Abstract
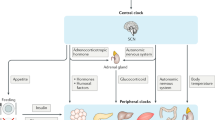
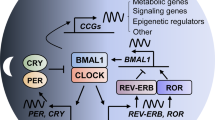
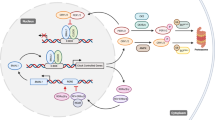
The circadian clock is a complex cellular mechanism that, through the control of diverse metabolic and gene expression pathways, governs a large array of cyclic physiological processes. Epidemiological and clinical data reveal a connection between the disruption of circadian rhythms and cancer that is supported by recent preclinical data. In addition, results from animal models and molecular studies underscore emerging links between cancer metabolism and the circadian clock. This has implications for therapeutic approaches, and we discuss the possible design of chronopharmacological strategies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Fu, L. & Lee, C. C. The circadian clock: pacemaker and tumour suppressor. Nat. Rev. Cancer 3, 350–361 (2003).
Sahar, S. & Sassone-Corsi, P. Metabolism and cancer: the circadian clock connection. Nat. Rev. Cancer 9, 886–896 (2009).
Masri, S., Kinouchi, K. & Sassone-Corsi, P. Circadian clocks, epigenetics, and cancer. Curr. Opin. Oncol. 27, 50–56 (2015).
Lie, J. A., Roessink, J. & Kjaerheim, K. Breast cancer and night work among Norwegian nurses. Cancer Causes Control 17, 39–44 (2006).
Papantoniou, K. et al. Night shift work, chronotype and prostate cancer risk in the MCC-Spain case-control study. Int. J. Cancer 137, 1147–1157 (2015).
Schernhammer, E. S. et al. Rotating night shifts and risk of breast cancer in women participating in the nurses’ health study. J. Natl. Cancer. Inst. 93, 1563–1568 (2001).
Straif, K. et al. Carcinogenicity of shift-work, painting, and fire-fighting. Lancet. Oncol. 8, 1065–1066 (2007).
Knutsson, A. et al. Breast cancer among shift workers: results of the WOLF longitudinal cohort study. Scand. J. Work. Environ. Health 39, 170–177 (2013).
Kakizaki, M. et al. Sleep duration and the risk of prostate cancer: the Ohsaki Cohort Study. Br. J. Cancer 99, 176–178 (2008).
Srour, B. et al. Circadian nutritional behaviours and cancer risk: new insights from the NutriNet-sante prospective cohort study: disclaimers. Int. J. Cancer 143, 2369–2379 (2018).
Fu, L., Pelicano, H., Liu, J., Huang, P. & Lee, C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111, 41–50 (2002).
Lee, S., Donehower, L. A., Herron, A. J., Moore, D. D. & Fu, L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One 5, e10995 (2010).
Papagiannakopoulos, T. et al. Circadian rhythm disruption promotes lung tumorigenesis. Cell. Metab. 24, 324–331 (2016).
Filipski, E. et al. Host circadian clock as a control point in tumor progression. J. Natl. Cancer. Inst. 94, 690–697 (2002).
Chen, S. T. et al. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis 26, 1241–1246 (2005).
Taniguchi, H. et al. Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res. 69, 8447–8454 (2009).
Zhu, Y. et al. Epigenetic impact of long-term shiftwork: pilot evidence from circadian genes and whole-genome methylation analysis. Chronobiol. Int. 28, 852–861 (2011).
Filipski, E. et al. Effects of chronic jet lag on tumor progression in mice. Cancer Res. 64, 7879–7885 (2004).
Kettner, N. M. et al. Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell. 30, 909–924 (2016).
Asher, G. & Sassone-Corsi, P. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161, 84–92 (2015).
Marcheva, B. et al. Circadian clocks and metabolism. Handb. Exp. Pharmacol. 217, 127–155 (2013).
Abbondante, S., Eckel-Mahan, K. L., Ceglia, N. J., Baldi, P. & Sassone-Corsi, P. Comparative circadian metabolomics reveal differential effects of nutritional challenge in the serum and liver. J. Biol. Chem. 291, 2812–2828 (2016).
Eckel-Mahan, K. L. et al. Reprogramming of the circadian clock by nutritional challenge. Cell 155, 1464–1478 (2013).
Hatori, M. et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell. Metab. 15, 848–860 (2012).
