropy of polygenic risk scores for schizophrenia in 106,160 patients across four health care systems. Am. J. Psychiatry 176, 846–855 (2019).
Mosley, J. D. et al. Predictive accuracy of a polygenic risk score compared with a clinical risk score for incident coronary heart disease. JAMA 323, 627–635 (2020).
Vassos, E. et al. An examination of polygenic score risk prediction in individuals with first-episode psychosis. Biol. Psychiatry 81, 470–477 (2017).
Perkins, D. O. et al. Polygenic risk score contribution to psychosis prediction in a target population of persons at clinical high risk. Am. J. Psychiatry 177, 155–163 (2020).
Davies, R. W. et al. Using common genetic variation to examine phenotypic expression and risk prediction in 22q11.2 deletion syndrome. Nat. Med. 26, 1912–1918 (2020).
World Health Organization. The ICD-10 Classification of Mental and Behavioural Disorders: Diagnostic Criteria for Research (World Health Organization, 1993).
Soysal, E. et al. CLAMP: a toolkit for efficiently building customized clinical natural language processing pipelines. J. Am. Med. Inform. Assoc. 25, 331–336 (2018).
Ruderfer, D. M. et al. Polygenic overlap between schizophrenia risk and antipsychotic response: a genomic medicine approach. Lancet Psychiatry 3, 350–357 (2016).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Gibbs, R. A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
O’Connell, J. et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 10, e1004234 (2014).
Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Rosvall, M. & Bergstrom, C. T. Maps of random walks on complex networks reveal community structure. Proc. Natl Acad. Sci. USA 105, 1118–1123 (2008).
Rosvall, M. & Bergstrom, C. T. Multilevel compression of random walks on networks reveals hierarchical organization in large integrated systems. PLoS ONE 6, e18209 (2011).
Choi, S. W. & O’Reilly, P. F. PRSice-2: polygenic risk score software for biobank-scale data. Gigascience 8, giz082 (2019).
Harrell, F. E. rms: Regression Modeling Strategies. R package version 6.1-1. https://CRAN.R-project.org/package=rms (2021).
Pedregosa, F. et al. Scikit-learn: Machine Learning in Python https://arxiv.org/abs/1201.0490 (2018).
Canty, A. & Ripley, B. boot: Bootstrap R (S-Plus) Functions. R package version 1.3-28 https://cran.r-project.org/web/packages/boot/index.html (2021).
Davison, A. C. & Hinkley, D. V. Bootstrap Methods and their Application (Cambridge University Press, 1997).
Champely, S. et al. pwr: Basic Functions for Power Analysis http://cran.r-project.org/web/packages/pwr/ (2020).
Acknowledgements
This study was supported by grant R01 MH121923 from the National Institute of Mental Health (NIMH). The authors thank A. Jain, A. Moscati, L. Zhou, Q. Song, S. Wenric and S. Ellis, all of whom are paid employees of the Icahn School of Medicine at Mount Sinai, for assisting with quality control and/or file handling for the BioMe exome sequencing and genome-wide genotyping data. The BioMe healthcare delivery cohort at Mount Sinai was established and maintained with a generous gift from the Andrea and Charles Bronfman Philanthropies. The authors also thank the Genomic Psychiatry Cohort (GPC) Investigators. The GPC was supported by grants R01 MH085548, R01 MH104964 and R01 MH123451-01 from the NIMH, and genotyping of samples was provided by the Stanley Center for Psychiatric Research at Broad Institute. T.B.B. is supported by a NARSAD Young Investigator Grant from the Brain and Behavior Research Foundation.
Author information
Authors and Affiliations
Contributions
A.W.C. conceived and supervised the study. A.W.C., I.L., D.A.K. and G.N.N. designed the study and supervised the modeling. A.W.C. and I.L. implemented and ran the analyses, interpreted the results, and wrote the paper. L.C. contributed to the creation of the BioMe clinical dataset for the present work. G.B. and M.P. substantially contributed to the BioMe genetic data used in this study. P.O.R. and N.D.B. extensively contributed to the discussions on methods and aim of the study. B.S.G. substantially edited the manuscript. M.T.P., C.N.P. and T.B.B. substantially contributed to the preparation of GPC clinical and genetic data for the present work. T.V.V. created the NLP concept extraction tool. All other authors (that is, R.J.F.L., E.K., E.E.S., E.D.A., P.F.B., D.L., D.P.M., S.A.M., M.H.R. and A.H.F.) extensively contributed to the creation of the BioMe or GPC datasets. All authors approved all versions of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Medicine thanks Carrie Bearden, Jose Rubio and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Anna Maria Ranzoni was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Grid-search for regularization parameter selection within repeated cross-validation framework.
Plots in each row display training and validation F2 scores for the prediction of different outcomes with regularization parameter C varying from 0.001 to 100. Plots in each column refer to different features included in the model, that is, clinical (a), (f), (k); clinical and genetic (b), (g), (l); clinical and binarized genetic (c), (h), (m); genetic (d), (i), (n); and binarized genetic (e), (j), (o). The dot marks the model with the highest validation score. In each plot, data is presented as mean values of n = 300 independent scores derived from subsets of n = 179 (BioMe) and n = 1,816 (GPC) observations in validation and n = 358 (BioMe) and n = 3,632 (GPC) observations in training. Validation scores are presented as mean values ± SD.
Extended Data Fig. 2 Models’ performance for BioMe and Genomic Psychiatry Cohort (GPC) datasets with different feature configurations.
