Abstract
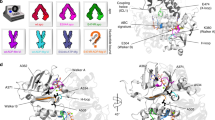
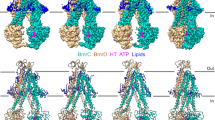
Substrate efflux by ATP-binding cassette (ABC) transporters, which play a major role in multidrug resistance, entails the ATP-powered interconversion between transporter intermediates. Despite recent progress in structure elucidation, a number of intermediates have yet to be visualized and mechanistically interpreted. Here, we combine cryogenic-electron microscopy (cryo-EM), double electron–electron resonance spectroscopy and molecular dynamics simulations to profile a previously unobserved intermediate of BmrCD, a heterodimeric multidrug ABC exporter from Bacillus subtilis. In our cryo-EM structure, ATP-bound BmrCD adopts an inward-facing architecture featuring two molecules of the substrate Hoechst-33342 in a striking asymmetric head-to-tail arrangement. Deletion of the extracellular domain capping the substrate-binding chamber or mutation of Hoechst-coordinating residues abrogates cooperative stimulation of ATP hydrolysis. Together, our findings support a mechanistic role for symmetry mismatch between the nucleotide binding and the transmembrane domains in the conformational cycle of ABC transporters and is of notable importance for rational design of molecules for targeted ABC transporter inhibition.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Structural data presented in this study are available for download from the Protein Data Bank and EMDB under accession codes 7M33 and EMD-23641, respectively. Raw cryo-EM micrographs/videos are also freely available to download from the Electron Microscopy Public Image Archive. Additional data can be provided on request from the authors.
References
Holland, I. B. Rise and rise of the ABC transporter families. Res. Microbiol. 170, 304–320 (2019).
Locher, K. P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016).
Thomas, C. et al. Structural and functional diversity calls for a new classification of ABC transporters. FEBS Lett. 594, 3767–3775 (2020).
Hofmann, S. et al. Conformation space of a heterodimeric ABC exporter under turnover conditions. Nature 571, 580–583 (2019).
Alam, A., Kowal, J., Broude, E., Roninson, I. & Locher, K. P. Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science 363, 753–756 (2019).
Kim, Y. & Chen, J. Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation. Science 359, 915–919 (2018).
Göddeke, H. & Schäfer, L. V. Capturing substrate translocation in an ABC exporter at the atomic level. J. Am. Chem. Soc. 142, 12791–12801 (2020).
Mishra, S. et al. Conformational dynamics of the nucleotide binding domains and the power stroke of a heterodimeric ABC transporter. eLife 3, e02740 (2014).
Johnson, Z. L. & Chen, J. ATP binding enables substrate release from Multidrug Resistance Protein 1. Cell 172, 81–89.e10 (2018).
Johnson, Z. L. & Chen, J. Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell 168, 1075–1085.e9 (2017).
Choudhury, H. G. et al. Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc. Natl Acad. Sci. USA 111, 9145–9150 (2014).
Ward, A., Reyes, C. L., Yu, J., Roth, C. B. & Chang, G. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc. Natl Acad. Sci. USA 104, 19005–19010 (2007).
Dawson, R. J. P. & Locher, K. P. Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185 (2006).
Verhalen, B. et al. Energy transduction and alternating access of the mammalian ABC transporter P-glycoprotein. Nature 543, 738–741 (2017).
Collauto, A., Mishra, S., Litvinov, A., Mchaourab, H. S. & Goldfarb, D. Direct spectroscopic detection of ATP turnover reveals mechanistic divergence of ABC exporters. Structure 25, 1264–1274.e3 (2017).
Dastvan, R., Mishra, S., Peskova, Y. B., Nakamoto, R. K. & Mchaourab, H. S. Mechanism of allosteric modulation of P-glycoprotein by transport substrates and inhibitors. Science 364, 689–692 (2019).
Siarheyeva, A., Liu, R. & Sharom, F. J. Characterization of an asymmetric occluded state of P-glycoprotein with two bound nucleotides: implications for catalysis. J. Biol. Chem. 285, 7575–7586 (2010).
Seelig, A. P-glycoprotein: one mechanism, many tasks and the consequences for pharmacotherapy of cancers. Front Oncol. 10, 576559 (2020).