Altman, B. J. et al. MYC disrupts the circadian clock and metabolism in cancer cells. Cell. Metab. 22, 1009–1019 (2015).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Gaucher, J., Montellier, E. & Sassone-Corsi, P. Molecular cogs: interplay between circadian clock and cell cycle. Trends. Cell Biol. 28, 368–379 (2018).
Jackson, E. L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001).
Ripperger, J. A. & Schibler, U. Rhythmic CLOCK–BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38, 369–374 (2006).
Walhout, A. J., Gubbels, J. M., Bernards, R., van der Vliet, P. C. & Timmers, H. T. c-Myc/Max heterodimers bind cooperatively to the E-box sequences located in the first intron of the rat ornithine decarboxylase (ODC) gene. Nucleic Acids Res. 25, 1493–1501 (1997).
Huber, A. L. et al. CRY2 and FBXL3 cooperatively degrade c-MYC. Mol. Cell 64, 774–789 (2016).
Shostak, A. et al. MYC/MIZ1-dependent gene repression inversely coordinates the circadian clock with cell cycle and proliferation. Nat. Commun. 7, 11807 (2016).
Chaix, A., Zarrinpar, A., Miu, P. & Panda, S. Time-restricted feeding is a preventative and therapeutic intervention against diverse nutritional challenges. Cell. Metab. 20, 991–1005 (2014).
Tognini, P. et al. Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell. Metab. 26, 523–538 e525 (2017).
Oishi, K., Uchida, D. & Itoh, N. Low-carbohydrate, high-protein diet affects rhythmic expression of gluconeogenic regulatory and circadian clock genes in mouse peripheral tissues. Chronobiol. Int. 29, 799–809 (2012).
Dyar, K. A. et al. Atlas of circadian metabolism reveals system-wide coordination and communication between clocks. Cell 174, 1571–1585. e11 (2018).
Kohsaka, A. et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell. Metab. 6, 414–421 (2007).
Vollmers, C. et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl. Acad. Sci. USA 106, 21453–21458 (2009).
Damiola, F. et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14, 2950–2961 (2000).
Stokkan, K. A., Yamazaki, S., Tei, H., Sakaki, Y. & Menaker, M. Entrainment of the circadian clock in the liver by feeding. Science 291, 490–493 (2001).
Dallmann, R., Viola, A. U., Tarokh, L., Cajochen, C. & Brown, S. A. The human circadian metabolome. Proc. Natl. Acad. Sci. USA 109, 2625–2629 (2012).
Sato, S., Parr, E. B., Devlin, B. L., Hawley, J. A. & Sassone-Corsi, P. Human metabolomics reveal daily variations under nutritional challenges specific to serum and skeletal muscle. Mol. Metab. 16, 1–11 (2018).
Wu, Y. et al. Reciprocal regulation between the circadian clock and hypoxia signaling at the genome level in mammals. Cell. Metab. 25, 73–85 (2017).
Adamovich, Y., Ladeuix, B., Golik, M., Koeners, M. P. & Asher, G. Rhythmic oxygen levels reset circadian clocks through HIF1α. Cell. Metab. 25, 93–101 (2017).
Peek, C. B. et al. Circadian clock interaction with HIF1α mediates oxygenic metabolism and anaerobic glycolysis in skeletal muscle. Cell. Metab. 25, 86–92 (2017).
Arendt, J., Bojkowski, C., Franey, C., Wright, J. & Marks, V. Immunoassay of 6-hydroxymelatonin sulfate in human plasma and urine: abolition of the urinary 24-h rhythm with atenolol. J. Clin. Endocrinol. Metab. 60, 1166–1173 (1985).
Zawilska, J. B., Skene, D. J. & Arendt, J. Physiology and pharmacology of melatonin in relation to biological rhythms. Pharmacol. Rep. 61, 383–410 (2009).
Reiter, R. J., Tan, D. X., Manchester, L. C. & Qi, W. Biochemical reactivity of melatonin with reactive oxygen and nitrogen species: a review of the evidence. Cell Biochem. Biophys. 34, 237–256 (2001).
Reiter, R. J., Tan, D. X., Manchester, L. C. & El-Sawi, M. R. Melatonin reduces oxidant damage and promotes mitochondrial respiration: implications for aging. Ann. NY Acad. Sci. 959, 238–250 (2002).