Average F2 scores in predicting different outcomes are displayed in panels (a), (c), and (e). Averages are computed on n = 300 independent scores from validation on subsets of n = 179 (BioMe) and n = 1,816 (GPC) observations. Boxplots’ center: median and mean; bound of box: 25th (Q1) and 75th (Q3) percentiles; minimum: Q1-1.5*(Q3-Q1); maximum: Q3+1.5*(Q3-Q1). Exact p-values from two-sided t-tests with Benjamini-Hochberg correction are displayed for significant comparisons. Validation scores for clinical, clinical and genetic, and clinical and binarized genetic are the same reported in manuscript’s Fig. 1 and replicated here to ease comparisons. Panels (b), (d), and (f) display precision-recall curves for linear regression models with different outcomes evaluated on test sets. Performance of random classifiers is displayed as reference.
Extended Data Fig. 3 Prediction performance for BioMe and Genomic Psychiatry Cohort (GPC) cohorts restricted to individuals of African (AFR) ancestry.
Training and validation F2 estimates for varying regularization parameters (C) are displayed within the ‘Cross-validated grid-search’ frame for each outcome and feature configuration of interest [that is, clinical, clinical and genetic (all), and clinical and binarized genetic (all binary)]. Data are presented as mean values for training and mean values ± SD for validation. Averages are computed on n = 300 independent scores derived from subsets of n = 82 (BioMe) and n = 822 (GPC) observations for validation and n = 164 (BioMe) and n = 1,644 (GPC) observations for training. The dot corresponds to the highest F2 score during validation. The best model, with related parameter C, is then trained on the entire training set and evaluated on the test set. The F2 validation score distributions obtained are enclosed in the ‘Performance evaluation’ frame. Boxplots’ center: median and mean; bound of box: 25th (Q1) and 75th (Q3) percentiles; minimum: Q1-1.5*(Q3-Q1); maximum: Q3+1.5*(Q3-Q1). Exact p-values from two-sided pairwise t-tests with Benjamini-Hochberg correction are displayed for significant comparisons. Precision-recall curves obtained from the models evaluated on test sets are reported on the right.
Extended Data Fig. 4 Prediction performance for BioMe and Genomic Psychiatry Cohort (GPC) cohorts restricted to individuals of admixed American (AMR) ancestry.
Training and validation F2 estimates for varying regularization parameters (C) are displayed within the ‘Cross-validated grid-search’ frame for each outcome and feature configuration of interest [that is, clinical, clinical and genetic (all), and clinical and binarized genetic (all binary)]. Data are presented as mean values for training and mean values ± SD for validation. Averages are computed on n = 300 independent scores derived from subsets of n = 74 (BioMe) and n = 144 (GPC) observations for validation and n = 148 (BioMe) and n = 290 (GPC) observations for training. The dot corresponds to the highest F2 score during validation. The best model, with related parameter C, is then trained on the entire training set and evaluated on the test set. The F2 validation score distributions obtained are enclosed in the ‘Performance evaluation’ frame. Boxplots’ center: median and mean; bound of box: 25th (Q1) and 75th (Q3) percentiles; minimum: Q1-1.5*(Q3-Q1); maximum: Q3+1.5*(Q3-Q1). Exact p-values from two-sided pairwise t-tests with Benjamini-Hochberg correction are displayed for significant comparisons. Precision-recall curves obtained from the models evaluated on test sets are reported on the right.
Extended Data Fig. 5 Prediction performance for BioMe and Genomic Psychiatry Cohort (GPC) cohorts restricted to individuals of European (EUR) ancestry.
Training and validation F2 estimates for varying regularization parameters (C) are displayed within the ‘Cross-validated grid-search’ frame for each outcome and feature configuration of interest [that is, clinical, clinical and genetic (all), and clinical and binarized genetic (all binary)]. Data are presented as mean values for training and mean values ± SD for validation. Averages are computed on n = 300 independent scores derived from subsets of n = 23 (BioMe) and n = 850 (GPC) observations for validation and n = 46 (BioMe) and n = 1,698 (GPC) observations for training. The dot corresponds to the highest F2 score during validation. The best model, with related parameter C, is then trained on the entire training set and evaluated on the test set. The F2 validation score distributions obtained are enclosed in the ‘Performance evaluation’ frame. Boxplots’ center: median and mean; bound of box: 25th (Q1) and 75th (Q3) percentiles; minimum: Q1-1.5*(Q3-Q1); maximum: Q3+1.5*(Q3-Q1). Exact p-values from two-sided pairwise t-tests with Benjamini-Hochberg correction are displayed for significant comparisons. Precision-recall curves obtained from the models evaluated on test sets are reported on the right.
Extended Data Fig. 6 Sensitivity power analysis.
Range of effect sizes and corresponding power for t-test model comparisons are displayed for validation (a) and test sets (b). Alpha level is set at 0.05 and sample size varies according to the cohort considered.
Supplementary information
Supplementary Information
Supplementary Tables 1-9
Rights and permissions
About this article
Cite this article
Landi, I., Kaji, D.A., Cotter, L. et al. Prognostic value of polygenic risk scores for adults with psychosis. Nat Med 27, 1576–1581 (2021). https://doi.org/10.1038/s41591-021-01475-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-021-01475-7
This article is cited by
-
Identification of schizophrenia symptom-related gene modules by postmortem brain transcriptome analysis
Translational Psychiatry (2023)
-
The molecular pathology of schizophrenia: an overview of existing knowledge and new directions for future research
Molecular Psychiatry (2023)
-
The genetic architecture of schizophrenia: review of large-scale genetic studies
Journal of Human Genetics (2023)
-
Genetics of bipolar disorder: insights into its complex architecture and biology from common and rare variants
Journal of Human Genetics (2023)
-
Addressing the ethical and societal challenges posed by genome-wide association studies of behavioral and brain-related traits
Nature Neuroscience (2023)