Robey, R. W. et al. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 18, 452–464 (2018).
Nosol, K. et al. Cryo-EM structures reveal distinct mechanisms of inhibition of the human multidrug transporter ABCB1. Proc. Natl Acad. Sci. USA 117, 26245–26253 (2020).
Bruggemann, E. P., Germann, U. A., Gottesman, M. M. & Pastan, I. Two different regions of phosphoglycoprotein are photoaffinity-labeled by azidopine. J. Biol. Chem. 264, 15483–15488 (1989).
S, D., M, R., I, P., MM, G. & SV, A. Evidence for two nonidentical drug-interaction sites in the human P-glycoprotein. Proc. Natl Acad. Sci. USA 94, 10594–10599 (1997).
HS, M., PR, S. & K, K. Toward the fourth dimension of membrane protein structure: insight into dynamics from spin-labeling EPR spectroscopy. Structure 19, 1549–1561 (2011).
Torres, C., Galián, C., Freiberg, C., Fantino, J.-R. & Jault, J.-M. The YheI/YheH heterodimer from Bacillus subtilis is a multidrug ABC transporter. Biochim. Biophys. Acta - Biomembr. 1788, 615–622 (2009).
Mi, W. et al. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237 (2017).
Ho, H. et al. Structural basis for dual-mode inhibition of the ABC transporter MsbA. Nature 557, 196–201 (2018).
Angiulli, G. et al. New approach for membrane protein reconstitution into peptidiscs and basis for their adaptability to different proteins. eLife 9, e53530 (2020).
Hohl, M. et al. Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc. Natl Acad. Sci. USA 111, 11025–11030 (2014).
Esser, L. et al. Structures of the multidrug transporter P-glycoprotein reveal asymmetric ATP binding and the mechanism of polyspecificity. J. Biol. Chem. 292, 446–461 (2017).
Hustedt, E. J., Stein, R. A. & McHaourab, H. S. Protein functional dynamics from the rigorous global analysis of DEER data: Conditions, components, and conformations. J. Gen. Physiol. 153, e201711954 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Holm, L. Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29–42 (2020).
Alexander, J. A. N. et al. Structural and kinetic analyses of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance in Staphylococcus aureus. J. Biol. Chem. 293, 19854–19865 (2018).
Cooley, R. B., Arp, D. J. & Karplus, P. A. Evolutionary origin of a secondary structure: π-helices as cryptic but widespread insertional variations of α-helices that enhance protein functionality. J. Mol. Biol. 404, 232–246 (2010).
Timachi, M. H. et al. Exploring conformational equilibria of a heterodimeric ABC transporter. eLife 6, e20236 (2017).
Debruycker, V. et al. An embedded lipid in the multidrug transporter LmrP suggests a mechanism for polyspecificity. Nat. Struct. Mol. Biol. 27, 829–835 (2020).
Jin, M. S., Oldham, M. L., Zhang, Q. & Chen, J. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature 490, 566–569 (2012).
Mittal, A., Böhm, S., Grütter, M. G., Bordignon, E. & Seeger, M. A. Asymmetry in the homodimeric ABC transporter MsbA recognized by a DARPin. J. Biol. Chem. 287, 20395–20406 (2012).
Lilkova, E. et al. The PyMOL molecular graphics system, version 2.0 (Schrödinger, LLC, 2015).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Zivanov, J., Nakane, T. & Scheres, S. H. W. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Grant, T., Rohou, A. & Grigorieff, N. CisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018).
Ramlaul, K., Palmer, C. M., Nakane, T. & Aylett, C. H. S. Mitigating local over-fitting during single particle reconstruction with SIDESPLITTER. J. Struct. Biol. 211, 10754 (2020).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 60, 2126–2132 (2004).
Wood, C. et al. Collaborative computational project for electron cryo-microscopy. Acta Crystallogr., Sect. D: Biol. Crystallogr. 71, 123–126 (2015).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. D Struct. Biol. 75, 861–877 (2019).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. Sect. D Struct. Biol. 74, 519–530 (2018).
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015).