Leon, J., Acuna-Castroviejo, D., Escames, G., Tan, D. X. & Reiter, R. J. Melatonin mitigates mitochondrial malfunction. J. Pineal Res. 38, 1–9 (2005).
Martin, M. et al. Melatonin-induced increased activity of the respiratory chain complexes I and IV can prevent mitochondrial damage induced by ruthenium red in vivo. J. Pineal Res. 28, 242–248 (2000).
Proietti, S., Cucina, A., Minini, M. & Bizzarri, M. Melatonin, mitochondria, and the cancer cell. Cell. Mol. Life Sci. 74, 4015–4025 (2017).
Bhatti, P., et al. Oxidative DNA damage during night shift work. Occup. Environ. Med. 74, 680–683. (2017).
Al-Zoughbi, W. et al. Tumor macroenvironment and metabolism. Semin. Oncol. 41, 281–295 (2014).
Lee, Y. M., Chang, W. C. & Ma, W. L. Hypothesis: solid tumours behave as systemic metabolic dictators. J. Cell. Mol. Med. 20, 1076–1085 (2016).
Rutkowski, M. R., Svoronos, N., Perales-Puchalt, A. & Conejo-Garcia, J. R. The tumor macroenvironment: cancer-promoting networks beyond tumor beds. Adv. Cancer Res. 128, 235–262 (2015).
Masri, S. et al. Lung adenocarcinoma distally rewires hepatic circadian homeostasis. Cell 165, 896–909 (2016).
Hojo, H. et al. Remote reprogramming of hepatic circadian transcriptome by breast cancer. Oncotarget 8, 34128–34140 (2017).
Brady, J. J. et al. Lung adenocarcinoma distally rewires hepatic circadian homeostasis. Cancer Cell. 29, 697–710 (2016).
Hensley, C. T. et al. Metabolic heterogeneity in human lung tumors. Cell 164, 681–694 (2016).
Faubert, B. et al. Lactate metabolism in human lung tumors. Cell 171, 358–371 e359 (2017).
Hui, S. et al. Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118 (2017).
Spinelli, J. B., et al. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science 358, 941–946 (2017).
Bass, J. & Takahashi, J. S. Circadian integration of metabolism and energetics. Science 330, 1349–1354 (2010).
Dallmann, R., Okyar, A. & Levi, F. Dosing-time makes the poison: circadian regulation and pharmacotherapy. Trends Mol. Med. 22, 430–445 (2016).
Svensson, R. U. et al. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat. Med. 22, 1108–1119 (2016).
Comerford, S. A. et al. Acetate dependence of tumors. Cell 159, 1591–1602 (2014).
Schug, Z. T. et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 27, 57–71 (2015).
Sahar, S. Circadian control of fatty acid elongation by SIRT1-mediated deacetylation of Acetyl-CoA synthetase 1. J. Biol. Chem. 289, 6091–6097 (2014).
Sulli, G. et al. Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature 553, 351–355 (2018).
Gui, D. Y. et al. Environment dictates dependence on mitochondrial complex i for nad+ and aspartate production and determines cancer cell sensitivity to metformin. Cell. Metab. 24, 716–727 (2016).
Asher, G. et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317–328 (2008).
Nakahata, Y. et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340 (2008).
Masri, S. et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158, 659–672 (2014).
Aguilar-Arnal, L., Katada, S., Orozco-Solis, R. & Sassone-Corsi, P. NAD + –SIRT1 control of H3K4 trimethylation through circadian deacetylation of MLL1. Nat. Struct. Mol. Biol. 22, 312–318 (2015).
Nakahata, Y., Sahar, S., Astarita, G., Kaluzova, M. & Sassone-Corsi, P. Circadian control of the NAD+salvage pathway by CLOCK-SIRT1. Science 324, 654–657 (2009).
Ramsey, K. M. et al. Circadian clock feedback cycle through NAMPT-mediated NAD+biosynthesis. Science 324, 651–654 (2009).
Fang, M., Guo, W. R., Park, Y., Kang, H. G. & Zarbl, H. Enhancement of NAD+-dependent SIRT1 deacetylase activity by methylselenocysteine resets the circadian clock in carcinogen-treated mammary epithelial cells. Oncotarget 6, 42879–42891 (2015).
Nahimana, A. et al. The NAD biosynthesis inhibitor APO866 has potent antitumor activity against hematologic malignancies. Blood 113, 3276–3286 (2009).