Mirdita, M. et al. ColabFold - Making protein folding accessible to all. bioRxiv https://doi.org/10.1101/2021.08.15.456425 (2021).
Staritzbichler, R., Anselmi, C., Forrest, L. R. & Faraldo-Gómez, J. D. GRIFFIN: a versatile methodology for optimization of protein-lipid interfaces for membrane protein simulations. J. Chem. Theory Comput. 7, 1167–1176 (2011).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 114, 7830–7843 (2010).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Jeschke, G. & Polyhach, Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys. Chem. Chem. Phys. 9, 1895–1910 (2007).
Jeschke, G. DEER distance measurements on proteins. Annu. Rev. Phys. Chem. 63, 419–446 (2012).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Qi, Y. et al. CHARMM‐GUI DEER facilitator for spin‐pair distance distribution calculations and preparation of restrained‐ensemble molecular dynamics simulations. J. Comput. Chem. 41, 415–420 (2020).
Tai, C. H., Paul, R., Dukka, K. C., Shilling, J. D. & Lee, B. SymD webserver: A platform for detecting internally symmetric protein structures. Nucleic Acids Res. 42, (2014).
Acknowledgements
This research was funded by the Division of Intramural Research of the National Heart, Lung and Blood Institute (W.Z. and J.D.F.G.), and grants from the National Institute of General Medicine Sciences and National Institute of Allergy and Infectious Disease awarded to T.M.T. (grant nos. R00 GM114245 and R01 AI156270) and H.S.M. (grant no. R01 GM128087). Computational resources were in part provided by the National Institutes of Health (NIH) HPC facility Biowulf. A portion of this research was supported by NIH grant no. U24GM129547 and performed at the PNCC at the Oregon Health Sciences University and accessed through Environmental Molecular Sciences Laboratory (grid.436923.9), a Department of Energy Office of Science User Facility sponsored by the Office of Biological and Environmental Research. We thank T. Humphries and other support staff at the PNCC for assistance with cryo-EM data collection. We thank D. Williams with assistance with cryo-EM data collection and resources supported by National Science Foundation funding (grant no. NSF1531991) awarded to the Eyring Materials Center at Arizona State University. This work was also supported by resources in the University of Arizona Imaging Cores – Life Sciences North (grant no. S10 OD011981). We thank members of the Tomasiak laboratory and D. Claxton from the Mchaourab laboratory for critical reading of this paper.
Author information
Authors and Affiliations
Contributions
T.M. Tomasiak and H.S.M. conceptualized this study. T.M. Thaker, S.M, W.Z., J.D.F.G., H.S.M. and T.M. Tomasiak conducted the experimental design, investigation and analyses. T.M. Thaker performed cryo-EM sample preparation, structure determination and analysis, and the construct design. S.M. performed DEER and biochemical experiments. W.Z. and J.D.F.G. performed MD simulation and docking experiments. M.M. and Q.T. assisted with biochemical experiments performed in revision. The initial manuscript was prepared by T. M. Thaker, T. M. Tomasiak and H.S.M., with further editing by S.M. and J.D.F.G. Data presentation and visualization were conducted by T.M. Thaker and S.M. Supervision of research and funding acquisition was carried out by T.M. Tomasiak and H.S.M.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Chemical Biology thanks Damian Ekiert, Lars Schäfer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 2 DEER decay signals for TMD/ECD spin-labeled BmrCD* variants.
a) Cartoon representations of BmrCD subdomains highlighting the position of spin-labeled cysteine pairs generated in the transmembrane domain (TMD) and the extracellular domain (ECD). DEER decay signals and distance distributions for spin-labeled pairs generated in the b) TMD and c) TMD/ECD spin-label pairs. The shaded regions in (B) and (C) represent confidence bands. Data shown are related to main text Figs. 1 and 3.