Thakur, B. K. et al. Involvement of p53 in the cytotoxic activity of the NAMPT inhibitor FK866 in myeloid leukemic cells. Int. J. Cancer 132, 766–774 (2013).
Puram, R. V. et al. Core circadian clock genes regulate leukemia stem cells in AML. Cell 165, 303–316 (2016).
Barberino, R. S. et al. Melatonin protects against cisplatin-induced ovarian damage in mice via the MT1 receptor and antioxidant activity. Biol. Reprod. 96, 1244–1255 (2017).
Gao, Y. Melatonin synergizes the chemotherapeutic effect of 5-fluorouracil in colon cancer by suppressing PI3K/AKT and NF-κB/iNOS signaling pathways. J. Pineal Res. 62, e12380 (2017).
Goncalves Ndo, N. et al. Effect of melatonin in epithelial mesenchymal transition markers and invasive properties of breast cancer stem cells of canine and human cell lines. PLoS One 11, e0150407 (2016).
Mao, L. et al. Circadian gating of epithelial-to-mesenchymal transition in breast cancer cells via melatonin-regulation of GSK3β. Mol. Endocrinol. 26, 1808–1820 (2012).
Lissoni, P., Chilelli, M., Villa, S., Cerizza, L. & Tancini, G. Five years survival in metastatic non-small cell lung cancer patients treated with chemotherapy alone or chemotherapy and melatonin: a randomized trial. J. Pineal Res. 35, 12–15 (2003).
Del Fabbro, E., Dev, R., Hui, D., Palmer, L. & Bruera, E. Effects of melatonin on appetite and other symptoms in patients with advanced cancer and cachexia: a double-blind placebo-controlled trial. J. Clin. Oncol. 31, 1271–1276 (2013).
Mayers, J. R. et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat. Med. 20, 1193–1198 (2014).
Nishiumi, S. et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS One 7, e40459 (2012).
Jobard, E. et al. A serum nuclear magnetic resonance-based metabolomic signature of advanced metastatic human breast cancer. Cancer Lett. 343, 33–41 (2014).
Kobayashi, T. et al. A novel serum metabolomics-based diagnostic approach to pancreatic cancer. Cancer Epidemiol. Biomarkers. Prev. 22, 571–579 (2013).
Wei, S. et al. Metabolomics approach for predicting response to neoadjuvant chemotherapy for breast cancer. Mol. Oncol. 7, 297–307 (2013).
Aviram, R. et al. Lipidomics analyses reveal temporal and spatial lipid organization and uncover daily oscillations in intracellular organelles. Mol. Cell 62, 636–648 (2016).
Eckel-Mahan, K. L. et al. Coordination of the transcriptome and metabolome by the circadian clock. Proc. Natl. Acad. Sci. USA 109, 5541–5546 (2012).
Haus, E., Lakatua, D. J., Swoyer, J. & Sackett-Lundeen, L. Chronobiology in hematology and immunology. Am. J. Anat. 168, 467–517 (1983).
Scheiermann, C., Kunisaki, Y. & Frenette, P. S. Circadian control of the immune system. Nat. Rev. Immunol. 13, 190–198 (2013).
Partch, C. L., Green, C. B. & Takahashi, J. S. Molecular architecture of the mammalian circadian clock. Trends. Cell Biol. 24, 90–99 (2014).
Bass, J. Circadian topology of metabolism. Nature 491, 348–356 (2012).
Gekakis, N. et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569 (1998).
Masri, S. & Sassone-Corsi, P. Plasticity and specificity of the circadian epigenome. Nat. Neurosci. 13, 1324–1329 (2010).
Panda, S. et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320 (2002).
Akhtar, R. A. et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 12, 540–550 (2002).
Koike, N. et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 (2012).
Rey, G. et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 9, e1000595 (2011).
Duong, H. A., Robles, M. S., Knutti, D. & Weitz, C. J. A molecular mechanism for circadian clock negative feedback. Science 332, 1436–1439 (2011).
Preitner, N. et al. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260 (2002).
Sato, T. K. et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43, 527–537 (2004).
Masri, S. & Sassone-Corsi, P. The circadian clock: a framework linking metabolism, epigenetics and neuronal function. Nat. Rev. Neurosci. 14, 69–75 (2013).