Extended Data Fig. 3 Correlative and orthogonal analysis of NBD distances in BmrCD-QQ*.
a) Cartoon representations of the cryo-EM structure of the BmrCD nucleotide binding domain (NBD) highlighting the position of spin-labeled cysteine pairs in the consensus (440/441) and degenerate (348/532) sites. b) Comparison of symmetry axis features determined using SymD60 in symmetric BmrCD (top) and asymmetric Mrp110 (bottom). The symmetry transformed structure (grey ribbon) for each model is shown superimposed to highlight the extent of asymmetry observed in each. c) Analyzed fits (left panel) of DEER decay signals (middle panel) are shown side-by-side with confidence bands (right panel) for data collected for BmrCD-QQ* samples prepared in digitonin containing buffer. Molecular dynamics distance simulations (MDDS) curves (dashed lines in (C)) calculated using the cryo-EM structure of BmrCD as a reference are shown superimposed for clarity. Lines numbered 1-4 in the consensus data and 1-3 in the degenerate data correspond to the mean peak distances summarized in Supplementary Table 4. Peak 2 in the consensus data and 3 in the degenerate data correspond to the same population based on our analysis. Data shown are related to main text Fig. 2. d) DEER decay and confidence band data for BmrCD-QQ* NBD spin label pairs collected in β-DDM buffer.
Extended Data Fig. 4 Examination of the Hoechst-33342 binding pose with MD simulations.
Data shown are for two independent MD trajectories (black, red) of a minimal construct of BmrCD-QQ* with two Hoechst molecules bound. a) Quantification of ionic and aromatic interactions between Hoechst molecule 1 (HT1) and residues in BrmC, in terms of probability distributions of the minimum distance between the ligand and each sidechain (excluding hydrogens). The only significant contact of HT1 with BrmD is also indicated. b) Same as in (A), for molecule 2 (HT2). A summary of these c) HT1 and (d) HT2 interactions together with those observed in the cryo-EM structure determined for BmrCD-QQ*H/ATP are shown in the schematics. Interacting residues are mapped onto the respective chains (BmrC in teal; BmrD in orange). Residues colored in red are observed interacting with both molecules of Hoechst-33342 in the cryo-EM structure. Residues with an asterisk were mutated in this study. Red spheres correspond to residues identified as interacting with HT1 or HT2 in both the cryo-EM structure and in the all-atom simulation. Data shown are related to main text Fig. 4.
Extended Data Fig. 5 Hoechst-33342 binding geometries in BmrCD compared to LmrP.
Cartoon representations of A) BmrCD-QQ*H/ATP and B) LmrP36 are shown with Hoechst molecules shown as spheres. C) RMSD analysis of Hoechst-33342 bound in BmrCD-QQ*H/ATP and LmrP, with Hoechst from each respective structure shown superposed to highlight diffefrences in ligand geometry.
Extended Data Fig. 6 Enzymatic activity in BmrCD* variants.
ATP turnover rates (Vmax) in C-less wild-type BmrCD (BmrCD*) and wild-type or QQ mutants of spin-labeled cysteine-pairs. Vmax data shown are the mean ± standard error (S.E.) of at least 2 biological repeats or 3-4 technical replicates. Data shown are related to main text Fig. 5.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10 and Tables 1–4.
Supplementary Data 1
Supplementary Fig. 2a uncropped SDS–PAGE gel.
Supplementary Data 2
Supplementary Fig. 2b representative cryo-EM micrograph uncropped.
Supplementary Data 3
Supplementary Fig. 3a representative cryo-EM micrograph uncropped.
Supplementary Data 4
Supplementary Fig. 3b representative cryo-EM micrograph uncropped.
Supplementary Data 5
Supplementary Fig. 10 uncropped SDS–PAGE gel.
Rights and permissions
About this article
Cite this article
Thaker, T.M., Mishra, S., Zhou, W. et al. Asymmetric drug binding in an ATP-loaded inward-facing state of an ABC transporter. Nat Chem Biol 18, 226–235 (2022). https://doi.org/10.1038/s41589-021-00936-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-021-00936-x
This article is cited by
-
Structural basis for autoinhibition by the dephosphorylated regulatory domain of Ycf1
Nature Communications (2024)
-
Spectroscopically Orthogonal Labelling to Disentangle Site-Specific Nitroxide Label Distributions
Applied Magnetic Resonance (2024)
-
Asymmetric conformations and lipid interactions shape the ATP-coupled cycle of a heterodimeric ABC transporter
Nature Communications (2023)
-
Structures of the CcmABCD heme release complex at multiple states
Nature Communications (2022)