Welsh, D. K., Takahashi, J. S. & Kay, S. A. Suprachiasmatic nucleus: cell autonomy and network properties. Annu. Rev. Physiol. 72, 551–577 (2010).
Yamazaki, S. et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science 288, 682–685 (2000).
Welsh, D. K., Logothetis, D. E., Meister, M. & Reppert, S. M. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14, 697–706 (1995).
Reppert, S. M. & Weaver, D. R. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 63, 647–676 (2001).
Welsh, D. K., Yoo, S. H., Liu, A. C., Takahashi, J. S. & Kay, S. A. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol. 14, 2289–2295 (2004).
Pando, M. P., Morse, D., Cermakian, N. & Sassone-Corsi, P. Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell 110, 107–117 (2002).
Thresher, R. J. et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science 282, 1490–1494 (1998).
Hirayama, J. et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 450, 1086–1090 (2007).
Nader, N., Chrousos, G. P. & Kino, T. Circadian rhythm transcription factor CLOCK regulates the transcriptional activity of the glucocorticoid receptor by acetylating its hinge region lysine cluster: potential physiological implications. FASEB J. 23, 1572–1583 (2009).
Doi, M., Hirayama, J. & Sassone-Corsi, P. Circadian regulator CLOCK is a histone acetyltransferase. Cell 125, 497–508 (2006).
Hung, H. C., Maurer, C., Kay, S. A. & Weber, F. Circadian transcription depends on limiting amounts of the transcription co-activator nejire/CBP. J. Biol. Chem. 282, 31349–31357 (2007).
Hosoda, H. et al. CBP/p300 is a cell type-specific modulator of CLOCK/BMAL1-mediated transcription. Mol. Brain 2, 34 (2009).
Feng, D. et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319 (2011).
Alenghat, T. et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature 456, 997–1000 (2008).
Chang, H. C. & Guarente, L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 153, 1448–1460 (2013).
Katada, S. & Sassone-Corsi, P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat. Struct. Mol. Biol. 17, 1414–1421 (2010).
Valekunja, U. K. et al. Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc. Natl. Acad. Sci. USA 110, 1554–1559 (2013).
Janich, P. et al. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature 480, 209–214 (2011).
Sato, S. et al. Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell 170, 664–677.e11 (2017).
Solanas, G. et al. Aged stem cells reprogram their daily rhythmic functions to adapt to stress. Cell 170, 678–692.e20 (2017).
Plikus, M. V. et al. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature 451, 340–344 (2008).
Mendez-Ferrer, S., Lucas, D., Battista, M. & Frenette, P. S. Haematopoietic stem cell release is regulated by circadian oscillations. Nature 452, 442–447 (2008).
Geyfman, M. et al. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc. Natl. Acad. Sci. USA 109, 11758–11763 (2012).
Hoffman, A. E. et al. CLOCK in breast tumorigenesis: genetic, epigenetic, and transcriptional profiling analyses. Cancer Res. 70, 1459–1468 (2010).
Hoffman, A. E. et al. The core circadian gene Cryptochrome 2 influences breast cancer risk, possibly by mediating hormone signaling. Cancer Prev. Res. (Phila) 3, 539–548 (2010).
Acknowledgements
The Masri laboratory is supported by a K22 Transition Career Development Award through the National Cancer Institute (NCI), the Concern Foundation, and the V Foundation for Cancer Research. Work in the Sassone-Corsi laboratory is supported by NIH (National Institutes of Health), INSERM (Institut National de la Sante et la Recherche Medicale, France), and KAUST (King Abdullah University of Science and Technology, Saudi Arabia).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflicts of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Masri, S., Sassone-Corsi, P. The emerging link between cancer, metabolism, and circadian rhythms. Nat Med 24, 1795–1803 (2018). https://doi.org/10.1038/s41591-018-0271-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0271-8
This article is cited by
-
Maternal circadian rhythm disruption affects neonatal inflammation via metabolic reprograming of myeloid cells
Nature Metabolism (2024)
-
Biologically informed deep learning for explainable epigenetic clocks
Scientific Reports (2024)
-
A review for the impacts of circadian disturbance on urological cancers
Sleep and Biological Rhythms (2024)
-
Gonadal dysfunction in women with diabetes mellitus
Endocrine (2024)
-
Oncogenic enhancers prime quiescent metastatic cells to escape NK immune surveillance by eliciting transcriptional memory
Nature Communications (2024)